Abstract
Approximately 25,000 ovarian cancers are diagnosed in the U.S. annually, and 75% are in the advanced stage and largely incurable. There is critical need for early detection tools and novel treatments. Proteasomal ubiquitin receptor ADRM1 is a protein that is encoded by the ADRM1 gene. Recently, we showed that among 20q13-amplified genes in ovarian cancer, ADRM1 overexpression was the most highly correlated with amplification and was significantly upregulated with respect to stage, recurrence, and metastasis. Its overexpression correlated significantly with shorter time to recurrence and overall survival. Array-CGH and microarray expression of ovarian cancer cell lines provided evidence consistent with primary tumor data that ADRM1 is a 20q13 amplification target. Herein, we confirm the ADRM1 amplicon in a second ovarian cancer cohort and define a minimally amplified region of 262 KB encompassing seven genes. Additionally, using RNAi knock-down of ADRM1 in naturally amplified cell line OAW42 and overexpression of ADRM1 via transfection in ES2, we show that (1) ADRM1 overexpression increases proliferation, migration, and growth in soft agar, and (2) knock-down of ADRM1 results in apoptosis. Proteomic analysis of cells with ADRM1 knock-down reveals dysregulation of proteins including CDK-activating kinase assembly factor MAT1. Taken together, the results indicate that amplified ADRM1 is involved in cell proliferation, migration and survival in ovarian cancer cells, supporting a role as an oncogene and novel therapeutic target for ovarian cancer.
Keywords: ADRM1, oncogene, ovarian cancer
1. Introduction
Ovarian cancer is the leading cause of death from gynecologic malignancy accounting for approximately 15,000 deaths in the US annually. The disease is identified primarily in its advanced stage when it is largely incurable. New approaches for detection and therapy are warranted.
Recently, we showed that out of the 214 known genes mapping to 20q13, ADRM1, a gene that encodes the proteasomal ubiquitin receptor ADRM1 protein, is the most likely amplification target at 20q13 in ovarian cancer with over-expression correlating significantly with a shorter time to recurrence and death [1]. ADRM1 encodes an integral plasma membrane protein involved in development, cell adhesion, deubiquitination, and proteolysis [2–5]. Overexpression of ADRM1 in 293T adenovirus-transformed human embryonic kidney cells reduces degradation of short-lived proteins [5]. It has been shown that ADRM1 binds directly to proteasome-associated deubiquitinating enzyme, UCH37, enhancing its isopeptidase activity [4,5]. Deubiquinating proteins in human cancer represent a newly emerging class of oncogenes and tumor suppressors [6]. In ADRM1 knock-out mice, the most notable phenotype is a defect in oogenesis, suggesting that ADRM1 normally plays a critical role in ovarian development [7]. The role ADRM1 plays in ovarian cancer is unknown but it has been shown to be upregulated in metastatic tumor cells, and its overexpression increased the propensity of cells to engage in cell-cell interactions [2,8]. A study of commonly overexpressed genes in solid tumors by microarray analysis shows that ADRM1 is consistently over-expressed in multiple tumor types, including lung adenocarcinoma, lung squamous cell carcinoma, bladder, colon, liver, kidney, and stomach cancer in addition to ovarian cancer [9], suggesting that this gene may be a potential target in many tumor types. Recently, it has been shown that knockdown of ADRM1 suppresses proliferation and inhibits colorectal cancer cell migration, survival and tumorigenicity [10,11].
We have shown that amplification in ovarian cancer tumors is a major mechanism of up-regulation of ADRM1 and that its expression increases with tumor stage and recurrence [1]. Consistent with these findings, using array-CGH and RNA expression analysis of ovarian cancer cell lines, we showed the amplification and overexpression of ADRM1 in 18% (7/39) of ovarian cancer cell lines [12]. Among these, ovarian cancer cell line OAW42 was found to be amplified and had the highest level of overexpression and was used as a model for knock-down of expression of ADRM1 in ADRM1-amplified ovarian cancer. Ovarian cancer cell line ES2 was not amplified and was used for transfection leading to in vitro overexpression of ADRM1. With a new ovarian cancer cohort in addition to these two ovarian cancer cell line models, herein we provide additional evidence that ADRM1 plays a role in ovarian cancer.
2. Results and Discussion
2.1. Confirmation of ADRM1 Amplification in a Second Ovarian Cancer Cohort
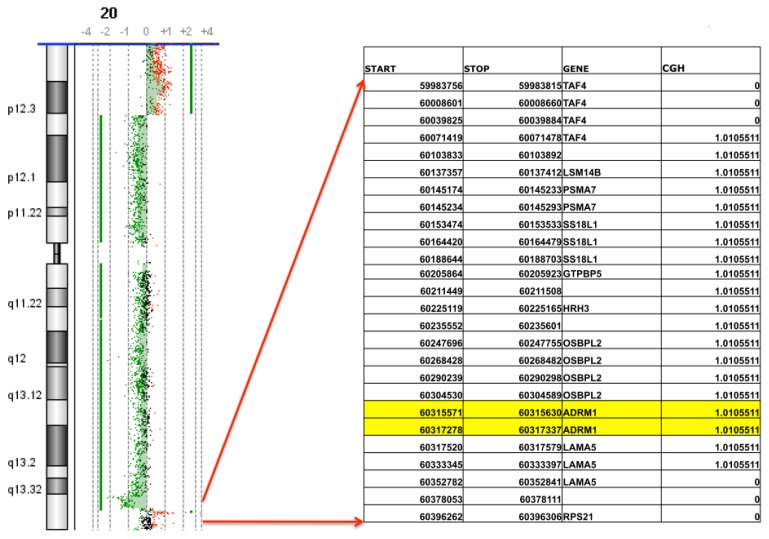
Array-CGH analysis of 68 primary ovarian carcinomas shows DNA amplification (CGH > 0.7X) of ADRM1 in 15% of tumors. Among the amplified tumors was a stage 3, grade 4 Endometrioid Ovarian Carcinoma with a 262 kilobase amplification of 1.01× mapping from 60,071,478 kb within the TAF4 gene on chromosome 20q13 extending to 60,333,397 kb within the LAMA5 gene on 20q13. The amplicon includes ADRM1 and six additional genes whose expression levels do not correlate as significantly with amplification [1]. There are no gains either proximal or distal to this region on chromosome 20q in this tumor (Figure 1).
Figure 1.
Array CGH used to define 262 KB ADRM1 amplicon. Array CGH on DNA isolated from ovarian endometrioid stage III, grade 4 tumor reveals a 1.01-fold amplification of seven genes including ADRM1 on 20q13.
2.2. ADRM1 Over-Expression Increases Cell Proliferation and Conversely, ADRM1 Knockdown Results in Significant Growth Inhibition
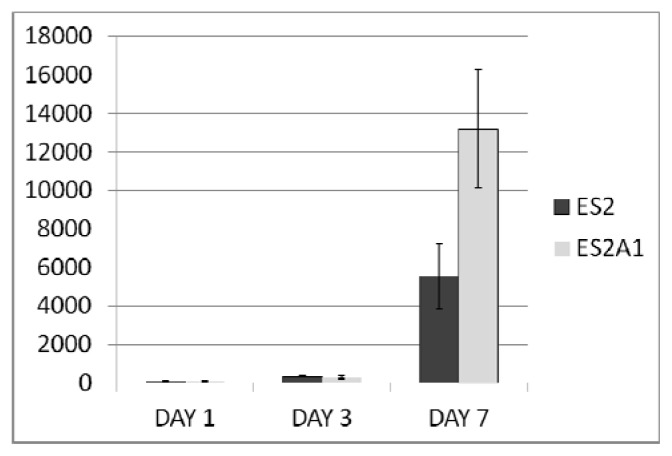
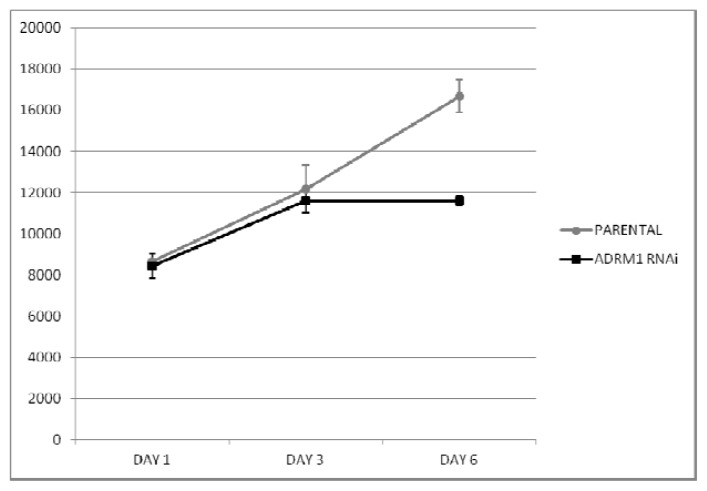
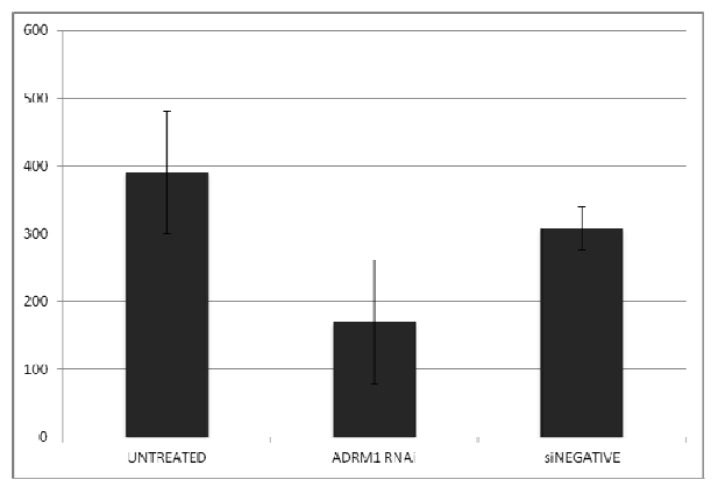
We speculated that the preferential amplification of ADRM1 gives overexpressing ovarian cancer cells a proliferative advantage. To determine whether or not this was the case, an ovarian cancer cell line that over-expresses ADRM1 (ES2A1) was created by stable transfection of an ADRM1 plasmid into ES2 cells. Cell counts in triplicate showed at least a 2.2-fold increase in cell growth on day 7 in ES2A1 cells compared to parental ES2 cells (Figure 2). In addition, naturally occurring ADRM1-amplified OAW42 cells treated with RNAi knock-down of ADRM1, resulted in a 1.4-fold growth inhibition compared to both parental ADRM1 untreated and non-targeting RNAi treated OAW42 cells by day 7 after transfection (Figure 3). Changes in cell cycle were not detected in the overexpressing and the knock-down cell line models.
Figure 2.
Cell counts (average of triplicate on 12-well) show a 2.2-fold increase in cell growth on day 7 in ADRM1-overexpressing clone ES2A1 compared to parental cell line ES2.
Figure 3.
RNAi interference of ADRM1 in OAW42 results in 1.4-fold growth inhibition compared to growth of PARENTAL (untreated) OAW42 cells.
2.3. ADRM1 Over-Expression Increases Growth in Soft-Agar and Conversely, Knockdown of ADRM1 Decreases Growth in Soft Agar
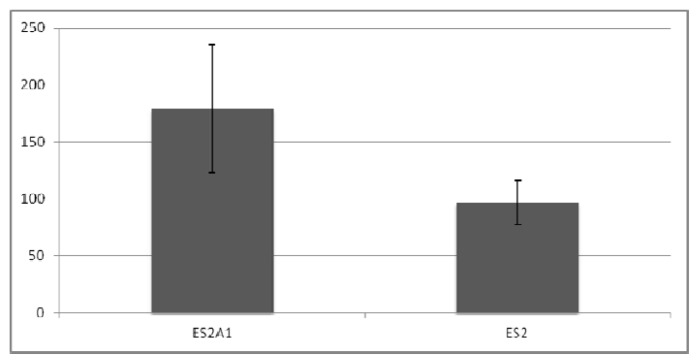
ADRM1 overexpression correlates with recurrence and metastasis, and therefore, we hypothesized that expression levels effect the malignant tendency of the cells. In soft agar experiments, colony-forming efficiency in ADRM1-overexpressing ES2A1 cells was 1.9-fold higher than parental ES2 cells (Figure 4), and colonies are larger (Figure 5). Conversely, parental OAW42 cells showed a 2.3-fold increase and non-targeting RNAi-treated OAW42 cells showed a 1.8-fold increase in colony-formation efficiency, when compared to ADRM1 RNAi-treated OAW42 cells (Figure 6).
Figure 4.
ADRM1-overexpressing cells are 1.9-fold more efficient in colony-formation (colony number) in soft agar than parental ES2 cells, and produce larger colonies.
Figure 5.
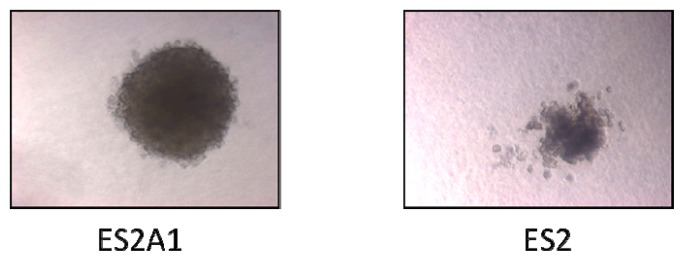
Largest ES2 colony in soft agar compared to typical ES2A1 colony in soft agar.
Figure 6.
Growth in soft agar (number of colonies) in parental (untreated) compared to ADRM1 siRNA-treated and non-targeting siRNA-treated OAW42 shows knock-down of ADRM1 inhibits colony formation.
2.4. ADRM1 Knockdown Results in a Decrease in Proliferation and/or Migration Using the Scratch Test
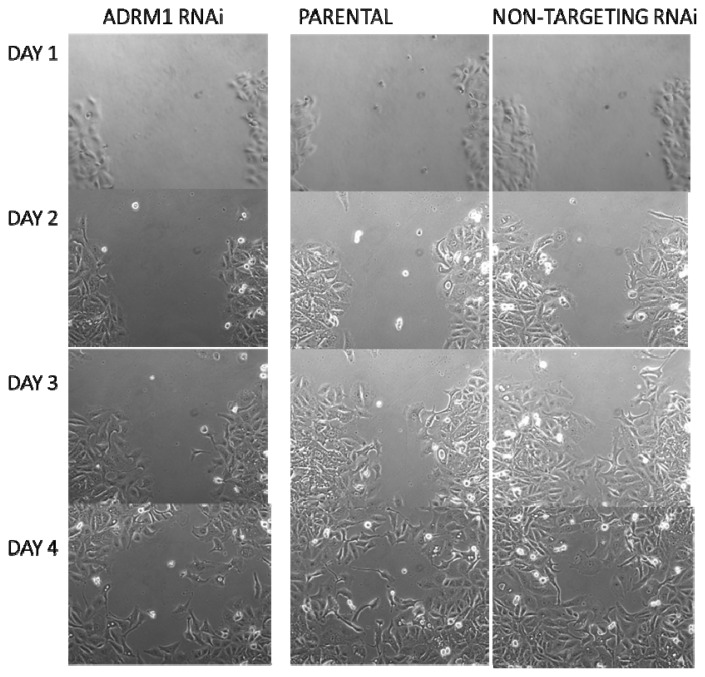
Additionally, a wound-healing assay was performed on ADRM1 knock-down OAW42 cells compared to both parental and non-targeting RNAi-treated OAW42 cells. A similar-sized wound was detected on the first day for each cell type. By day 3, the ADRM1-knock-down cells were only 50% healed, compared to 75% closing of the scratch wound in the parental and 88% in the non-targeting control cells (Figure 7).
Figure 7.
Wound-healing assay using the scratch test shows decrease in proliferation and/or migration in ADRM1-silenced OAW42 cells compared to non-targeting-RNAi treated OAW42 cells and parental untreated OAW42 cells.
2.5. Knockdown of ADRM1 Increases Apoptosis in Ovarian Cells
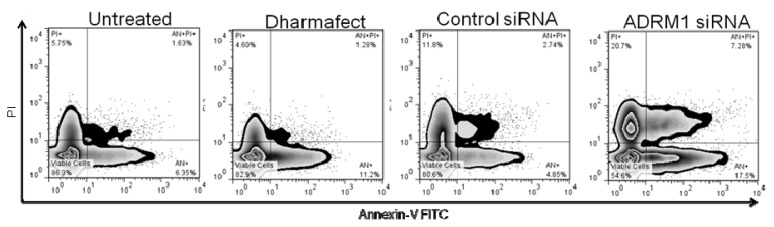
Cell cycle and apoptosis experiments were performed to determine whether ADRM1 expression has an effect on cell cycle and whether blocking ADRM1 results in apoptosis. Cell cycle and apoptosis were examined by flow cytometry for ADRM1 over expressing clone ES2A1 and parental ES2, and no significant differences were detected (data not shown). Additionally, no significant differences in the cell cycle were detected between OAW42 cells with knock down of ADRM1 compared to parental untreated OAW42, nor dharmafect-treated OAW42 (nor OAW42 treated with either of two non-specific RNAi controls) (data not shown). However, there was a 20% increase in apoptosis, both early and late apoptosis (AN+/AN+PI+), and a 10% increase in cell death (PI+ cells) in ADRM1 RNAi-treated OAW42 cells compared to parental untreated OAW42, nor dharmafect-treated OAW42 and non-specific RNAi controls (Figure 8).
Figure 8.
Effects on apoptosis of silencing ADRM1 in OAW42 ovarian cells. ADRM1 siRNA cells shows an increase in the percentage of apoptotic cells (25% Annexin-V positive) and dead cells (21% PI+) cells when compared with untreated control (8% Annexin-V positive/6% PI+), Dharmafect (13% Annexin-V positive/5% PI+) or scramble control siRNA (8% Annexin-V positive/12% PI+). No significant apoptosis or death was seen in Dharmafect or control siRNA compared to untreated OAW42 cells.
2.6. ADRM1 Knockdown Results in the Down-Regulation of 15 Proteins Including MNAT1 and the Up-Regulation of Seven Proteins
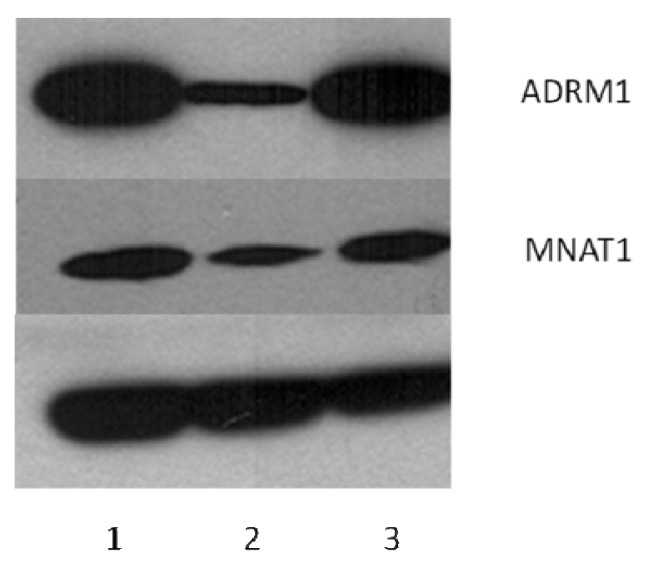
ADRM1 is a proteasomal ubiquitin receptor and therefore we predicted knock-down of ADRM1 might affect the regulation of other proteins. Therefore, we performed Difference Gel Electrophoresis (DiGE) analysis on OAW42 cells with RNAi knock-down of ADRM1 and compared protein levels to parental untreated and non-targeting RNAi-treated OAW42 cells. We identified 15 proteins that were down-regulated and 7 proteins that were upregulated when ADRM1 is knocked-down (Table 1). These proteins were excised from the gel and identified with mass spectrometry. Three of the proteins were confirmed to be differentially expressed by Western Blot Analysis on the original lysates, including MNAT1, HAX1, and ST1A3 (Figure 9).
Table 1.
Difference Gel Electrophoresis (DIGE) and mass spectrometry results in identification of 15 down-regulated and 7 up-regulated proteins in ADRM1 RNAi-treated OAW42 cells compared to untreated parental and non-targeting siRNA-treated OAW42 cells (not including ADRM1).
| Av. Ratio | t-test | ID | Protein | pI | MW |
|---|---|---|---|---|---|
| 1.43 | 0.0019 | LAMB1_HUMAN | Laminin subunit beta-1 | 4.83 | 205150 |
| 1.41 | 0.00043 | LAMC1_HUMAN | Laminin subunit gamma-1 | 5.01 | 183191 |
| 1.28 | 0.00016 | ITAV_HUMAN | Integrin alpha-V | 5.45 | 117048 |
| 1.22 | 0.00011 | IDHC_HUMAN | Isocitrate dehydrogenase (NADP) cytoplasmic | 6.53 | 46915 |
| 1.21 | 0.00017 | PLST_HUMAN | Plastin-3 | 5.41 | 71279 |
| 1.21 | 0.00024 | ST1A3_HUMAN | Sulfotransferase 1A3/1A4 | 5.68 | 34288 |
| 1.2 | 2.30E−05 | DC1I2_HUMAN | Cytoplasmic dynein 1 intermediate chain 2 | 5.08 | 71811 |
| −1.17 | 0.00012 | PURA2_HUMAN | Adenylosuccinate synthetase isozyme 2 | 6.13 | 50465 |
| −1.18 | 0.0053 | CAPZB_HUMAN | F-actin-capping protein subunit beta | 5.36 | 31616 |
| −1.2 | 0.00059 | CSDE1_HUMAN | Cold shock domain-containing protein E1 | 5.88 | 89684 |
| −1.21 | 0.00021 | KAP0_HUMAN | cAMP-dependent protein kinase type I-alpha regulatory subunit | 5.27 | 43183 |
| −1.25 | 8.20E−05 | SUGT1_HUMAN | Suppressor of G2 allele of SKP1 homolog | 5.07 | 41284 |
| −1.26 | 0.0083 | HS90B_HUMAN | Heat shock protein HSP 90-beta | 4.97 | 83554 |
| −1.31 | 0.033 | HS90A_HUMAN | Heat shock protein HSP 90-alpha | 4.94 | 85006 |
| −1.33 | 0.031 | H90B3_HUMAN | Putative heat shock protein HSP 90-beta-3 | 4.71 | 68624 |
| −1.33 | 0.0038 | H90B3_HUMAN | Putative heat shock protein HSP 90-beta-3 | 4.71 | 68624 |
| −1.39 | 0.0098 | HS90A_HUMAN | Heat shock protein HSP 90-alpha | 4.94 | 85006 |
| −1.44 | 0.0024 | H90B3_HUMAN | Putative heat shock protein HSP 90-beta-3 | 4.71 | 68626 |
| −1.47 | 0.00026 | H90B3_HUMAN | Putative heat shock protein HSP 90-beta-3 | 4.71 | 68626 |
| −1.48 | 1.50E−06 | CALU_HUMAN | Calumenin | 4.47 | 37198 |
| −1.5 | 0.0053 | HAX1_HUMAN | HCLS1-associated protein X-1 | 4.76 | 31601 |
| −1.59 | 1.30E−06 | MAT1_HUMAN | CDK-activating kinase assembly factor MAT1 | 5.79 | 36256 |
| −1.78 | 4.00E−07 | ADRM1_HUMAN | Proteasomal ubiquitin receptor ADRM1 | 4.96 | 42412 |
| −2.11 | 1.40E−09 | ADRM1_HUMAN | Proteasomal ubiquitin receptor ADRM1 | 4.96 | 42412 |
Figure 9.
Western Blotting confirms that gene-silencing of ADRM1 results in dysregulation of MNAT1 (and HAX1 and ST1A3, not shown). (1) Dharmafect-treated OAW42; (2) ADRM1 RNAi-treated OAW42; (3) siRNA non-targeting treated OAW42.
2.7. Discussion
Recently, we showed that among 225 ovarian samples, ADRM1 is a likely 20q13-amplification target in ovarian cancer. It was amplified and overexpressed in 23% of tumors, was significantly upregulated with respect to stage, recurrence and metastasis, and its overexpression correlates significantly with shorter time to recurrence and overall survival [1]. Consistent with these findings, using array-CGH and RNA expression analysis of ovarian cancer cell lines, we showed the amplification and overexpression of ADRM1 in 18% (7/39) of ovarian cancer cell lines [12]. Herein, CGH analysis of ADRM1 in a separate cohort of 68 primary ovarian carcinomas, including one with high level amplification of a 262 kilobase region containing ADRM1, provide additional evidence that ADRM1 is the driver of a 20q13 amplicon in ovarian cancer.
The role of this gene was explored using ADRM1-upregulated and ADRM1 down-regulated in vitro models. Evidence suggests that ADRM1 has traits of an oncogene as amplification leads to increased cell proliferation and increased colony-formation efficiency. Conversely, down-regulation of ADRM1 at the RNA and protein levels results in growth inhibition, decreased colony-formation efficiency, slower wound-healing, decreased cell viability, and significantly increased apoptosis. Knock-down of ADRM1 resulted in the dysregulation of multiple proteins already known to play a role in cancer. In particular, multiple HSP90 fragments were identified. A recent study showed HSP90 inhibition leads to simultaneous inactivation of multi-RTKs including EGFR, ERBB2, MET, and AXL and suppresses the downstream survival/proliferation signaling in multiple ovarian cancer cell lines [13] making it a promising target for ovarian cancer therapy. Drugs targeting heat shock proteins are currently of intense focus, and are involved in several clinical trials in ovarian and other cancers [14].
SULT1A1 (ST1A3) is up regulated with the knock-down of ADRM1 and encodes a protein that catalyzes the sulfate conjugation of many hormones. Variants in ST1A3 are linked to increased hormone-dependant ovarian cancer risk [15,16].
The gene HAX1 was shown in this study to be down regulated with ADRM1 and is known to associate with hematopoeitc cell-specific Lyn substrate 1, a substrate of the Src family of tyrosine kinases [17]. HAX1 promotes cell survival via inhibition of CASP9, responsible for apoptosis execution [18]. Therefore its down regulation may explain the significant increase in apoptosis in ADRM1 RNAi-treated ovarian cancer in this study. It also potentiates GNA13-mediated cell migration and invasion [19,20], and thus its down regulation may contribute to the decrease in wound healing in ADRM1 knock-down cells shown in this study. Thus one can hypothesize that the correlation between ADRM1 over expression and recurrence and metastasis shown previously in ovarian cancer may be through regulation of HAX1.
Finally, the most significantly dysregulated gene, MNAT1 (MAT1), stabilizes the cyclin H-CDK7 complex to form a functional CDK-activating kinase (CAK) enzymatic complex, which in turn activates the cyclin-associated kinases CDK1, CDK2, CDK4 and CDK6 by threonine phosphorylation [21]. Interestingly, in a mouse model for pancreatic cancer lymphatic metastasis, ADRM1 was one of 30 genes found to be upregulated in the metastatic cell line compared to the parental cell line [22]. In another study, direct RNAi silencing of MNAT1 suppressed the growth of pancreatic cancer cells in vitro, and significantly achieved an anti-tumor effect on the subcutaneously transplanted pancreatic tumor in vivo [23]. Thus, down regulation of MNAT1 in ADRM1-silenced ovarian cancer may explain the similar anti-tumor effects shown in this study. Additionally, MNAT1 has been shown to interact with Cyclin H, Estrogen receptor alpha, MTA1, and cyclin-dependent kinase 7, all linked previously to ovarian cancer [24–26].
3. Experimental Section
3.1. Tissue
Sixty-eight primary ovarian carcinomas were snap frozen in liquid nitrogen and prepared for DNA and RNA processing as described below. The samples consisted of 38 ovarian endometrioid carcinomas (19 stage I and II, 19 stage III and IV), 16 ovarian clear cell carcinomas (5 stage I and II, 11 stage III and IV), 11 mucinous ovarian tumors (10 stage I, 1 stage III), and 3 mixed ovarian epithelial carcinomas (1 stage I, 2 stage III and IV).
3.2. Cell Lines
The cell line OAW42 was obtained from the European Collection of Cell Cultures (Salisbury, UK). The cell line ES2 was obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). OAW42 and cells was cultured in DMEM (ATCC) plus 10% FBS. ES2 was cultured in McCoy’S 5A medium (ATCC) plus 10% FBS.
3.3. Array CGH
Genomic DNA was extracted from frozen cell pellets using the DNeasy Blood and Tissue Kit (Qiagen) with the following modifications. Seventy percent ethanol was substituted for Buffer AW2 in the final wash, and DNA was eluted in 50 mL of sterile water (Invitrogen, Carlsbad, CA, USA). The concentration and quality of the DNA were measured with the NanoDrop Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and by electrophoresis in 1% agarose.
Labeling and hybridization of Agilent 105K oligonucleotide CGH arrays was performed according to the manufacturer’s protocol for Human Genome CGH 105A Oligo Microarray Kit (version 5.0; Agilent Technologies, Santa Clara, CA, USA, 2008). Labeled tumor and reference DNAs were combined, annealed with COT-1 DNA (Invitrogen, Carlsbad, CA, USA) and 10× Blocking Agent (Agilent, Santa Clara, CA, USA) for 30 min at 37 °C after boiling, then hybridized to Agilent 105A arrays for 40 h at 65 °C according to the manufacturer’s instructions. After hybridization, arrays were washed according to Procedure B (which includes an ozone blocking wash), and scanned using an Agilent Scanner (G2565BA). Files were extracted using Agilent Feature Extraction software (version 9.5; Agilent, Santa Clara, CA, USA, 2007) with the default CGH protocol. Extracted arrays with a DRL Spread< 0.3 were included in the analysis.
CGH Analytics software (version 4.0; Agilent Technologies, Santa Clara, CA, USA, 2008) was used for copy number analysis, employing the ADM1 algorithm (Threshold 5), with Fuzzy Zero and Centralization corrections to minimize background noise. All map positions were based on the March 2006 NCBI 36/hg18 genome assembly. A minimum of 3 consecutive probes was required to define a region as amplified or deleted. The data was also filtered by requiring a minimum absolute average log2 ratio of 0.4. All data was inspected visually using the interactive view. Log2 ratios >0.4 (1.3-fold) were considered gain and log2 ratios >1 (2-fold) were considered amplified.
3.4. Analyses of Cell Proliferation
ES2 and ES2A1 cells were counted and seeded in triplicate on 12-well plates and were harvested by trypsinization on days 1, 3, and 7 and counted using a particle counter (Z1; Beckman Coulter, Inc., Brea, CA, USA). OAW42 cells were counted and seeded in triplicate on 6-well plates and treated with dharmafect only, dharmafect and ADRM1 siRNA, or dharmafect and si-negative control as explained in the RNA interference section. Cells were harvested by trypsinization on days 1, 2, 3, and 6 after transfection and counted using a particle counter (Z1; Beckman Coulter, Inc., Brea, CA, USA). Standard deviations were calculated from triplicate experiments and shown in bar graphs.
3.5. In Vitro Apoptosis and Cell Cycle Assays
The effects of silencing ADRM1 on apoptosis and cell cycle were evaluated using Annexin V-FITC and Propidium Iodide (PI) staining and DAPI DNA staining respectively. Cells were plated evenly and allowed to grow to log phase before being transfected with siRNA to knockdown ADRM1 or control. Also, a vehicle for transfection Dharmafect was used as a control. For apoptosis analysis supernatant was collected, cells were washed with PBS, and trypsin was applied to release cells, which were then centrifuged at 2000 rpm for 5 min. Supernatant was aspirated and cells were then suspended in 200 and allowed to grow to log ained with 10 μL of Annexin V-FITC and 5 min. Supernatant was aspirated and cell (Medical & Biological Laboratories, Co., Woburn, MA, USA). The effects of siRNA ADRM1 on the cell cycle were assessed using Nim-DAPI staining (NPE Systems, Pembroke Pines, FL, USA). Flow cytometry was performed using a CellQuanta (Beckman Coulter, Brea, CA, USA) flow machine, and the results were analyzed using Flow Jo software (version 7.6.5; Tree Star Inc., Ashland, OR, USA, 2011).
3.6. Soft Agar Assay for Colony Formation
ES2, ES2A1, OAW42, and siRNA-treated (ADRM1 siRNA and non-targeting control siRNA) OAW42 cells were counted and seeded in triplicate to 10,000 cells per well in a 24-well culture dish containing 0.4% low-melting agarose over a 0.6% agarose layer, both in culture medium, and incubated for 3–5 weeks at 37 °C. Colonies were stained with 10% Neutral Red (Fisher Scientific, Waltham, MA, USA) and counted. Standard deviations were calculated from triplicate experiments and shown in bar graphs. Experiment was repeated twice to confirm findings. Knock-down of ADRM1 in cells from siRNA experiments was confirmed by Western Blot with protein lysates isolated seven days after seeding.
3.7. Antibodies and Western Blotting
Transfected and untransfected cells were isolated two days after transfection. Cells were washed in PBS and lysed at 4 °C in lysis buffer. Insoluble material was cleared by centrifugation at 10,000× g for 10 min. Protein was quantitated using BCA (Pierce Biochemicals, Rockford, IL, USA), resolved by SDS-PAGE, and transferred to nitrocellulose membranes (Invitrogen, Carlsbad, CA, USA). ADRM1, MNAT1, and ST1A3 were detected, respectively with antibodies to ADRM1 monoclonal 3C6, MNAT1 anti-MNAT antibody produced in rabbit, ST1A3 anti-SULT1A1 monoclonal 1F8 (Sigma-Aldrich, St. Louis, MO, USA). Monoclonal Anti-HAX1 antibody produced in mouse was purchased from BD Biosciences, San Jose, CA, USA. Detection was performed using chemifluorescent reagent (SuperSignal West Pico Chemiluminescent Substrate, Thermo Scientific, Rockford, IL, USA). Antialpha-tubulin mouse antibody (Calbiochem, San Diego, CA, USA) 0.1 μg/mL was used as a loading control.
3.8. RNA Interference
Four interfering RNAs specific for ADRM1 were purchased from Dharmacon (ThermoFisher Scientific, Lafayette, CO, USA) as a Smartpool in addition to two non-targeting siRNA oligos to use as a controls. Transfection was performed using 0.4% Dharmafect transfection reagent and 10 nM siRNA according to the manufacturer (Dharmacon, Lafayette, CO, USA). OAW42 cells were seeded and transfected at 60% confluency in antibiotic-free media (DMEM (Cellgro, Lawrence KS, USA) supplemented with 1% l-glutamine, 0.91 μg/mL bovine insulin, 4.5 g/1 L glucose, and 3.7 g/L sodium bicarbonate). After 24 h at 37 °C, growth media with 10% fetal bovine serum and antibiotic was added. Knock-down of ADRM1 was confirmed by Western Blot at day 2, 3, 4, and 7 after transfection.
3.9. Plasmid Constructs and Transfection
The ADRM1 Ultimate Human ORF Clone from IOH13967 was purchased from Invitrogen (Carlsbad, CA, USA) and cloning and transfection were performed following Invitrogen’s protocol for transfection of a mammalian expression clone. Kanamycin resistant IOH13967 clones were isolated. The plasmid DNA was subcloned into amp-resistant pcDNA 6.2/V5-DEST Mammalian expression vector. True expression clone was confirmed by testing ampicillin resistance and chloramphenicol sensitivity. Pure DNA was isolated with the Purelink HQ kit and the clone was sequenced to confirm it was in frame. Ovarian cell line ES2 was tested for Blasticidin sensitivity and a concentration of 0.015 mg/mL Blasticidin kills all ES2 parental cells. ES2 was transfected with the ADRM1 expression vector with lipofectamine. Blasticidin-resistant clones grown in 0.015 mg/mL Blasticidin were tested for increased ADRM1 expression by Western blot. The highest ADRM1 expressing clone ES2A1 was used in experiments in this study.
3.10. Scratch Test
For the scratch assay, OAW42 cells treated with ADRM1-specific RNAi, non-targeting RNAi, and Dharmafect only (parental), were grown in complete growth medium until 90%–100% confluency was reached. A 3 mm cross-wound was introduced across the diameter of each plate. Cell migration was observed by microscopy and photographed at 24 h, 48 h, 72 h and 96 h later.
3.11. Cell Growth and Protein Extraction
OAW42 ovarian cell lines were independently grown in eighteen 100 mm regular Petri dishes in complete medium for 24 h and then each of six OAW42 ovarian cell lines were treated either with 10 nM ADRM1-specific RNAi or non-targeting RNAi (scrambled RNAi) and the control cells with only 0.4% of Dharmafect transfection reagent (parental) in antibiotic-free medium. After 24 h, cells were incubated in complete medium for another 24 h. Then, cells were washed two times with low-serum medium at 4 °C, followed by 2 times with PBS at 4 °C, and directly lysed in 0.3 mL of ice cold labeling buffer (7 M urea, 2 M thiourea, 4% CHAPS and 20 mM Tris-HCl, pH 8.8) and then briefly sonicated in Bioruptor (Diagenade) on ice cold water bath for 30 s on and 1 min off for a total six times at high settings. Cell lysates were centrifuged at 13,500 rpm for 15 min at 4 °C and extracted proteins were stored at −80 °C.
3.12. DIGE Labeling, Analysis and Mass Spectrometry
DIGE labeling, analysis and mass spectrometry were performed as described previously [27]. Briefly, protein extracts used for the dige experiments were cleaned using the methanol/chloroform method and protein pellets were resuspended in the labeling buffer and centrifuged. For the dige analysis, protein samples were labeled with the N-hydroxysuccinimidyl ester derivates of Cy2, 3 and 5 Dyes according to the manufactures protocol (GE Healthcare, Piscataway, NJ, USA) for 30 min on ice water in the dark and then excess Cy Dyes were quenched by adding 10 mM Lysine solution and incubated 10 min. Briefly, 50 micrograms of protein from each experimental group repeated 6 times (ADRM1-specific RNAi, non-targeting RNAi treated, and control cells) were labeled with the 400 pmol of Cy3 and Cy5 Dyes, respectively, to avoid any Cy Dye labeling biases. Internal standard sample was prepared by pooling equal (25 micrograms) amount of proteins from all eighteen samples and labeled with the 400 pmol of Cy2. After the labeling, 50 micrograms of Cy2, Cy3 and Cy5 labeled samples were combined with the 300 micrograms of unlabeled protein mix from the all samples and the rehydration solution was added (7 M urea, 2 M thiourea, 4% CHAPS, 1% DTT. 0.5% pH 4–7 IPG buffer, 5% glycerol, 10% isopropanol). Samples were incubated for another 20 min, briefly centrifuged for 5 min at 11,000 rpm and then applied to 24 cm pH 4–7 IPG strips (GE Healthcare), passively rehydrated overnight at room temperature. Proteins were focused according with the following steps at 200 V (hold) for 2 h, 500 V (hold) for 2 h, 1500 V (gradient) for 2 h, 8000 V (gradient) for 4 h 30 min and 8000 V (hold) for 6 h 30 min for the total of 77,000 Vh. After the IEF, IPG strips were equilibrated in a 10 mL of reducing solution (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, 1% DTT, 0.002% bromophenol blue) for 15 min and then a second equilibration step was followed with 4.5% (w/v) iodoacetamide in the same solution without DTT. After reduction and alkylation of proteins, IPG strips were placed on the top of the 12.5% SDS polyacrylamide gels and sealed with the 1% agarose solution. A second dimension SDS gel electrophoresis was run in Ettan DALTtwelve system (GE healthcare) at 50 V for 3 h at 24 °C and then continued at 1 W per gel at 24 °C for overnight until bromphenol blue runs out of the gels. The next day, gels were scanned using the Typhoon Trio Variable Mode Imager (GE Healthcare) at 100 micron resolution using 488 nm laser/520BP40 filter for Cy2, 532 nm laser/580BP30 nm filter for Cy3, 633 nm laser/670BP30 for filter Cy5. Gel images were cropped using Image Quant Software (GE Healthcare, Piscataway, NJ, USA). The Decyder 2D Differential Analysis software (version 6.5; GE Healthcare: Piscataway, NJ, USA) was used for the identification of statistically significantly differentially expressed proteins. After the analysis, proteins of interests were selected based on the fold changes and student’s t test. Gels were fixed and then stained by Sypro-Ruby. Sypro-Ruby stained images were re-matched to the dige images and protein spots were picked from three gels using the Ettan Spot Picker (GE Healthcare) and digested with modified trypsin (Trypsin Gold, Mass Spectrometry grade, Promega, (Madison, WI, USA) using the ProGest protein Digestion Robotic Station system (Genomic Solutions, Inc, (Marlborough, MA, USA). MALDI-TOF/TOF mass spectrometry analysis was performed on an Ultraflex TOF/TOF instrument (Bruker Daltonics, Bremen, Germany) [28].
4. Conclusions
Thus this study provides evidence that ADRM1 plays a role in ADRM1-amplified ovarian cancer. The results indicate that amplified ADRM1 is involved in cell proliferation, migration and survival in ovarian cancer cells, supporting a role as an oncogene and novel therapeutic target for ovarian cancer. Because ADRM1 is a proteasomal ubiquitin receptor, future work may focus on the susceptibility to proteasome inhibitors in ADRM1-amplified ovarian cancer. Additionally, small molecule inhibitors targeting ADRM1 directly may represent a novel beneficial therapy for 20q13-amplified ovarian cancer, and may act through altering regulation of multiple ovarian cancer targets including HSP90s, MNAT1 and respective cyclins, ST1A3 and hormone levels, and HAX1 and apoptosis.
Acknowledgements
This work was supported by the Adelson Medical Research Foundation (AMRF) and Revlon/UCLA Women’s Cancer Research Program (recipient Dennis Slamon).
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Fejzo M.S., Dering J., Ginther C., Anderson L., Ramos L., Walsh C., Karlan B., Slamon D.J. Comprehensive analysis of 20q13 genes in ovarian cancer identifies ADRM1 as amplification target. Genes Chromosomes Cancer. 2008;47:873–883. doi: 10.1002/gcc.20592. [DOI] [PubMed] [Google Scholar]
- 2.Lamerant N., Kieda C. Adhesion properties of adhesion-regulating molecule 1 protein on endothelial cells. FEBS J. 2005;272:1833–1844. doi: 10.1111/j.1742-4658.2005.04613.x. [DOI] [PubMed] [Google Scholar]
- 3.Cherix N., Froquet R., Charette S.J., Blanc C., Letourneur F., Cosson P. A Phg2-ADRM1 pathway participates in the nutrient-controlled developmental response in Dictyostelium. Mol. Biol. Cell. 2006;17:4982–4987. doi: 10.1091/mbc.E06-07-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamazaki J., Iemura S., Natsume T., Yashiroda H., Tanaka K., Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu X.B., Ouyang S.Y., Li C.J., Miao S., Wang L., Goldberg A.L. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J.M. Emerging roles of deubiquitinating enzymes in human cancer. Acta Pharmacol. Sin. 2007;28:1325–1330. doi: 10.1111/j.1745-7254.2007.00687.x. [DOI] [PubMed] [Google Scholar]
- 7.Al-Shami A., Jhaver K.G., Vogel P., Wilkins C., Humphries J., Davis J.J., Xu N., Potter D.G., Gerhardt B., Mullinax R., et al. Regulators of the proteasome pathway, Uch37 and Rpn13, play distinct roles in mouse development. PLoS One. 2010;5:e13654. doi: 10.1371/journal.pone.0013654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simins A.B., Weighardt H., Weidner K.M., Weidle U.H., Holzmann B. Functional cloning of ARM-1, an adhesion-regulating molecule upregulated in metastatic tumor cells. Clin. Exp. Metastasis. 1999;17:641–648. doi: 10.1023/a:1006790912877. [DOI] [PubMed] [Google Scholar]
- 9.Pilarsky C., Wenzig M., Specht T., Saeger H.D., Grützmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–750. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W., Hu X.T., Shi Q.L., Zhang F.B., He C. Silencing of ADRM1 by RNA interference suppresses proliferation of colorectal cancer cells. Zhonghua ZhongLiu ZaZhi. 2009;31:815–819. [PubMed] [Google Scholar]
- 11.Chen W., Hu X.T., Shi Q.L., Zhang F.B., He C. Knockdown of the novel proteasome subunit ADRM1 located on the 20q13 amplicon inhibits colorectal cancer cell migration, survival and tumorigenicity. Oncol. Rep. 2009;21:531–537. [PubMed] [Google Scholar]
- 12.Fejzo M.S., Ginther C., Dering J., Anderson L., Venkatessen N., Karlan B., Slamon D.J. Knock-down of ovarian cancer amplification target ADRM1 leads to down regulation of GIPC1 and up-regulation of RECK. Genes Chromosomes Cancer. 2011;50:434–441. doi: 10.1002/gcc.20868. [DOI] [PubMed] [Google Scholar]
- 13.Jiao Y., Ou W., Meng F., Zhou H., Wang A. Targeting HSP90 in ovarian cancers with multiple receptor tyrosine kinase coactivation. Mol. Cancer. 2011;10:125. doi: 10.1186/1476-4598-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peyrat J.F., Messaoudi S., Brion J.D., Alami M. Inhibitors of the heat shock protein 90: From cancer clinical trials to neurodegenerative diseases. Atlas Genet. Cytogenet. Oncol. Haematol. 2010. [(accessed on 25 January 2013]. Available online: http://AtlasGeneticsOncology.org/Deep/HSP90inCancerTreatmentID20086.html.
- 15.Mikhailova O.N., Gulyaeva L.F., Prudnikov A.V., Gerasimov A.V., Krasilnikov S.E. Estrogen-metabolizing gene polymorphisms in the assessment of female hormone-dependent cancer risk. Pharmacogenomics J. 2006;6:189–193. doi: 10.1038/sj.tpj.6500365. [DOI] [PubMed] [Google Scholar]
- 16.Gulyaeva L.F., Mikhailova O.N., PustyInyak V.O., Kim I.V., IV, Gerasimov A.V., Krasilnikov S.E., Filipenko M.L., Pechkovsky E.V. Comparative analysis of SNP in estrogen-metabolizing enzymes for ovarian, endometrial, and breast cancers in Novosibirsk, Russia. Adv. Exp. Med. Biol. 2008;617:359–366. doi: 10.1007/978-0-387-69080-3_34. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki Y., Demoliere C., Kitamura D., Takeshita H., Deuschle U., Watanabe T. HAX-1, a novel intracellular protein, localized on mitochondria, directly associates with HS1, a substrate of Src family tyrosine kinases. J. Immunol. 1997;158:2736–2744. [PubMed] [Google Scholar]
- 18.Han Y., Chen Y.S., Liu Z., Bodyak N., Rigor D., Bisping E., Pu W.T., Kang P.M. Overexpression of HAX-1 protects cardiac myocytes from apoptosis through caspase-9 inhibition. Circ. Res. 2006;99:415–423. doi: 10.1161/01.RES.0000237387.05259.a5. [DOI] [PubMed] [Google Scholar]
- 19.Radhika V., Onesime D., Ha J.H., Dhanasekaran N. Galpha13 stimulates cell migration through cortactin-interacting protein Hax-1. J. Biol. Chem. 2004;279:49406–49413. doi: 10.1074/jbc.M408836200. [DOI] [PubMed] [Google Scholar]
- 20.Ramsay A.G., Keppler M.D., Jazayeri M., Thomas G.J., Parsons M., Violette S., Weinreb P., Hart I.R., Marshall J.F. HS1-associated protein X-1 regulates carcinoma cell migration and invasion via clathrin-mediated endocytosis of integrin alphavbeta6. Cancer Res. 2007;67:5275–5284. doi: 10.1158/0008-5472.CAN-07-0318. [DOI] [PubMed] [Google Scholar]
- 21.Lolli G., Johnson L.N. CAK-Cyclin-dependent activating kinase: A key kinase in cell cycle control and a target for drugs? Cell Cycle. 2005;4:572–577. [PubMed] [Google Scholar]
- 22.Long J., Luo G., Liu C., Cui X., Satoh K., Xiao Z., Zhang B., Xu J., Ni Q., Li M., et al. Development of a unique mouse model for pancreatic cancer lymphatic metastasis. Int. J. Oncol. 2012;41:1662–1668. doi: 10.3892/ijo.2012.1613. [DOI] [PubMed] [Google Scholar]
- 23.Liu J.P., Yuan S.Z., Zhang S.N. Experimental study of MAT1 gene silencing mediated by siRNA in pancreatic cancer. Zhonghua Yi Xue Za Zhi. 2007;87:2719–2723. [PubMed] [Google Scholar]
- 24.Sawiris G.P., Sherman-Baust C.A., Becker K.G., Cheadle C., Teichberg D., Morin P.J. Development of a highly specialized cDNA array for the study and diagnosis of epithelial ovarian cancer. Cancer Res. 2002;62:2923–2928. [PubMed] [Google Scholar]
- 25.Darb-Esfahani S., Wirtz R.M., Sinn B.V., Budczies J., Noske A., Weichert W., Faggad A., Scharff S., Sehouli J., Oskay-Ozcelik G., et al. Estrogen receptor 1 mRNA is a prognostic factor in ovarian carcinoma: Determination by kinetic PCR in formalin-fixed paraffin-embedded tissue. Endocr. Relat. Cancer. 2009;16:1229–1239. doi: 10.1677/ERC-08-0338. [DOI] [PubMed] [Google Scholar]
- 26.Prisco M.G., Zannoni G.F., de Stefano I., Vellone V.G., Tortorella L., Fagotti A., Mereu L., Scambia G., Gallo D. Prognostic role of metastasis tumor antigen 1 in patients with ovarian cancer: A clinical study. Hum. Pathol. 2012;43:282–288. doi: 10.1016/j.humpath.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Jung E.J., Avliyakulov N.K., Boontheung P., Loo J.A., Nel A.E. Pro-oxidative DEP chemicals induce heat shock proteins and an unfolding protein response in a bronchial epithelial cell line as determined by DIGE analysis. Proteomics. 2007;3:3906–3918. doi: 10.1002/pmic.200700377. [DOI] [PubMed] [Google Scholar]
- 28.Mosessian S., Avliyakulov N.K., Mulholland D.J., Boontheung P., Loo J.A., Wu H. Analysis of PTEN complex assembly and identification of heterogeneous nuclear ribonucleoprotein C as a component of the PTEN-associated complex. J. Biol. Chem. 2009;284:30159–30166. doi: 10.1074/jbc.M109.027995. [DOI] [PMC free article] [PubMed] [Google Scholar]