Abstract
The delivery of a macromolecule to the cytosol of human cells is assessed by using a pendant di-O-glycosylated derivative of fluorescein. Its fluorescence is unmasked by Escherichia coli β-galactosidase installed in the cytosol. Background is diminished by using RNAi to suppress the expression of GLB1, which encodes a lysosomal β-galactosidase. This strategy was used to quantify the cytosolic entry of a highly cationic protein, ribonuclease A.
The cytosol of mammalian cells is the intended destination for many proteins and other macromolecules of interest.1 Typically, delivery entails endocytosis, followed by translocation from an endosomal compartment to the cytosol. Uptake into endosomes can be facilitated by a cationic appendage2 or graft,3,4 and can be assessed with a pendant fluorogenic probe that is unmasked by an endosomal esterase.5-8 But it is the next step, entailing traversal of a phospholipid bilayer, that is the major hurdle to cytosolic delivery.9
Endosome-to-cytosol translocation is a rare event and thus difficult to assess.10,11 Two strategies are evident for distinguishing the cytosolic pool of internalized molecules from the endosomal pool. One is to separate membrane-bound compartments from the cytosol by subcellular fractionation. This method requires cell lysis, and is thus inapplicable to live cells or high-throughput screens. A second method relies on the modification of a latent probe, such that macromolecules that reach the cytosol are differentiated from those in endosomes. Previous strategies for measuring translocation to the cytosol have relied on fluorescence resonance energy transfer (FRET), either by monitoring FRET between a preinstalled cytosolic fluorescent protein and a fluorophore-tagged peptide,12 or by monitoring the unquenching of fluorescence of a pair of disulfide-linked fluorophores.13 Although these strategies did overcome the difficulty of excluding signals from endocytic compartments, they suffer from a high background.
To avoid the background that accompanies chemical reactions, we sought an enzyme–substrate pair that was orthogonal to human cells. We reasoned that macromolecules tagged with fluorescein di-β-D-galactopyranoside (FDG) could be unmasked by Escherichia coli β-galactosidase (β-gal) produced by heterologous expression in the cytosol (Fig. 1).14 As a model protein, we chose bovine pancreatic ribonuclease (RNase A), which has well-established physicochemical properties.15-17 Like other highly cationic proteins,3,4 RNase A (pI = 9.33 18) is taken up readily by mammalian cells.19-22
Fig. 1.
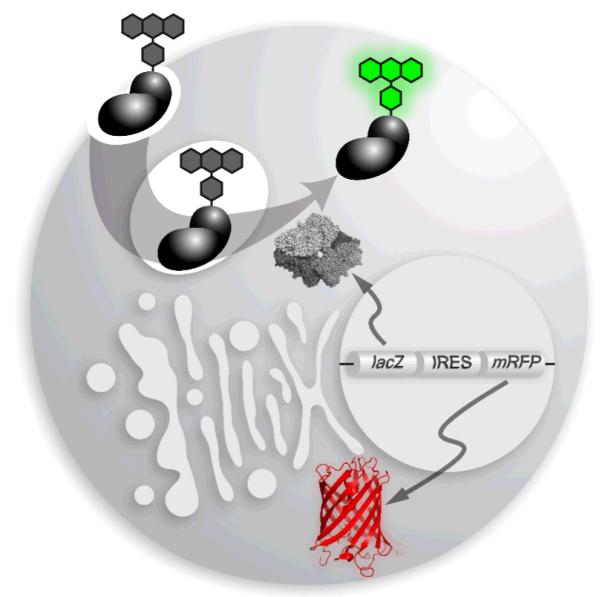
Strategy for detecting the entry of an exogenous protein into the cytosol of a human cell. A protein with a pendant fluorogenic probe is internalized by endocytosis. The conjugate is not fluorescent until hydrolyzed by β-galactosidase installed in the cytosol by transfection of a plasmid, which directs the concomitant expression of the monomeric red fluorescent protein (mRFP).
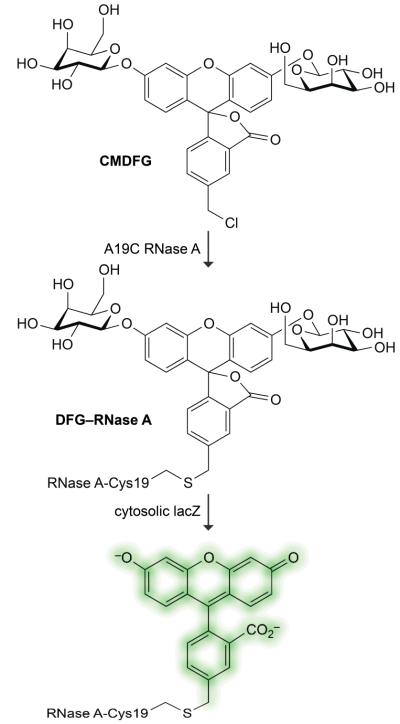
We sought a means to conjugate FDG to RNase A. Sulfhydryl groups are readily alkylated by a variety of electrophiles. Wild-type RNase A has no sulfhydryl groups, so we used site-directed mutageneis to create the A19C variant, which has a cysteine residue in a solvent-exposed surface loop.23 We alkylated its nascent sulfhydryl group with 5-chloromethylfluorescein di-β-D-galactopyranoside (CMDFG) to create FDG–RNase A (Fig. 2).
Fig. 2.
Strategy for conjugating fluorescein di-β-D-galactopyranoside (FDG) to bovine pancreatic ribonuclease (RNase A). The linkage is made via a cysteine residue installed at position 19 of RNase A by site-directed mutagenesis.
We assessed the hydrolytic stability of FDG–RNase A and its ability to be a substrate for β-gal. We found that the fluorescence of FDG–RNase A remains masked for ≥12 h at pH 7, as well as at pH 4 (Fig. S1A in the Supplementary Information), which is close to that found in late endosomes. We also found that the fluorescence of FDG–RNase A increases by 102-fold upon addition of β-gal (Fig. S1B in the Supplementary Information), validating the conjugate as a substrate.
Next, we examined the hydrolytic stability of FDG–RNase A in unmodified HeLa cells, which are lacZ−. Surprisingly, extended incubation (24 h) resulted in substantial background fluorescence, suggesting that an endogenous human enzyme can catalyze the unmasking of FDG–RNase A. We suspected that the undesired activity could arise from a human β-galactosidase, GLB1, which is a 76-kDa lysosomal enzyme that is not homologous to β-gal.24,25
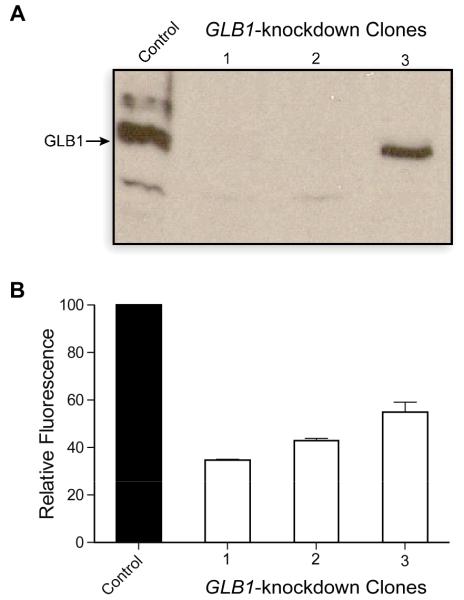
We used RNAi to suppress the activity of GLB1. Specifically, we created HeLa cell clones that stably express a short hairpin RNA (shRNA) that targets GLB1.25 To assess success, we incubated FDG–RNase A with three clones having decreased GLB1 expression (Fig. 3A) and measured the ensuing fluorescence with flow cytometry (Fig. 3B). The GLB1-knockdown clones had 50–65% less background fluorescence than unmodified HeLa cells, indicating that GLB1 activity is responsible for most of the unmasking of FDG–RNase A in unmodified HeLa cells.
Fig. 3.
Stable HeLa GLB1 knockdown clones with reduced background fluorescence. (A) Cell lysates from unmodified HeLa cells (control) and cells stably transfected with GLB1-targeting shRNA were analyzed by immunoblotting with an anti-GLB1 antibody. (B) FDG–RNase A was incubated with unmodified HeLa cells and GLB1 knockdown clones for 24 h at 37°C. Mean values (±SE) are from flow cytometry of 20,000 live cells (≥3 populations).
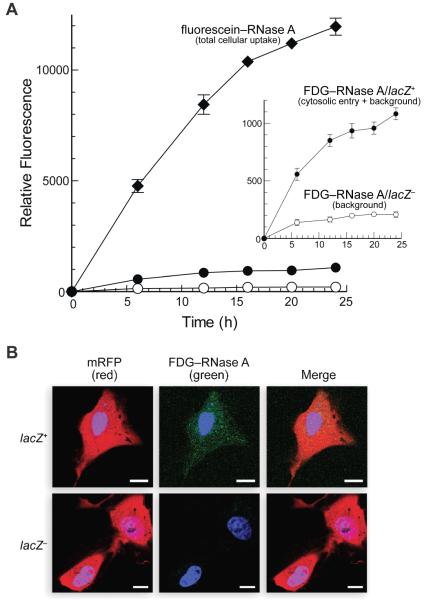
Then, we transfected GLB1-knockdown HeLa cells (clone 1) with a plasmid containing a lacZ–IRES–mRFP construct (Fig. 1). Here, the internal ribosomal entry site (IRES) correlates the expression of the monomeric red fluorescent protein (mRFP) and the lacZ gene. Thus, lacZ+ cells can be identified definitively by their mRFP fluorescence. The cytosolic entry of RNase A was determined by incubating FDG–RNase A with lacZ+ cells. In a control experiment, the same amount of ribonuclease conjugate was incubated with cells transfected with a plasmid containing IRES–mRFP but lacking lacZ.26 The cytosolic influx of RNase A increases rapidly in lacZ+ cells (Fig. 4A). Levels of background fluorescence, as measured within lacZ− cells, accounts for ~20% of the fluorescence of lacZ+ cells. These data are consistent with observations made by using fluorescence microscopy. As expected, we observed diffuse fluorescence in lacZ+ clone-1 HeLa cells, but little or no fluorescence in lacZ− cells (Fig. 4B). Punctations likely arise from the ~20% background fluorscence. These results illustrate that cytosolic entry of a protein can be assessed by using pendant FDG with lacZ+ human cells.
Fig. 4.
Cellular uptake and cytosolic entry of RNase A. lacZ+ clone-1 HeLa cells were transfected with the lacZ–IRES–mRFP construct (Fig. 1). lacZ− clone-1 HeLa cells were transfected with the same vector but containing an IRES–mRFP construct. (A) FDG–RNase A (5 μM) was added to lacZ+ (●) and lacZ− (○) cells. Mean values (±SE) are for green fluorescence from flow cytometry of 10,000 live cells (≥6 populations) that emit red fluorescence. Fluorescein–RNase A (5 μM) was added to cells treated with bafilomycin A1 (0.10 μM). Mean values (±SE) are for green fluorescence from flow cytometry of 10,000 live cells (≥6 populations). Inset: Close-up of FDG–RNase A data. (B) FDG–RNase A (5 μM) was incubated with lacZ+ and lacZ− cells for 24 h at 37°C. Blue dye, Hoechst 33342; scale bars, 10 μm.
Next, we employed a simple fluorescein–RNase A conjugate19 to discern the fraction of internalized molecules that reach the cytosol. The conjugate acid of the fluorescein anion has pKa = 6.30,27 and loses fluorescence upon protonation. Accordingly, we used bafilomycin A1, which is a specific inhibitor of the vacuolar ATPase responsible for endosome acidification,28,20 to neutralize endosomes and enable an accurate measure of total fluorescein–RNase A uptake. The ensuing data indicate that the vast majority of this cationic protein is entrapped in endocytic vesicles (Fig. 4A). We estimate that only ~7% of the protein that enters cells is able to translocate to the cytosol in 24 h.
Finally, we extended our analysis by quantifying the RNase A molecules that reach the cytosol. Ribonucleases are promising chemotherapeutic agents. They have an innate ability to enter the cytosol, where they catalyze RNA degradation if they can evade the resident ribonuclease inhibitor protein, which binds to RNase A with femtomolar affinity.29-33 Evasive RNase A variants inhibit the proliferation of human cancer cells with IC50 values in the high nanomolar range.34 Using quantum dot calibration beads,35 we found that the total fluorescence in lacZ+ cells upon incubation with 5 μM FDG–RNase A for 24 h derives from (3.5 ± 0.2) × 104 molecules/cell. As noted above, ~80% of these fluorescent molecules are in the cytosol, and the remaining ~20% are in the large pool of endosomal FDG–RNase A (Fig. 4A). The volume of the cytosol in a HeLa cell is 940 μm3.36 Accordingly, a 5 μM dose of FDG–RNase A for 24 h results in a cytosolic concentration of ~50 nM. As the concentration of ribonuclease inhibitor in the cytosol is 4 μM,37 these data explain the inability of low concentrations of wild-type RNase A to manifest cytotoxicity—the efficiency of its translocation to the cytosol is too low to overcome the ribonuclease inhibitor protein.
Supplementary Material
Acknowledgments
This work was supported by Grants R01 CA073808 and R01 GM044783 (NIH). T.-Y.C. was supported by the Dr. James Chieh-Hsia Mao Wisconsin Distinguished Graduate Fellowship. We are grateful to L. D. Lavis and J. E. Lomax for advice, and to D. C. DiMaio and B. Sugden for plasmids.
Footnotes
Electronic supplementary information (ESI) available: Experimental procesures. See DOI: 10.1039/xx.
Notes and references
- 1.Torchilin VP. Recent approaches to intracellular delivery of drugs and DNA and organelle targeting. Annu. Rev. Biophys. Biomol. Eng. 2006;8:343–375. doi: 10.1146/annurev.bioeng.8.061505.095735. [DOI] [PubMed] [Google Scholar]
- 2.Wender PA, Galliher WC, Goun EA, Jones LR, Pillow TH. The design of guanidinium-rich transporters and their internalization mechanisms. Adv. Drug Deliv. Rev. 2008;60:452–472. doi: 10.1016/j.addr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs SM, Raines RT. Arginine grafting to endow cell permeability. ACS Chem. Biol. 2007;2:167–170. doi: 10.1021/cb600429k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson DB, Cronican JJ, Liu DR. Engineering and identifying supercharged proteins for macromolecule delivery into mammalian cells. Methods Enzymol. 2012;503:293–319. doi: 10.1016/B978-0-12-396962-0.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang TL, Székács A, Uematsu T, Kuwano E, Parkinson A, Hammock BD. Hydrolysis of carbonates, thiocarbonates, carbamates, and carboxylic esters of α-naphthol, β-naphtol, and p-nitrophenol by human, rat, and mouse liver carboxylesterases. Pharm. Res. 1993;10:639–648. doi: 10.1023/a:1018987111362. [DOI] [PubMed] [Google Scholar]
- 6.Lavis LD, Chao T-Y, Raines RT. Fluorogenic label for biomolecular imaging. ACS Chem. Biol. 2006;1:252–260. doi: 10.1021/cb600132m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavis LD, Raines RT. Bright ideas for chemical biology. ACS Chem. Biol. 2008;3:142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian L, Yang Y, Wysocki LW, Arnold AC, Hu A, Ravichandran B, Sternson SM, Looger LL, Lavis LD. Selective esterase–ester pair for targeting small molecules with cellular specificity. Proc. Natl. Acad. Sci. USA. 2012;109:4756–4761. doi: 10.1073/pnas.1111943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larson GM, Lee KD. Macromolecular cytosolic delivery: Cell membranes as the primary obstacle. Arch. Pharm. Res. 1998;21:621–628. doi: 10.1007/BF02976747. [DOI] [PubMed] [Google Scholar]
- 10.Jensen KD, Nori A, Tijerina M, Kopeckova P, Kopecek J. Cytoplasmic delivery and nuclear targeting of synthetic macromolecules. J. Control. Release. 2003;87:89–105. doi: 10.1016/s0168-3659(02)00352-8. [DOI] [PubMed] [Google Scholar]
- 11.Henriques ST, Melo MN, Castanho M. How to address CPP and AMP translocation? Methods to detect and quantify peptide internalization in vitro and in vivo. Mol. Membr. Biol. 2007;24:173–184. doi: 10.1080/09687860601102476. [DOI] [PubMed] [Google Scholar]
- 12.Adams SR, Tsien RY. Langel U, editor. Imaging the influx of cell-penetrating peptides into the cytosol of individual live cells. Handbook of Cell-Penetrating Peptides. (2nd edn) 2006:505–512. [Google Scholar]
- 13.Cheung JC, Chiaw PK, Deber CM, Bear CE. A novel method for monitoring the cytosolic delivery of peptide cargo. J. Control. Release. 2009;137:2–7. doi: 10.1016/j.jconrel.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 14.Plovins A, Alvarez AM, Ibanez M, Molina M, Nombela C. Use of fluorescein-di-β-D-galactopyranoside (FDG) and C12-FDG as substrates for β-galactosidase detection by flow cytometry in animal, bacterial, and yeast cells. Appl. Environ. Microbiol. 1994;60:4638–4641. doi: 10.1128/aem.60.12.4638-4641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raines RT. Ribonuclease A. Chem. Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 16.Marshall GR, Feng JA, Kuster DJ. Back to the future: Ribonuclease A. Biopolymers. 2008;90:259–277. doi: 10.1002/bip.20845. [DOI] [PubMed] [Google Scholar]
- 17.Cuchillo CM, Nogués MV, Raines RT. Biochemistry. 2011;50:7835–7841. doi: 10.1021/bi201075b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ui N. Isolectric points and conformation of proteins. I. Effect of urea on the behavior of some proteins in isoelectric focusing. Biochim. Biophys. Acta. 1971;229:567–581. [PubMed] [Google Scholar]
- 19.Haigis MC, Raines RT. Secretory ribonucleases are internalized by a dynamin-independent endocytic pathway. J. Cell Sci. 2003;116:313–324. doi: 10.1242/jcs.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodríguez M, Torrent G, Bosch M, Rayne F, Dubremetz J-F, Ribó M, Benito A, Vilanova M, Beaumelle B. Intracellular pathway of Onconase that enables its delivery to the cytosol. J. Cell Sci. 2007;120:1405–1411. doi: 10.1242/jcs.03427. [DOI] [PubMed] [Google Scholar]
- 21.Chao T-Y, Raines RT. Mechanism of ribonuclease A endocytosis: Analogies to cell-penetrating peptides. Biochemistry. 2010;50:8374–8382. doi: 10.1021/bi2009079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao T-Y, Lavis LD, Raines RT. Cellular uptake of ribonuclease A relies on anionic glycans. Biochemistry. 2010;49:10666–10673. doi: 10.1021/bi1013485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kothandaraman S, Hebert MC, Raines RT, Nibert ML. No role for pepstatin-A–sensitive acidic proteinase in reovirus infections of L or MDCK cells. Virology. 1998;251:264–272. doi: 10.1006/viro.1998.9434. [DOI] [PubMed] [Google Scholar]
- 24.Norden AGW, Tennant LL, Og’Brien JS. GM1 ganglioside β-galactosidase A. J. Biol. Chem. 1974;249:7969–7976. [PubMed] [Google Scholar]
- 25.Lee BY, Han JA, Im JS, Morrone A, Johung K, DiMaio D, Hwang ES. Senescence-associated β-galactosidase is lysosomal β-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 26.Pratt ZL, Zhang J, Sugden B. The latent membrane protein 1 (LMP1) oncogene of Epstein–Barr virus can simultaneously induce and inhibit apoptosis in B cells. J. Virol. 2012;86:4380–4393. doi: 10.1128/JVI.06966-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavis LD, Rutkoski TJ, Raines RT. Tuning the pKa of fluorescein to optimize binding assays. Anal. Chem. 2007;79:6775–6782. doi: 10.1021/ac070907g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crider BP, Xie X-S, Stone DK. Bafilomycin inhibits proton flow through the H+ channel of vacuolar proton pumps. J. Biol. Chem. 1994;269:17379–17381. [PubMed] [Google Scholar]
- 29.Leland PA, Schultz LW, Kim B-M, Raines RT. Ribonuclease A variants with potent cytotoxic activity. Proc. Natl. Acad. Sci. USA. 1998;95:10407–10412. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold U, Ulbrich-Hofmann R. Natural and engineered ribonucleases as potential cancer therapeutics. Biotechnol. Lett. 2006;28:1615–1622. doi: 10.1007/s10529-006-9145-0. [DOI] [PubMed] [Google Scholar]
- 31.Lee JE, Raines RT. Ribonucleases as novel chemotherapeutics: The ranpirnase example. BioDrugs. 2008;22:53–58. doi: 10.2165/00063030-200822010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutkoski TJ, Raines RT. Evasion of ribonuclease inhibitor as a determinant of ribonuclease cytotoxicity. Curr. Pharm. Biotechnol. 2008;9:185–199. doi: 10.2174/138920108784567344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lomax JE, Eller CH, Raines RT. Rational design and evaluation of mammalian ribonuclease cytotoxins. Methods Enzymol. 2012;502:273–290. doi: 10.1016/B978-0-12-416039-2.00014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutkoski TJ, Kurten EL, Mitchell JC, Raines RT. Disruption of shape-complementarity markers to create cytotoxic variants of ribonuclease A. J. Mol. Biol. 2005;354:41–54. doi: 10.1016/j.jmb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, Campos SK, Lopez GP, Ozbun MA, Sklar LA, Buranda T. The development of quantum dot calibration beads and quantitative multicolor bioassays in flow cytometry and microscopy. Anal. Biochem. 2007;364:180–192. doi: 10.1016/j.ab.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujioka A, Terai K, Itoh RE, Aoki K, Nakamura T, Kuroda S, Nishida E, Matsuda M. Mechanisms of signal transduction. J. Biol. Chem. 2006;281:8917–8926. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- 37.Haigis MC, Kurten EL, Raines RT. Ribonuclease inhibitor as an intracellular sentry. Nucleic Acids Res. 2003;31:1024–1032. doi: 10.1093/nar/gkg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.