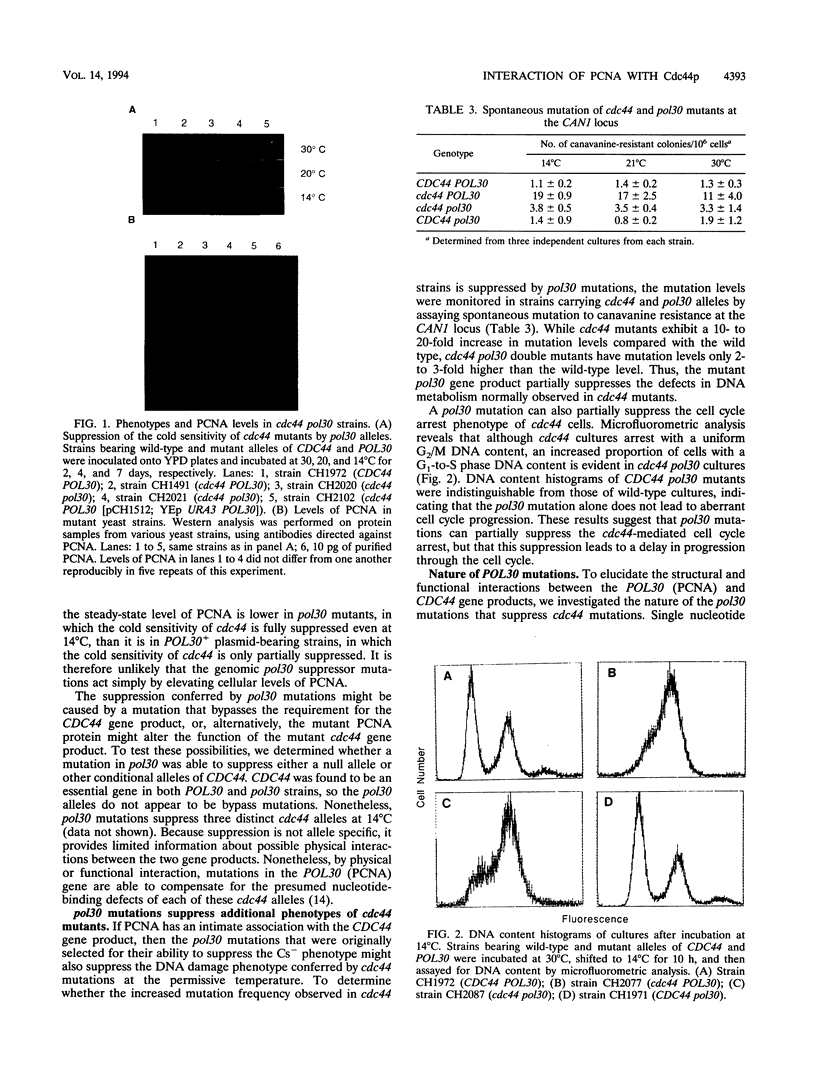
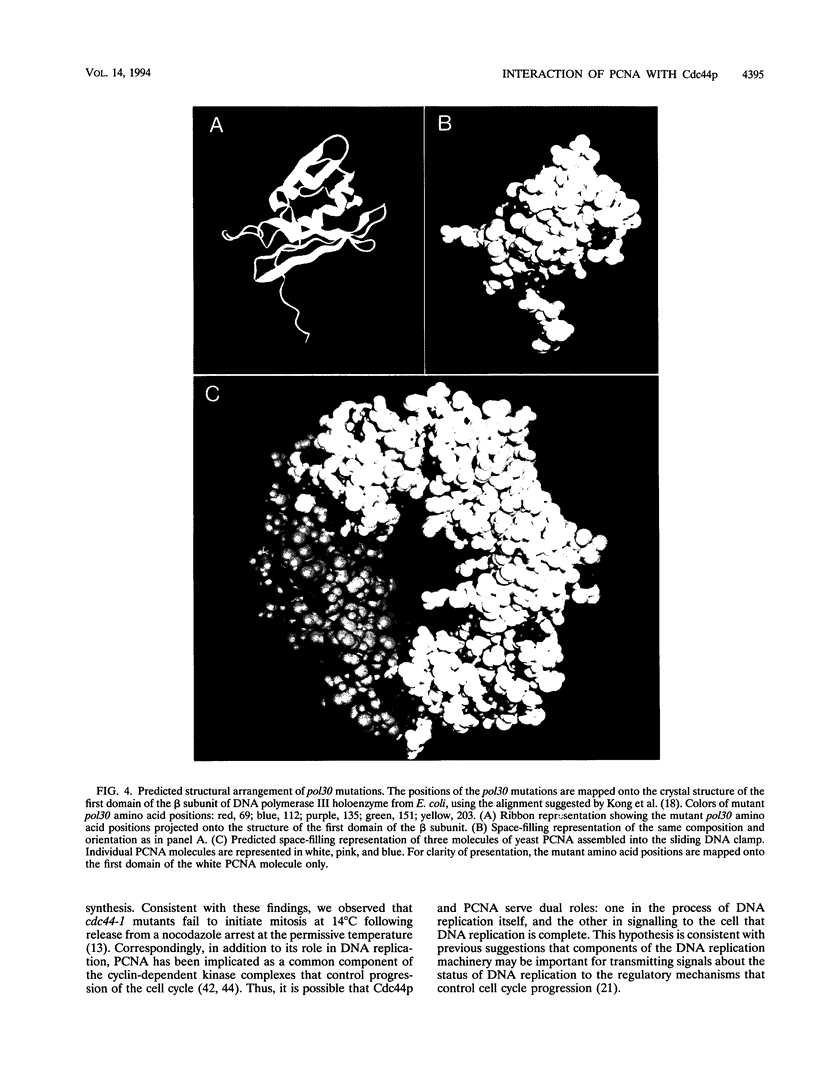
Abstract
In addition to its role as a processivity factor in DNA replication, proliferating cell nuclear antigen (PCNA) may function in the regulation of cell cycle progression. We present genetic evidence that PCNA interacts with the gene product of CDC44, an essential nucleotide-binding protein that encodes the large subunit of yeast replication factor C (K. Fien and B. Stillman, personal communication). Mutations in POL30 (PCNA) suppress cold-sensitive alleles of cdc44 that contain mutations in or near nucleotide-binding consensus domains, but they do not suppress a null allele. Thus, it appears that PCNA interacts with Cdc44p but cannot substitute for its function. pol30 mutations suppress additional phenotypes of cdc44 mutations, including the cold sensitivity that they were selected to suppress. This observation suggests an intimate association between PCNA and Cdc44p. Each of five independent pol30 mutants contains a unique single mutation that maps to a localized region on one face of the predicted three-dimensional structure of PCNA. This face identifies a region likely to be important for functional interaction between the CDC44 and POL30 gene products.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer G. A., Burgers P. M. Molecular cloning, structure and expression of the yeast proliferating cell nuclear antigen gene. Nucleic Acids Res. 1990 Jan 25;18(2):261–265. doi: 10.1093/nar/18.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R., Celis J. E. A search for differential polypeptide synthesis throughout the cell cycle of HeLa cells. J Cell Biol. 1980 Mar;84(3):795–802. doi: 10.1083/jcb.84.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R., Frank R., Blundell P. A., Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature. 1987 Apr 2;326(6112):515–517. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- Brill S. J., Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991 Sep;5(9):1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- Burgers P. M. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases delta and epsilon. J Biol Chem. 1991 Nov 25;266(33):22698–22706. [PubMed] [Google Scholar]
- Chen M., Pan Z. Q., Hurwitz J. Sequence and expression in Escherichia coli of the 40-kDa subunit of activator 1 (replication factor C) of HeLa cells. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2516–2520. doi: 10.1073/pnas.89.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Pan Z. Q., Hurwitz J. Studies of the cloned 37-kDa subunit of activator 1 (replication factor C) of HeLa cells. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5211–5215. doi: 10.1073/pnas.89.12.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch T., Nurse P. Coupling M phase and S phase: controls maintaining the dependence of mitosis on chromosome replication. Cell. 1991 Jun 14;65(6):921–923. doi: 10.1016/0092-8674(91)90542-7. [DOI] [PubMed] [Google Scholar]
- Fien K., Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol Cell Biol. 1992 Jan;12(1):155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten U. B., Erlich H. A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell E. A., McAlear M. A., Rose D., Holm C. CDC44: a putative nucleotide-binding protein required for cell cycle progression that has homology to subunits of replication factor C. Mol Cell Biol. 1994 Jan;14(1):255–267. doi: 10.1128/mcb.14.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter K. J., Eipel H. E. Microbial determinations by flow cytometry. J Gen Microbiol. 1979 Aug;113(2):369–375. doi: 10.1099/00221287-113-2-369. [DOI] [PubMed] [Google Scholar]
- Jarvik J., Botstein D. Conditional-lethal mutations that suppress genetic defects in morphogenesis by altering structural proteins. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2738–2742. doi: 10.1073/pnas.72.7.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. M., Snyder M., Chang L. M., Davis R. W., Campbell J. L. Isolation of the gene encoding yeast DNA polymerase I. Cell. 1985 Nov;43(1):369–377. doi: 10.1016/0092-8674(85)90042-x. [DOI] [PubMed] [Google Scholar]
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Lee S. H., Hurwitz J. Mechanism of elongation of primed DNA by DNA polymerase delta, proliferating cell nuclear antigen, and activator 1. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5672–5676. doi: 10.1073/pnas.87.15.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Ishimi Y., Kenny M. K., Bullock P., Dean F. B., Hurwitz J. An inhibitor of the in vitro elongation reaction of simian virus 40 DNA replication is overcome by proliferating-cell nuclear antigen. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9469–9473. doi: 10.1073/pnas.85.24.9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. J., Deshaies R. J. Exercising self-restraint: discouraging illicit acts of S and M in eukaryotes. Cell. 1993 Jul 30;74(2):223–226. doi: 10.1016/0092-8674(93)90413-k. [DOI] [PubMed] [Google Scholar]
- Li X., Burgers P. M. Molecular cloning and expression of the Saccharomyces cerevisiae RFC3 gene, an essential component of replication factor C. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):868–872. doi: 10.1073/pnas.91.3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo I. A., Hough P. V., Wall J. S., Dodson M., Dean F. B., Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989 Apr 20;338(6217):658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- Moir D., Botstein D. Determination of the order of gene function in the yeast nuclear division pathway using cs and ts mutants. Genetics. 1982 Apr;100(4):565–577. doi: 10.1093/genetics/100.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir D., Stewart S. E., Osmond B. C., Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics. 1982 Apr;100(4):547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Araki H., Clark A. B., Hamatake R. K., Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990 Sep 21;62(6):1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell M., Onrust R., Dean F. B., Chen M., Hurwitz J. Homology in accessory proteins of replicative polymerases--E. coli to humans. Nucleic Acids Res. 1993 Jan 11;21(1):1–3. doi: 10.1093/nar/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D. Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics. 1949 Sep;34(5):607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G., Tan C. K., Kostura M., Mathews M. B., So A. G., Downey K. M., Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987 Apr 2;326(6112):517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitney K. C., Budd M. E., Campbell J. L. DNA polymerase III, a second essential DNA polymerase, is encoded by the S. cerevisiae CDC2 gene. Cell. 1989 Feb 24;56(4):599–605. doi: 10.1016/0092-8674(89)90582-5. [DOI] [PubMed] [Google Scholar]
- So A. G., Downey K. M. Eukaryotic DNA replication. Crit Rev Biochem Mol Biol. 1992;27(1-2):129–155. doi: 10.3109/10409239209082561. [DOI] [PubMed] [Google Scholar]
- Stukenberg P. T., Studwell-Vaughan P. S., O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991 Jun 15;266(17):11328–11334. [PubMed] [Google Scholar]
- Tan C. K., Castillo C., So A. G., Downey K. M. An auxiliary protein for DNA polymerase-delta from fetal calf thymus. J Biol Chem. 1986 Sep 15;261(26):12310–12316. [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991 Jan 25;266(3):1950–1960. [PubMed] [Google Scholar]
- Waseem N. H., Labib K., Nurse P., Lane D. P. Isolation and analysis of the fission yeast gene encoding polymerase delta accessory protein PCNA. EMBO J. 1992 Dec;11(13):5111–5120. doi: 10.1002/j.1460-2075.1992.tb05618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988 Jul 15;241(4863):317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Wold M. S., Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Zhang H., Beach D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992 Oct 30;71(3):505–514. doi: 10.1016/0092-8674(92)90518-h. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Zhang H., Beach D. Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation. Genes Dev. 1993 Aug;7(8):1572–1583. doi: 10.1101/gad.7.8.1572. [DOI] [PubMed] [Google Scholar]
- Yoder B. L., Burgers P. M. Saccharomyces cerevisiae replication factor C. I. Purification and characterization of its ATPase activity. J Biol Chem. 1991 Nov 25;266(33):22689–22697. [PubMed] [Google Scholar]
- Zhang H., Xiong Y., Beach D. Proliferating cell nuclear antigen and p21 are components of multiple cell cycle kinase complexes. Mol Biol Cell. 1993 Sep;4(9):897–906. doi: 10.1091/mbc.4.9.897. [DOI] [PMC free article] [PubMed] [Google Scholar]