Abstract
Ventricular septal defect (VSD) is one of the most common types of congenital heart defects (CHD). There are vivid multifactorial causes for VSD in which both genetic and environmental risk factors are consequential in the development of CHD. Methionine synthase reductase (MTRR) and methylenetetrahydrofolate reductase (MTHFR) are two of the key regulatory enzymes involved in the metabolic pathway of homocysteine. Genes involved in homocysteine/folate metabolism may play an important role in CHDs. In this study; we determined the association of A66G and C524T polymorphisms of the MTRR gene and C677T polymorphism of the MTHFR gene in Iranian VSD subjects. A total of 123 children with VSDs and 125 healthy children were included in this study. Genomic DNA was extracted from the buccal cells of all the subjects. The restriction fragment length polymorphism polymerase chain reaction (PCR-RFLP) method was carried out to amplify the A66G and C524T polymorphism of MTRR and C677T polymorphism of MTHFR genes digested with Hinf1, Xho1 and Nde1 enzymes, respectively. The genotype frequencies of CC, CT and TT of MTRR gene among the studied cases were 43.1%, 40.7% and 16.3%, respectively, compared to 52.8%, 43.2% and 4.0%, respectively among the controls. For the MTRR A66G gene polymorphism, the genotypes frequencies of AA, AG and GG among the cases were 33.3%, 43.9% and 22.8%, respectively, while the frequencies were 49.6%, 42.4% and 8.0%, respectively, among control subjects. The frequencies for CC and CT genotypes of the MTHFR gene were 51.2% and 48.8%, respectively, in VSD patients compared to 56.8% and 43.2% respectively, in control subjects. Apart from MTHFR C677T polymorphism, significant differences were noticed (p < 0.05) in C524T and A66G polymorphisms of the MTRR gene between cases and control subjects.
Keywords: MTHFR, MTRR, polymorphism, congenital heart disease, ventricular septal defect
1. Introduction
Congenital heart disease (CHD) is one of the most commonly occurring noninfectious diseases and birth defects among children. CHD is a condition whereby there is a malformation in the cardiovascular system whilst the child is still at an embryo stage [1]. The frequency of this disease is around 1 in 100 live births, but this figure is different throughout the world [1]. It constitutes one third of all congenital anomalies and is the primary cause of birth defects leading to infant mortality [2]. Congenital heart defects are constituted of several malformations such as hypoplasia, obstruction defects, septal defect and cyanotic defects [3].
Ventricular septal defect (VSD) is the most common form of CHD occurring in children at a range of 1.56 to 53.2 per 1000 live births [4]. In spite of recent advances in the CHD diagnosis and treatment, experts are yet to fully understand the underlying causes of most CHDs. Several genetic association studies have been conducted by comparing the genetic variations of DNA between cases and control subjects [5]. The commonest forms of human genetic pool variation are the single nucleotide polymorphisms (SNPs), which are responsible for 90% of DNA variation [6]. Homocysteine (Hcy), an intermediary in the production of cysteine from methionine, is a non-essential sulfur-containing amino acid. Both environmental and genetic factors affect plasma homocysteine levels. Low consumption of folate, vitamin B12 and B6, smoking and obesity are said to be environmental factors, while the genetic factors include several variants of enzymes involved in the homocysteine metabolic pathway. One genetic factor, which is the change of alanine to valine caused by C677T polymorphism of methylene tetrahydrofolate reductase (MTHFR), a thermolabile variant with reduced activity, and has been associated with elevated plasma homocysteine concentrations, especially when there is insufficient folate [7]. Nevertheless, other authors have failed to find a significant relationship in analogous studies where contrasting results were evident [8,9].
Methionine synthase reductase (MTRR) is one of the main regulatory enzymes in the homocysteine metabolic pathway. This flavoprotein has an important role in maintaining sufficient levels of activated cobalamin, which acts as a carrier for methyl during the remethylation of homocysteine to methionine. Therefore, a lack of this enzyme may cause hyperhomocysteinemia. Wilson et al. found an MTRR gene polymorphism that is a “missense” mutation (A66G), which leads to an isoleucine being substituted by a methionine residue at codon 22 [10]. Some researchers identified the A66G polymorphism as a contributing element for high plasma Hcy levels leading to incidents of vascular disease [11–13]. However, other studies have failed to provide evidence that MTRR polymorphism is one of the causes of alterations in either plasma Hcy levels and/or vascular disease [14,15]. A study carried out in Ahwaz (Iran) from 1998–2007 found that the mean prevalence of CHD was 12.30 per 1000 live births of the total 3061 instances of live births and an annual prevalence ranging from 7.93 to 17.51 per 1000 live births [16]. The folate metabolism genes were highly studied in various populations [17–22]. Apart from that, transcription factor genes were also commonly studied in relation to coronary artery disease (CAD), CHD and VSD in various populations with conflicting results (Table 1).
Table 1.
Conflicting results of genetic variants found in different populations.
| Gene Variants | Disease | Population/Reference | No. of Subjects | p-value |
|---|---|---|---|---|
| MTRR A66G | Vascular disease | Italy [17] | 114 | NS |
| MTRR A66G | CAD | Morocco [18] | 151 | S |
| MTHFR C677T | CAD | South Indians [19] | 108 | S |
| MTHFR C677T | Venous Thrombosis | Iran [20] | 200 | NS |
| MTHFR C677T | CHD | Japan [21] | 233 | S |
| MTHFR C677T | CHD | Taiwan [22] | 231 | S |
|
| ||||
| GATA4 | CHD | USA [23] | 157 | NS |
| GATA4 | CHD | Canada [24] | 120 | S |
| GATA4, NKX2.5 | CHD | China [25] | 62 | S |
| NKX2.5 | CHD | China [26] | 230 | NS |
| TBX5 | VSD | China [1] | 192 | S |
NS: Non-Significant (p > 0.05), S: Significant (p < 0.05). MTRR: methionine synthase reductase; MTHFR: methylene tetrahydrofolate reductase; CAD: coronary artery disease; CHD: congenital heart disease.
The aim of this study was to understand the etiology of the genes responsible for VSD in the Iranian population by investigating the MTHFR and MTRR gene polymorphisms for the development of VSD in Iranian subjects through a case-control study. To our knowledge, no prior studies have been conducted in Iran regarding the association of MTHFR and MTRR gene polymorphisms and VSD. Taking this into an account, our main objective was to determine whether the MTHFR and MTRR gene polymorphisms are risk factors or not for the development of VSD in Iranian subjects.
2. Results and Discussion
Methionine synthase is critical for homocysteine metabolism, and methionine synthase reductase is required to maintain methionine synthase in an active state. Both enzymes are associated with frequent polymorphisms that alter the primary structure of the proteins, and both have been subject to extensive analysis of metabolite and disease associations. Studies have suggested that the MTHFR and MTRR gene polymorphisms have become known as eventual contributors to elevated homocysteine plasma concentrations. Previous reports have reported that those genetic polymorphisms are not consistent in coronary artery disease (CAD) subjects in many populations [11,12,27]. These contradictory findings were suggested as the plausible differences in ethnicities. To our knowledge, there are no other reports on the prevalence of MTHFR and MTRR gene polymorphisms, and CAD has been reported in Asian population. In this study we examined the association of three SNPs; a common MTHFR gene polymorphisms C677T (rs1801133), and two MTRR gene polymorphisms A66G (rs1801394) and C524T (rs1532268) in Iranian VSD subjects. Homozygosity (TT) for this MTHFR single nuclear polymorphism is associated with higher homocysteine levels and lower serum folate concentrations [28,29], than heterozygosity (CT) or wild type genotype (CC), hence it is considered as a genetic risk factor for diseases associated with hyperhomocysteinemia [30,31], although some authors failed to show this interrelation [32,33]. Attempting to confirm the existence of an association between MTHFR and CHD, several studies with conflicting outcomes have been published [34–39], later studies, however, do not support the theory of MTHFR acting as a risk factor for the development of CHD.
To the best of our knowledge, this is the first study to examine the association of C677T polymorphism of the MTHFR gene and A66G and C524T polymorphisms of the MTRR gene among Iranian VSD subjects. This study was designed to compare the genotypes and allele frequencies of polymorphisms between cases and controls. Case control studies are the most common method used for testing associations of genetic polymorphisms with traits [40]. In addition, this study design was selected because it is inexpensive and relatively quick and suitable for relatively small sample sizes. After excluding participants who did not meet the inclusion criteria, an adequate sample size of 248 subjects (123 cases and 125 controls), which was collected from the Mofid Children Hospital (Tehran, Iran) was included in this study.
2.1. Socio-Demographic Characteristics
The percentage of males and females among the VSD cases was 46.3% and 53.7% respectively, whereas the percentage of males and females among the control subjects was 44.8% and 55.2%, respectively, in this study. The mean age of case subjects was 4.51 ± 2.39 whereas the mean age of controls was 5.43 ± 0.51. There was a significant difference of mean group of age between cases and controls (p < 0.05).
2.2. C677T Polymorphism of MTHFR Gene
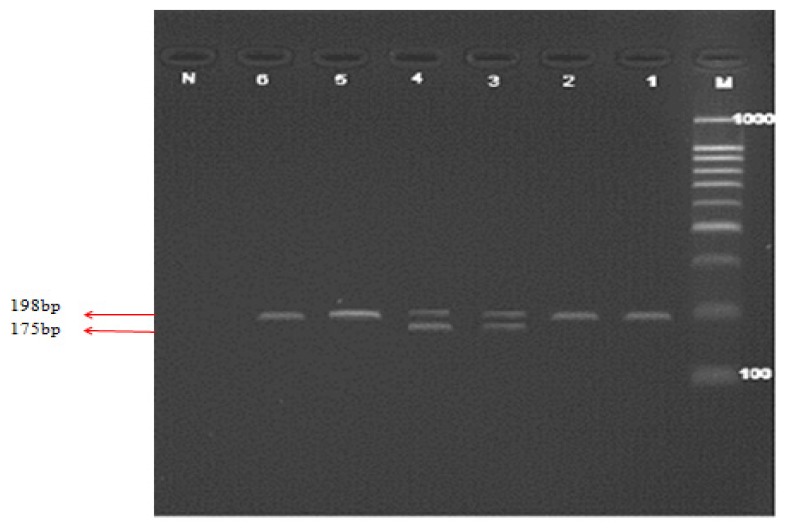
Genotypes of C677T polymorphism were determined by incubating the PCR product with Hinf1 restriction enzyme. The fragments were resolved on 4% agarose gel. Figure 1 shows the PCR product and PCR-RFLP of MTHFR C677T gene polymorphism. The genotype and allele frequencies of C677T polymorphism of MTHFR are shown in Table 2. The Chi-square test did not show any significant differences between genotype and allele frequencies of C677T polymorphism among cases and controls (p > 0.05) and (p > 0.05), respectively. The percentage of CC and CT genotypes among cases was 51.2% and 48.8%, respectively, compared to 56.8% and 43.2%, respectively among control subjects. The derived allele frequencies of C and T alleles were 75.6% and 24.4%, respectively, among cases and 78.4% and 21.6%, respectively, among controls.
Figure 1.
Polymerase chain reaction (PCR) products for methylene tetrahydrofolate reductase (MTHFR) C677T polymorphism and restricted fragments on 4% agarose gel electrophoresis.
Table 2.
Genotypic and allelic distribution of MTHFR gene polymorphism.
| Gene | Genotypes and Alleles | Case (%) | Control (%) |
|---|---|---|---|
| C677T Genotypes | CC | 63 (51.2) | 71 (56.8) |
| CT | 60 (48.8) | 54 (43.2) | |
| p value | 0.38 * | ||
| Odds Ratio (95% CI) | 0.82 (0.49–1.31) | ||
|
| |||
| Alleles | C | 186 (75.6) | 196 (78.4) |
| T | 60 (24.4) | 54 (21.6) | |
| p value | 0.47 * | ||
| Odds Ratio (95% CI) | 0.85 (0.56–1.30) | ||
p value > 0.05.
Lane M illustrates the 100bp DNA ladder. Lanes 1 and 2 show the PCR products (198bp), Lanes 3 and 4 show the heterozygote fragments (198 and 1475 bp), Lanes 5 and 6 shows the wild type fragment (198 bp) and Lane N represents the negative test control.
Genotype and allele frequencies of C677T polymorphism of MTHFR were compared between cases and controls. There were no significant differences between genotype and allele frequencies of C677T polymorphism among cases and controls (p > 0.05). The percentage of CC and CT genotypes among cases was 51.2% and 48.8%, respectively, compared to 56.8% and 43.2%, respectively, among control subjects. The derived allele frequencies of C and T alleles were 75.6% and 24.4%, respectively, among cases and 78.4% and 21.6%, respectively, among controls. As there was no TT genotype in cases and controls, we failed to show an association between MTHFR C677T gene polymorphism and the occurrence of VSD. The results obtained from a study done by Zhu et al. on a Chinese population suggested that the MTHFR C677T locus variation is associated with the occurrence of atrial septal defect (ASD) and patent ductus arteriosus (PDA), and carriers of mutant homozygote TT and allele T had a high risk of these two types of CHDs [41]. Junker et al. studied the relationship of MTHFR C677T gene polymorphism with CHDs, and the results showed that TT homozygoty was significantly related to congenital heart structure teratogenesis, especially with pulmonary artery stenosis, left heart dysplasia syndrome and coarctation of aorta [35]. Subsequent studies [36–39] failed to show significant results, which could indicate an association between CHD and the MTHFR gene polymorphism, which is in consistence to our study but in contrast to the findings of Bennouar et al. in a Moroccan population [42].
2.3. C524T and A66G Polymorphisms of MTRR Gene
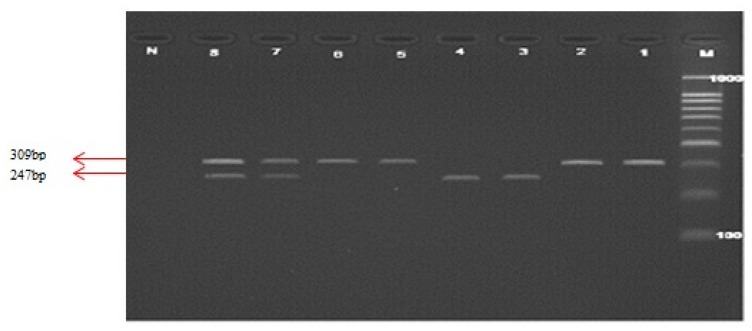
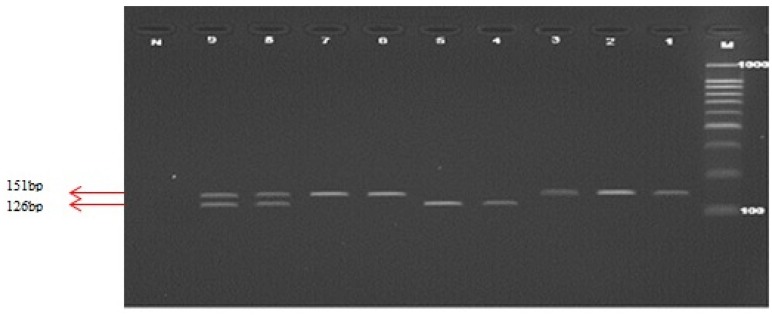
All genotypes were determined using PCR-RFLP using respective restriction enzymes. In order to genotype C524T polymorphism of MTRR, the amplified PCR product (as shown in Figure 2) was digested with Xho1 restriction enzyme. The resolved fragments on 4% agarose gel showed 247 and 62 bp in C524C homozygote, 309, 247, and 62 bp in C524T heterozygote and 309bp in T524T homozygote. Genotypes of A66G polymorphism were determined by digestion of the PCR product with Nde I restriction enzyme. This digestion produced fragments of 126, 25 bp for MTRR A66A homozygote, 151, 126 and 25 bp in MTRR A66G heterozygote, and 151 bp in MTRR G66G homozygote. Figure 3 shows the PCR product and RFLP analysis of MTRR A66G polymorphism respectively.
Figure 2.
PCR products for methionine synthase reductase (MTRR) C524T polymorphism and restricted fragments on 4% agarose gel electrophoresis.
Figure 3.
PCR products for MTRR A66G polymorphism and restriction fragments on 4% agarose gel electrophoresis.
In Figure 2, Lane M illustrates the 100 bp DNA ladder. Lanes 1 and 2 show the PCR products (309 bp), Lanes 3 and 4 show the wild type fragments (247 bp), Lanes 5 and 6 show the mutant type fragment (309 bp), Lanes 7 and 8 show the heterozygote fragments (309 and 247 bp) and Lane N represents the negative test control.
In Figure 3, Lane M illustrates the 100 bp DNA ladder. Lanes 1–3 show the PCR products (151 bp), Lane 4,5 shows the wild type fragments (126 bp), Lane 6,7 shows the mutant type fragment (151 bp), Lane 8,9 shows the heterozygote fragments (151 and 126 bp) and Lane N represents the negative test control.
For MTRR C524T polymorphism, the percentage of genotypes C524C, C524T and T524T among cases was 43.1%, 40.7%, and 16.3%, respectively, whereas the frequencies among controls were 52.8%, 43.2% and 4%, respectively (Table 3). There was a significant difference between cases and controls (p < 0.05). Derived allele frequencies of C allele and T allele were 63.4% and 36.6% among cases and 74.4% and 25.6% among controls. There was a significant difference between the allele frequency among cases and controls (p < 0.05). For the A66G polymorphism of MTRR, the genotype and frequencies differed significantly between cases and controls (p < 0.05). The genotypes of A66A, A66G and G66G were 33.3%, 43.9%, and 22.8%, respectively, in cases compared to 49.6%, 42.4% and 8%, respectively, among control subjects. The allele frequencies as shown in Table 3 also showed a significance difference between cases and controls (p < 0.05). C677T genotype frequencies were in agreement with those predicted by the Hardy Weinberg distribution, whereas, MTRR gene polymorphisms did not meet the Hardy Weinberg assumption in neither cases nor controls.
Table 3.
Genotypic and allelic distribution of MTRR gene polymorphisms.
| Gene | Genotypes and Alleles | Case (%) | Control (%) |
|---|---|---|---|
| C524T Genotypes | CC | 53 (43.1) | 66 (52.8) |
| CT | 50 (40.6) | 54 (43.2) | |
| TT | 20 (16.3) | 5 (4.0) | |
| p value | 0.00 * | ||
|
| |||
| CC vs. CT, p value | 0.60 ** | ||
| Odds Ratio (95% CI) | 0.87 (0.51–1.47) | ||
| CC vs. TT, p value | 0.00 * | ||
| Odds Ratio (95% CI) | 0.20 (0.07–0.57) | ||
| CT vs. TT, p value | 0.00 * | ||
| Odds Ratio (95% CI) | 0.23 (0.08–0.66) | ||
|
| |||
| Alleles | C | 156 (63.4) | 186 (74.4) |
| T | 90 (36.6) | 64 (25.6) | |
| p value | 0.00 * | ||
| Odds Ratio (95% CI) | 0.596 (0.40–0.87) | ||
|
| |||
| Gene | Genotypes and Alleles | Case (%) | Control (%) |
|
| |||
| A66G Genotypes | AA | 41 (33.3) | 62 (49.6) |
| AG | 54 (43.9) | 53 (42.4) | |
| GG | 28 (22.8) | 10 (8.0) | |
| p value | 0.00 * | ||
|
| |||
| AA vs. AG, p value | 0.12 * | ||
| Odds Ratio (95% CI) | 0.65 (0.38–1.12) | ||
| AA vs. GG, p value | 0.00 * | ||
| Odds Ratio (95% CI) | 0.24 (0.10–0.54) | ||
| AG vs. GG, p value | 0.00 * | ||
| Odds Ratio (95% CI) | 0.36 (0.16–0.82) | ||
|
| |||
| Alleles | A | 136 (55.3) | 177 (70.8) |
| G | 110 (44.7) | 73 (29.2) | |
| p value | 0.00 * | ||
| Odds Ratio (95% CI) | 0.51 (035–0.73) | ||
p value < 0.05,
p value > 0.05.
MTRR is a central regulatory enzyme in the metabolic pathway of homocysteine/folate. The malfunction of this enzyme has been considered as a modifiable risk factor for heart and vascular disease [43]. In this case-control study, significant differences were observed for A66G and C524T polymorphisms of MTRR gene between cases and controls (p < 0.05), which is in accordance with some studies [44–47]. We found a modest association between the A66G and C524T alleles of the MTRR gene and CHDs in the Iranian subjects. In contrast, a difference in homocysteine concentration between the 66 MTRR genotypes in CAD and ischemic cerebrovascular disease was not detected in some studies [14,16]. The conflicting results found in the associated studies were due to many confounding factors, such as study design, sample size bias, gen-environment interactions, population heterogeneity, mismatched phenotypes and population stratification. Collectively, the findings from this study provide adequate evidence that the risks of VSD are influenced by the variants of the three polymorphisms of MTHFR and MTRR genes in Iranian subjects. It is important to consider the need for future studies of folate-pathway genes other than those found in the current study, and it is also potentially complex to study the gene-gene and gene-environment interactions and VSD in the Iranian population.
2.4. DNA Sequencing
For further confirmation, random samples were selected and repeated with the same PCR conditions to confirm the genotyping results. Those samples were purified and DNA sequencing was performed with the Applied Biosystems 3730xl DNA Sequencer provided by Medigene Sdn. Bhd, Selangor, Malaysia.
2.5. Study Limitations
In this study, a number of limitations have to be considered. The present study has provided only a genetic association for MTHFR and MTRR gene polymorphisms among Iranian VSD patients as compared to control subjects. However, we failed to analyze the MTHFR and MTRR mRNA levels in both VSD patients and control subjects. Apart from the MTHFR and MTRR gene polymorphisms other polymorphisms such as TBX5, NKX2.5 and GATA4 need to be analyzed to determine the association of the other candidate genes with VSD and other defects. Replication studies with a larger number of samples and assessing the genotypic and allelic frequencies of mothers are also needed in order to understand the etiological factors.
3. Materials and Methods
3.1. Ethical Approval
Ethical approval was obtained from the Shaheed Beheshti University of Medical Sciences and also from the Faculty of Medicine and Health Sciences, Universiti Putra Malaysia. A written informed consent was obtained from the parents of the children with CHD. Similarly, consent forms were obtained from all of the control subjects enrolled in this study.
3.2. Study Subjects
A total of 150 pediatric patients were approached, and 123 pediatric patients were recruited as the other subjects were excluded due to the inconsistent results and extreme values. All the patients were recruited from the pediatric infectious research center (PIRC), Mofid children hospital, Tehran, Iran. All the subjects were visited during July 2010 to Jun 2011 in the hospital from the different parts of the districts. The diagnosis of non-syndromic CHD and classification of the type of the cardiac defect had been done by a pediatric cardiologist based on the clinical and echocardiography findings with or without the diagnostic cardiac catheterization findings and surgical notes. Whereas for the control subjects, a total of 125 healthy control subjects with no history of congenital heart disease were recruited in for this study.
3.3. Genomic DNA Extraction
Buccal cell samples were collected on a sterile cytobrush (Qiagen Inc., Chatsworth, CA, USA) Genomic DNA extraction was carried out using the DNA isolation kit (Qiagen Inc., Chatsworth, CA, USA). The purity of extracted DNA was quantified using Eppendorf UVette® in Biophotometer (Eppendorf, Hamburg, Germany).
3.4. Genotyping MTHFR and MTRR Gene Polymorphisms
To determine the genotypes of MTHFR and MTRR genes, genomic DNA was amplified first by the respective primers using the polymerase chain reaction (PCR) technique. The PCR amplification for all the respective polymorphisms was performed in a total volume of a 25 μL reaction mixture consisting of 10 pmol of each primer and the Mastermix (i-DNA Biotechnology (M) Sdn Bhd, Kuchai lama, Kuala Lumpur, Malaysia) and the template DNA. A negative control containing no genomic DNA and a positive control of known genotype were always included in the set of reactions. All the PCR cycling conditions were carried out on an iCycler machine (BioRad Laboratories, Hercules, CA, USA). The amplified PCR products for all the three gene polymorphisms were separated at 2%–4% agarose gel (Bioline, London, UK). The agarose gel was stained in ethidium bromide and visualized using Alpha Imager (Alpha Innotech, San Leandro, CA, USA). The PCR products of the respective genes were digested with 2–4 units of the respective restriction enzymes (Thermo Fisher Scientific, Inc, provided by Research Instruments Sdn Bhd, Petaling Jaya, Malaysia) with 10× Fast Digest Green Buffer in a final volume of 30 μL reaction mixture. Table 4 shows the primers used for the RFLP method, restriction endonucleases and the digested restricted fragment size products. Identical results were obtained when genotyping was performed for 10% of the samples on two separate occasions.
Table 4.
PCR conditions for the three studied gene polymorphisms using restriction fragment length polymorphism polymerase chain reaction (PCR-RFLP) method.
| Gene Polymorphism | Forward Primer (FP) Reverse Primer (RP) |
Restriction Endonuclease Enzymes | PCR Products (bp) | Restriction Fragment Size (bp) |
|---|---|---|---|---|
| MTHFR C677T | FP-5′-TGAAGGAGAAGGT GTCTGCGGGA-3′ RP-5′AGGACGGTGCGGT GCGGTGAGAGTG-3′ |
Hinf1 | 198 | Wild type: 198 Mutant: 175, 23 Heterozygote: 198,175, 23 |
| MTRR C524T | FP-5′-GTCAAGCAGAGGACA AGAG-3′ RP-5′AGAGACTCCTGCAGAT GTAC-3′ |
Xho1 | 309 | Wild type: 247, 62 Mutant: 309 Heterozygote: 309, 247, 62 |
| MTRR A66G | FP-5′-CAGGCAAAGGCCAT CGCAGAAGACAT-3′ RP-5′CACTTCCCAACCAAAA TTCTTCAAAG-3′ |
Nde1 | 151 | Wild type: 126, 25 Mutant: 151 Heterozygote: 151, 126, 25 |
3.5. Automated Sequencing
Purified PCR products were sent to Research Biolabs Malaysia to confirm the nucleotide sequence. The sequencing results were aligned with the respective reference gene sequence from the NCBI-GeneBank sequences using the sequence alignment MEGA4 software [48].
3.6. Statistical Analysis
Data analysis was done by SPSS version 18.00 (SPSS Inc, South Wacker Drive, Chicago, IL, USA). Alleles and genotype distribution were tested for deviation from the Hardy-Weinberg by a Chi Square test. The odds ratio (OR) and its 95% confidence intervals (CI) were used to illustrate the association, with p < 0.05 considered in all tests to be statistically significant.
4. Conclusions
This study failed to show an association between the MTHFR gene and VSD subjects. However, the MTRR gene polymorphisms (C524T and A66G) can be considered as risk factors for the development for VSD in Iranian subjects.
Acknowledgments
The authors thank all the volunteers participated in the study.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Liu C.X., Shen A.D., Li X.F., Jiao W.W., Bai S., Yung F., Guan X., Zhang X.G., Zhang G.R., Li Z.Z. Association of TBX5 gene polymorphism with ventricular septal defect in the Chinese Han population. Chin. Med. J. 2009;122:30–34. [PubMed] [Google Scholar]
- 2.Van Beynum I.M., Blom H.J., Kapusta L., den Heijer M. The MTHFR 677C→T polymorphism and the risk of congenital heart defects: A literature review and meta-analysis. QJM. 2007;100:743–753. doi: 10.1093/qjmed/hcm094. [DOI] [PubMed] [Google Scholar]
- 3.Shanley T.P., Wheeler D.S., Wong H.R. Pediatric Critical Care Medicine: Basic Science and Clinical Evidence. Springer; New York, NY, USA: 2007. [Google Scholar]
- 4.Minette M.S., Sahn D.J. Congenital heart disease for the adult cardiologist, ventricular septal defects. Am. Heart Assoc. 2006;114:2190–2197. doi: 10.1161/CIRCULATIONAHA.106.618124. [DOI] [PubMed] [Google Scholar]
- 5.Little J., Higgins J.P., Ioannidis J.P., Moher D., Gagnon F., von Elm E., Khoury M.J., Cohen B., Scheet P., Hutc K. Association of strengthening the reporting of genetic association studies (STREGA): An extension of the STROBE statement the C677T methylenetetrahydrofolate reductase mutation with congenital heart diseases. Eur. J. Epidemiol. 2009;24:37–55. doi: 10.1007/s10654-008-9302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins F.S., Brooks L.D., Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- 7.Gupta S.K., Kotwal J., Kotwal A., Dhall A., Garg S. Role of homocysteine & MTHFR C677T gene polymorphism as risk factors for coronary artery disease in young Indians. Indian J. Med. Res. 2012;135:506–512. [PMC free article] [PubMed] [Google Scholar]
- 8.Domagala T.B., Adamek L., Nizankowska E., Sanak M., Szczeklik A. Mutations C677T and A1298C of the 5,10-methylenetetrahydrofolate reductase gene and fasting plasma homocysteine levels are not associated with the increased risk of venous thromboembolic disease. Blood Coagul. Fibrinolysis. 2002;13:423–431. doi: 10.1097/00001721-200207000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Frederiksen J., Juul K., Grande P., Jensen G.B., Schroeder T.V., Tybjaerg-Hansen A., Nordestgaard B.G. Methylenetetrahydrofolate Reductase polymorphism (C677T), hyperhomocysteinemia, and risk of ischemic cardiovascular disease and venous thromboembolism: Prospective and case–control studies from the Copenhagen City Heart Study. Blood. 2004;104:3046–3051. doi: 10.1182/blood-2004-03-0897. [DOI] [PubMed] [Google Scholar]
- 10.Wilson R.P., Leclerc D., Christensen B., Yang H., Gravel R.A., Rozen R. A common variant in Methionine Synthase Reductase combined with low cobalamin (Vitamin B12) increases risk for spina bifida. Mol. Genet. Metab. 1999;67:317–323. doi: 10.1006/mgme.1999.2879. [DOI] [PubMed] [Google Scholar]
- 11.Gaughan D.J., Kluijtmans L.A., Barbaux S., McMaster D., Young I.S., Yarnell J.W., Evans A., Whitehead A.S. The Methionine Synthase Reductase (MTRR) A66G polymorphism is a novel genetic determinant of plasma homocysteine concentrations. Atherosclerosis. 2001;157:451–456. doi: 10.1016/s0021-9150(00)00739-5. [DOI] [PubMed] [Google Scholar]
- 12.Guéant-Rodriguez R.M., Juillirrem Y., Candito M., Adjalla C.E., Gibelin P., Herbeth B., van Obberghen E., Gueánt J.L. Association of MTRRA66G polymorphism (but not of MTHFR C677T and A1298C, MTRA2756G, TCN C776G) with homocysteine and coronary artery disease in the French population. Thromb. Haemost. 2005;94:510–515. doi: 10.1160/TH05-04-0262. [DOI] [PubMed] [Google Scholar]
- 13.Laraqui A., Allami A., Carrié A., Coiffard A.S., Benkouka F., Benjouad A., Bendriss A., Kadiri N., Bennouar N., Benomar A., et al. Influence of methionine synthase (A2756G) and Methionine Synthase Reductase (A66G) polymorphisms on plasma homocysteine levels and relation to risk of coronary artery disease. Acta Cardiol. 2006;61:51–61. doi: 10.2143/AC.61.1.2005140. [DOI] [PubMed] [Google Scholar]
- 14.Jacques P.F., Bostom A.G., Selhub J., Rich S., Ellison R.C., Eckfeldt J.H., Gravel R.A., Rozen R. Effects of polymorphisms of methionine synthase and Methionine Synthase Reductase on total plasma homocysteine in the NHLBI Family Heart Study. Atherosclerosis. 2003;166:49–55. doi: 10.1016/s0021-9150(02)00204-6. [DOI] [PubMed] [Google Scholar]
- 15.Brilakis E.S., Berger P.B., Ballman K.V., Rozen R. Methylenetetrahydrofolate Reductase (MTHFR) 677C>T and Methionine Synthase Reductase (MTRR) 66A>G polymorphisms: association with serum homocysteine and angiographic coronary artery disease in the era of flour products fortified with folic acid. Atherosclerosis. 2003;168:315–322. doi: 10.1016/s0021-9150(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 16.Rahim F., Ebadi A., Saki G., Remazani A. Prevalence of congenital heart disease in Iran: A clinical study. J. Med. Sci. 2008;8:547–552. [Google Scholar]
- 17.Scazzone C., Acuto S., Guglielmini E., Campisi G., Bono A. Methionine Synthase Reductase (MTRR) A66G polymorphism is not related to plasma homocysteine concentration and the risk for vascular disease. Exp. Mol. Pathol. 2009;86:131–133. doi: 10.1016/j.yexmp.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Laraqui A., Allami A., Carrié A., Raisonnier A., Coiffard A.-S., Benkouka F., Bendriss A., Benjouad A., Bennouar N., El Kadiri N., et al. Relation between plasma homocysteine, gene polymorphisms of homocysteine metabolism-related enzymes, and angiographically proven coronary artery disease. Eur. J. Int. Med. 2007;18:474–483. doi: 10.1016/j.ejim.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Vinukonda G., Shaik Mohammad N., Md Nurul Jain J., Prasad Chintakindi K., Rama Devi Akella R. Genetic and environmental influences on total plasma homocysteine and coronary artery disease (CAD) risk among South Indians. Clin. Chim. Acta. 2009;405:127–131. doi: 10.1016/j.cca.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 20.Soltanpour M.S., Soheili Z., Pourfathollah A.A., Samiei S., Meshkani R., Deyhim M.R., Safa M., Ataei Z. The association between common C677T mutation in Methylenetetrahydrofolate reductase gene and the risk of venous thrombosis in an Iranian population. Lab. Med. 2008;39:97–100. [Google Scholar]
- 21.Ou T., Yamakawa K., Arinami T., Amemiya H., Fujiwara H., Kawata K., Saito M., Kikuchi S., Noguchi Y., Sugishita Y., et al. Methylenetetrahydrofolate reductase and apolipoprotein E polymorphisms are independent risk factors for coronary heart disease in Japanese: A case-control study. Atherosclerosis. 1998;137:23–28. doi: 10.1016/s0021-9150(97)00244-x. [DOI] [PubMed] [Google Scholar]
- 22.Lee C.N., Su Y.N., Cheng W.F., Lin M.T., Wang J.K., Wu M.H., Hsieh F.J. Association of the C677T methylenetetrahydrofolate reductase mutation with congenital heart diseases. Acta Obstet. Gynecol. Scand. 2005;84:1134–1140. doi: 10.1111/j.0001-6349.2005.00611.x. [DOI] [PubMed] [Google Scholar]
- 23.Schluterman M.K., Krisiyak A., Kathiriya I.S., Abate N., Chandalia M., Srivastava D., Garg V. Screening and biochemical analysis of GATA4 sequence variations identified in patients with congenital heart disease. Am. J. Med. Genet. 2007;143A:817–823. doi: 10.1002/ajmg.a.31652. [DOI] [PubMed] [Google Scholar]
- 24.Nemer G., Fadlalah F., Usta J., Nemer M., Dbaibo G., Obeid M., Bitar F. A novel mutation in the GATA4 gene in patients with Tetralogy of Fallot. Human Mutat. 2006;27:293–294. doi: 10.1002/humu.9410. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W.M., Li X., Ma Z.Y., Zhang J., Zhou S.H., Li T., Shi L., Li Z.Z. GATA4 and NKX2.5 gene analysis in Chinese Uygur patients with congenital heart disease. Chin. Med. 2009;122:416–419. [PubMed] [Google Scholar]
- 26.Zhang W.M., Li X., Shen A., Jiao W., Guan X., Li Z. Screening NKX2.5 mutation in a sample of 230 Han Chinese children with congenital heart diseases. Genet. Test Mol. Biomark. 2009;13:159–162. doi: 10.1089/gtmb.2008.0044. [DOI] [PubMed] [Google Scholar]
- 27.Kluijtmans L.A., Young I., Boreham C.A., Murray L., McMaster D., McNulty H., Strain J.J., McPartlin J., Scott J.M., Whitehead A.S. Genetic and nutritional factors contributing to hyperhomocysteinemia in young adults. Blood. 2003;101:2483–2488. doi: 10.1182/blood.V101.7.2483. [DOI] [PubMed] [Google Scholar]
- 28.Van der Put N.M., Steegers-Theunissen R.P., Frosst P., Trijbels F.J., Eskes T.K., van den Heuvel L.P., Mariman E.C., den Heyer M., Rozen R., Blom H.J. Mutated Methylenetetrahydrofolate Reductase as a risk factor for spina bifida. Lancet. 1995;346:1070–1071. doi: 10.1016/s0140-6736(95)91743-8. [DOI] [PubMed] [Google Scholar]
- 29.Jacques P.F., Bostom A.G., Williams R.R., Ellison R.C., Eckfeldt J.H., Rosenberg I.H., Selhub J., Rozen R. Relation between folate status, a common mutation in Methylenetetrahydrofolate Reductase, and plasma homocysteine concentrations. Circulation. 1996;93:7–9. doi: 10.1161/01.cir.93.1.7. [DOI] [PubMed] [Google Scholar]
- 30.Botto L.D., Yang Q.H. 5,10-Methylenetetrahydrofolate Reductase gene variations and congenital anomalies: A huge review. Am. J. Epidemiol. 2000;151:862–877. doi: 10.1093/oxfordjournals.aje.a010290. [DOI] [PubMed] [Google Scholar]
- 31.Shields D.C., Krike P., Mills J.L., Ramsbottom D., Molly A.M., Bruke H., Weir D.G., Scott J.M., Whitehead A.S. The “thermolabile” variant of methylenetetrahydrofolate reductase and neural tube defects: An evaluation of genetic risk and relative importance of genotypes of embryo and the mother. Am. J. Hum. Genet. 1999;64:1045–1055. doi: 10.1086/302310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch M.C., Stegmann K., Ziegler A., Schroter B., Ermert A. Evaluation of the MTHFR C677T allele and the MTHFR gene locus in a German spina bifida population. Eur. J. Pediatr. 1998;157:487–492. doi: 10.1007/s004310050860. [DOI] [PubMed] [Google Scholar]
- 33.Molloy A.M., Mills J., Kirke P.N., Ramsbottom D., McPartlin J.M., Burke H., Conley M., Whitehead A.S., Weir D.G., Scott J.M. Low blood folates in NTD pregnancies are only partly explained by thermolabile 5,10-methylenetetrahydrofolate reductase: Low folate status alone may be the critical factor. Am. J. Med. Genet. 1988;78:155–159. [PubMed] [Google Scholar]
- 34.Wenstrom K.D., Johanning G.L., Johnston K.E., DuBard M. Association of the C677T methylenetetrahydrofolate reductase mutation and elevated homocysteine levels with congenital cardiac malformations. Am. J. Obstet. Gynecol. 2001;184:806–812. doi: 10.1067/mob.2001.113845. [DOI] [PubMed] [Google Scholar]
- 35.Junker R., Kotthoff S., Vielhaber H., Halimeh S., Kosch A., Koch H.G., Kassenbohmer R., Heineking B., Nowak-Gottl U. Infant methylenetetrahydrofolate reductase 667TT genotype is a risk factor for congenital heart disease. Cardiovasc. Res. 2001;51:251–254. doi: 10.1016/s0008-6363(01)00286-3. [DOI] [PubMed] [Google Scholar]
- 36.Storti S., Vittorini S., Lascone M.R., Sacchelli M., Collavoli A., Ripoli A., Cocchi G., Biagini A., Clerico A. Association between 5,10-Methylenetetrahydrofolate Reductase C677T and A1298C polymorphisms and conotruncal heart defects. Clin. Chem. Lab. Med. 2003;41:276–280. doi: 10.1515/CCLM.2003.043. [DOI] [PubMed] [Google Scholar]
- 37.McBride K.L., Fernbach S., Menesses A., Molinari L., Quay E., Pignatelli R., Towbin J.A., Belmont J.W. A family-based association study of congenital left-sided heart malformations and 5,10-Methylenetetrahydrofolate Reductase. Birth. Defects Res. A. 2004;78:825–830. doi: 10.1002/bdra.20049. [DOI] [PubMed] [Google Scholar]
- 38.Pereira A.C., Xavier N.J., Mesquita S.M., Mota G.F., Lopes A.A., Krieger J.E. Lack of evidence of association between MTHFR C677T polymorphism and congenital heart disease in a TDT study design. Int. J. Cardiol. 2005;105:15–18. doi: 10.1016/j.ijcard.2004.10.049. [DOI] [PubMed] [Google Scholar]
- 39.Shaw G.M., Iovannisci D., Yang W., Finnell R.H., Carmichael S.L., Cheng S., Lammer E.J. Risks of human conotruncal heart defects associated with 32 single nucleotide polymorphisms of selected cardiovascular disease-related genes. Am. J. Med. Genet. 2005;138:21–26. doi: 10.1002/ajmg.a.30924. [DOI] [PubMed] [Google Scholar]
- 40.Cardon L.R., Bell J.I. Association study designs for complex diseases. Nat. Rev. Genet. 2001;2:91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- 41.Zhu W.L., Li Y., Yan L., Dao J., Li S. Maternal and offspring MTHFR gene C677T polymorphism as predictors of congenital atrial septal defect and patent ductus arteriosus. Mol. Hum. Reprod. 2006;12:51–54. doi: 10.1093/molehr/gah252. [DOI] [PubMed] [Google Scholar]
- 42.Bennouar N., Allami A., Azeddoug H., Bendris A., Laraqui A., Jaffali A.E., Kadiri N.E., Benzidia R., Benomar A., Fellat S., et al. Thermolabile Methylenetetrahydrofolate Reductase C677T polymorphism and homocysteine are risk factors for coronary artery disease in Moroccan population. J. Biomed. Biotechnol. 2007;2007:80687–80696. doi: 10.1155/2007/80687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verkleij-Hagoort A.C., Verlinde M., Ursem N.T., Lindemans J., Helbing W.A., Ottenkamp J., Siebel F.M., Gittenberger-de Groot A.C., de Jonge R., Bartelings M.M., et al. Maternal hyperhomocysteinaemia is a risk factor for congenital heart disease. BJOG. 2006;113:1412–1418. doi: 10.1111/j.1471-0528.2006.01109.x. [DOI] [PubMed] [Google Scholar]
- 44.Van Beynum I.M., Kouwenberg M., Kapusta L., den Heijer M., van der Linde I.J., Daniels O., Blom H.J. MTRR 66A>G polymorphism in relation to congenital heart defects. Clin. Chem. Lab. Med. 2006;44:1317–1323. doi: 10.1515/CCLM.2006.254. [DOI] [PubMed] [Google Scholar]
- 45.Fredriksen A., Meyer K., Ueland P.M., Vollset S.E., Grotmol T. Large-scale population-based metabolic phenotyping of thirteen genetic polymorphisms related to one-carbon metabolism. Hum. Mutat. 2007;28:856–865. doi: 10.1002/humu.20522. [DOI] [PubMed] [Google Scholar]
- 46.Verkleij-Hagoort A., van Driel L.M., Lindemans J., Isaacs A., Steegers E.A., Helbing W.A., Uitterlinden A.G., Steegers-Theunissen R.P. Genetic and lifestyle factors related to the periconception vitamin B12 status and congenital heart defects: A dutch case-control study. Mol. Genet. Metab. 2008;94:112–119. doi: 10.1016/j.ymgme.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Shaw G.M., Iovannisc D., Yang W., Finnell R.H., Carmichael S.L., Cheng S., Lammer E.J. 118 SNPs of folate-related genes and risks of spina bifida and conotruncal heart defects. BMC Med. Genet. 2009;10:49. doi: 10.1186/1471-2350-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]