Abstract
The title compound, C11H10O3, is a phenyl-subsituted dihydropyrandione in which the heterocycle adopts a boat conformation with the phenyl substituent canted 72.14 (5)° relative to the mean plane of the heterocycle.
Related literature
For the crystal structure of methyl 4-methyl-3,5-dioxo-1-phenyl-2-oxaspiro[5.5]-4-carboxylate, see: Kirillov et al. (2010 ▶) and of trans-5,6-diphenylperhydropyran-2,4-dione, see: de Souza et al. (2009 ▶). For the synthesis, see: Andersh et al. (2008 ▶). For the biological activity of the title compound and its derivatives, see: Aguiar Amaral et al. (2005 ▶); Souza et al. (2004 ▶); Tait et al. (1997 ▶); Wang et al. (1999 ▶). For a description of the Cambridge Structural Database, see: Allen (2002 ▶). A geometry check was performed using Mogul, see: Bruno et al. (2004 ▶). For puckering parameters, see: Cremer & Pople (1975 ▶).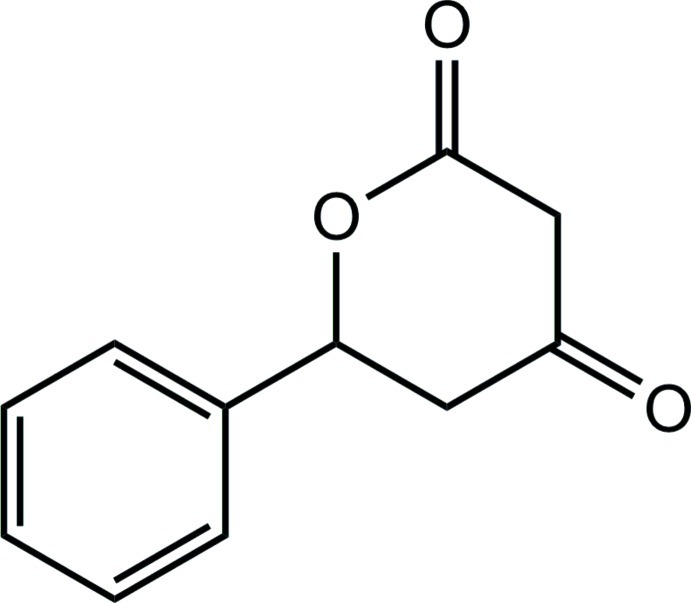
Experimental
Crystal data
C11H10O3
M r = 190.19
Orthorhombic,

a = 16.9888 (6) Å
b = 5.4501 (2) Å
c = 19.7350 (8) Å
V = 1827.28 (12) Å3
Z = 8
Mo Kα radiation
μ = 0.10 mm−1
T = 100 K
0.17 × 0.14 × 0.03 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2008 ▶) T min = 0.662, T max = 0.746
17960 measured reflections
1804 independent reflections
1322 reflections with I > 2σ(I)
R int = 0.071
Refinement
R[F 2 > 2σ(F 2)] = 0.047
wR(F 2) = 0.114
S = 1.06
1804 reflections
127 parameters
H-atom parameters constrained
Δρmax = 0.29 e Å−3
Δρmin = −0.25 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: APEX2 and SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SUPERFLIP (Palatinus & Chapuis, 2007 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶) and Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶), PLATON (Spek, 2009 ▶) and publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812049781/bx2433sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812049781/bx2433Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812049781/bx2433Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank the NSF–CHE (grant No. 1039689) for funding the X-ray diffractometer.
supplementary crystallographic information
Comment
The title compound has a diverse array of biological effects, including reducing sensitivity to pain (Aguiar Amaral et al., 2005) and killing mollusks (Souza et al., 2004). Derivatives of this compound have anti-fungal properties (Wang et al., 1999) and are effective HIV protease inhibitors (Tait et al., 1997).
The molecular structure (Fig. 1.) is the singular moiety in the asymmetric unit. A Mogul (Bruno et al., 2004) geometry check showed all non-H bond angles and distances to be normal. Ring puckering analysis of the dihydropyrandione ring using PLATON (Spek, 2009; Cremer & Pople, 1975) indicates Φ = 297.5 (2)° and θ = 84.76 (18)° for the O3—C1—C2—C3—C4—C5 ring. These parameters are consistent with a formal conformational assignment close to an idealized BC2,C5 boat with C2 at the bow and C5 at the stern. The plane of the phenyl ring attached to C5 may be described as a rudder canted 72.14 (5)° relative to the mean plane of the six core atoms of the heterocycle. The 106.6 (2)° C6—C5—O3 bond angle compared to the 112.8 (2)° C6—C5—C4 bond angle indicates a small steer to said rudder; however, whether it is to port or starboard depends upon which enantiomer is considered.
Based upon a CSD search (Allen, 2002), two structures containing similar lactone ring motifs have been reported in the crystallographic literature. These include the spiro compound methyl 4-methyl-3,5-dioxo-1-phenyl-2-oxaspiro[5.5]-4-carboxylate with CSD refcode IRITIN (Kirillov et al., 2010) and trans-5,6-diphenylperhydropyran-2,4-dione with CSD refcode PONVAQ (de Souza et al., 2009). In all three cases the pyran rings adopt the boat conformation.
Experimental
The title compound 6-(phenyl)-dihydro-2H-pyran-2,4-(3H)-dione, (also named 5-phenyl-3-oxo-delta-lactone), was prepared by the literature method (Andersh et al., 2008). Benzaldehyde (2 mmol), ethanol (2 ml), ethylacetoacetate (2 mmol), and potassium carbonate (4 mmol) were heated overnight under nitrogen at 318 K. The solution was diluted with ethylacetate, treated with 1 M HCl, and the combined organic layer extracts were dried, filtered, concentrated, and purified by flash chromatography.
Crystals suitable for X-Ray analysis were grown by vapor diffusion of pentane into a concentrated solution of the lactone in dichloromethane.
Refinement
All non-H atoms were refined anisotropically. All H atoms were included in the refinement in the riding-model approximation (C–H = 0.95, 0.99, and 1.00 Å for Ar–H, CH2, and CH; Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
The molecular structure of the compound with the atomic numbering scheme. Displacement ellipsoids are drawn at the 50% probability level.
Crystal data
| C11H10O3 | F(000) = 800 |
| Mr = 190.19 | Dx = 1.383 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 2535 reflections |
| a = 16.9888 (6) Å | θ = 2.4–23.5° |
| b = 5.4501 (2) Å | µ = 0.10 mm−1 |
| c = 19.7350 (8) Å | T = 100 K |
| V = 1827.28 (12) Å3 | Prism, colourless |
| Z = 8 | 0.17 × 0.14 × 0.03 mm |
Data collection
| Bruker APEXII CCD diffractometer | 1322 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.071 |
| φ and ω scans | θmax = 26.0°, θmin = 2.1° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −20→20 |
| Tmin = 0.662, Tmax = 0.746 | k = −6→6 |
| 17960 measured reflections | l = −24→24 |
| 1804 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.047 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.114 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.039P)2 + 1.8272P] where P = (Fo2 + 2Fc2)/3 |
| 1804 reflections | (Δ/σ)max < 0.001 |
| 127 parameters | Δρmax = 0.29 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.15992 (11) | 0.2655 (4) | 0.22819 (11) | 0.0198 (5) | |
| C2 | 0.13487 (12) | 0.0646 (4) | 0.27556 (11) | 0.0208 (5) | |
| H2A | 0.1203 | 0.1376 | 0.3198 | 0.025* | |
| H2B | 0.1799 | −0.0474 | 0.2833 | 0.025* | |
| C3 | 0.06630 (12) | −0.0808 (4) | 0.24883 (12) | 0.0200 (5) | |
| C4 | 0.06699 (12) | −0.1241 (4) | 0.17377 (11) | 0.0203 (5) | |
| H4A | 0.0222 | −0.0357 | 0.1529 | 0.024* | |
| H4B | 0.0598 | −0.3015 | 0.1649 | 0.024* | |
| C5 | 0.14301 (12) | −0.0391 (4) | 0.14093 (11) | 0.0197 (5) | |
| H5 | 0.1869 | −0.1489 | 0.1557 | 0.024* | |
| C6 | 0.13875 (12) | −0.0384 (4) | 0.06490 (11) | 0.0196 (5) | |
| C7 | 0.17318 (12) | −0.2284 (4) | 0.02863 (12) | 0.0240 (5) | |
| H7 | 0.2002 | −0.3554 | 0.052 | 0.029* | |
| C8 | 0.16830 (13) | −0.2336 (4) | −0.04141 (12) | 0.0289 (6) | |
| H8 | 0.1921 | −0.364 | −0.0659 | 0.035* | |
| C9 | 0.12897 (13) | −0.0501 (4) | −0.07580 (12) | 0.0283 (5) | |
| H9 | 0.1257 | −0.0542 | −0.1238 | 0.034* | |
| C10 | 0.09416 (13) | 0.1406 (4) | −0.03986 (12) | 0.0284 (6) | |
| H10 | 0.0672 | 0.2673 | −0.0634 | 0.034* | |
| C11 | 0.09869 (12) | 0.1464 (4) | 0.03021 (11) | 0.0242 (5) | |
| H11 | 0.0745 | 0.2763 | 0.0546 | 0.029* | |
| O1 | 0.17979 (9) | 0.4702 (3) | 0.24675 (8) | 0.0240 (4) | |
| O2 | 0.01417 (8) | −0.1540 (3) | 0.28609 (8) | 0.0237 (4) | |
| O3 | 0.16073 (8) | 0.2144 (3) | 0.16170 (7) | 0.0211 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0139 (10) | 0.0164 (11) | 0.0290 (13) | 0.0004 (8) | −0.0010 (9) | −0.0005 (10) |
| C2 | 0.0213 (10) | 0.0181 (11) | 0.0230 (11) | 0.0013 (8) | 0.0009 (9) | −0.0004 (9) |
| C3 | 0.0190 (10) | 0.0102 (9) | 0.0307 (12) | 0.0038 (8) | 0.0010 (10) | 0.0024 (9) |
| C4 | 0.0183 (10) | 0.0147 (10) | 0.0279 (12) | −0.0013 (9) | 0.0001 (9) | −0.0002 (9) |
| C5 | 0.0193 (10) | 0.0125 (10) | 0.0274 (12) | 0.0011 (8) | −0.0004 (9) | −0.0019 (9) |
| C6 | 0.0151 (10) | 0.0168 (10) | 0.0269 (12) | −0.0038 (8) | 0.0001 (9) | −0.0007 (10) |
| C7 | 0.0221 (11) | 0.0184 (11) | 0.0314 (13) | −0.0003 (9) | 0.0002 (9) | −0.0002 (10) |
| C8 | 0.0305 (12) | 0.0234 (12) | 0.0327 (14) | −0.0010 (10) | 0.0030 (10) | −0.0079 (11) |
| C9 | 0.0332 (13) | 0.0285 (13) | 0.0232 (12) | −0.0066 (10) | −0.0002 (10) | −0.0006 (11) |
| C10 | 0.0315 (13) | 0.0210 (12) | 0.0326 (14) | −0.0021 (10) | −0.0039 (10) | 0.0030 (11) |
| C11 | 0.0258 (11) | 0.0179 (11) | 0.0289 (13) | −0.0008 (9) | 0.0017 (10) | −0.0029 (10) |
| O1 | 0.0257 (8) | 0.0159 (7) | 0.0303 (9) | −0.0028 (6) | −0.0007 (7) | −0.0015 (7) |
| O2 | 0.0230 (8) | 0.0158 (7) | 0.0324 (9) | −0.0014 (6) | 0.0062 (7) | 0.0014 (7) |
| O3 | 0.0229 (7) | 0.0152 (7) | 0.0253 (9) | −0.0048 (6) | −0.0002 (6) | 0.0004 (7) |
Geometric parameters (Å, º)
| C1—O1 | 1.222 (3) | C5—H5 | 1 |
| C1—O3 | 1.342 (2) | C6—C7 | 1.388 (3) |
| C1—C2 | 1.501 (3) | C6—C11 | 1.395 (3) |
| C2—C3 | 1.505 (3) | C7—C8 | 1.385 (3) |
| C2—H2A | 0.99 | C7—H7 | 0.95 |
| C2—H2B | 0.99 | C8—C9 | 1.381 (3) |
| C3—O2 | 1.218 (2) | C8—H8 | 0.95 |
| C3—C4 | 1.500 (3) | C9—C10 | 1.391 (3) |
| C4—C5 | 1.517 (3) | C9—H9 | 0.95 |
| C4—H4A | 0.99 | C10—C11 | 1.385 (3) |
| C4—H4B | 0.99 | C10—H10 | 0.95 |
| C5—O3 | 1.472 (2) | C11—H11 | 0.95 |
| C5—C6 | 1.502 (3) | ||
| O1—C1—O3 | 118.69 (19) | C6—C5—H5 | 109.2 |
| O1—C1—C2 | 123.9 (2) | C4—C5—H5 | 109.2 |
| O3—C1—C2 | 117.43 (18) | C7—C6—C11 | 119.4 (2) |
| C1—C2—C3 | 112.66 (18) | C7—C6—C5 | 119.53 (19) |
| C1—C2—H2A | 109.1 | C11—C6—C5 | 121.03 (19) |
| C3—C2—H2A | 109.1 | C8—C7—C6 | 120.3 (2) |
| C1—C2—H2B | 109.1 | C8—C7—H7 | 119.8 |
| C3—C2—H2B | 109.1 | C6—C7—H7 | 119.8 |
| H2A—C2—H2B | 107.8 | C9—C8—C7 | 120.3 (2) |
| O2—C3—C4 | 123.38 (19) | C9—C8—H8 | 119.9 |
| O2—C3—C2 | 121.6 (2) | C7—C8—H8 | 119.9 |
| C4—C3—C2 | 115.03 (18) | C8—C9—C10 | 119.8 (2) |
| C3—C4—C5 | 112.35 (17) | C8—C9—H9 | 120.1 |
| C3—C4—H4A | 109.1 | C10—C9—H9 | 120.1 |
| C5—C4—H4A | 109.1 | C11—C10—C9 | 120.2 (2) |
| C3—C4—H4B | 109.1 | C11—C10—H10 | 119.9 |
| C5—C4—H4B | 109.1 | C9—C10—H10 | 119.9 |
| H4A—C4—H4B | 107.9 | C10—C11—C6 | 120.0 (2) |
| O3—C5—C6 | 106.59 (17) | C10—C11—H11 | 120 |
| O3—C5—C4 | 109.98 (16) | C6—C11—H11 | 120 |
| C6—C5—C4 | 112.73 (17) | C1—O3—C5 | 117.72 (16) |
| O3—C5—H5 | 109.2 | ||
| O1—C1—C2—C3 | 139.8 (2) | C11—C6—C7—C8 | 0.4 (3) |
| O3—C1—C2—C3 | −40.6 (3) | C5—C6—C7—C8 | 178.5 (2) |
| C1—C2—C3—O2 | −141.94 (19) | C6—C7—C8—C9 | −0.1 (3) |
| C1—C2—C3—C4 | 37.3 (2) | C7—C8—C9—C10 | 0.0 (3) |
| O2—C3—C4—C5 | −173.43 (19) | C8—C9—C10—C11 | −0.2 (3) |
| C2—C3—C4—C5 | 7.4 (2) | C9—C10—C11—C6 | 0.5 (3) |
| C3—C4—C5—O3 | −50.7 (2) | C7—C6—C11—C10 | −0.6 (3) |
| C3—C4—C5—C6 | −169.46 (17) | C5—C6—C11—C10 | −178.68 (19) |
| O3—C5—C6—C7 | 137.70 (18) | O1—C1—O3—C5 | 174.97 (17) |
| C4—C5—C6—C7 | −101.5 (2) | C2—C1—O3—C5 | −4.6 (3) |
| O3—C5—C6—C11 | −44.2 (2) | C6—C5—O3—C1 | 173.64 (17) |
| C4—C5—C6—C11 | 76.5 (2) | C4—C5—O3—C1 | 51.1 (2) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BX2433).
References
- Aguiar Amaral, P., Bergold, A. M. & Eifler-Lima, V. L. (2005). J. Pharm. Pharm. Sci. 8, 69–75. [PubMed]
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. [DOI] [PubMed]
- Andersh, B., Gereg, J., Amanuel, M. & Stanley, C. (2008). Synth. Commun. 38, 482–488.
- Bruker (2008). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruno, I. J., Cole, J. C., Kessler, M., Luo, J., Motherwell, W. D. S., Purkis, L. H., Smith, B. R., Taylor, R., Cooper, R. I., Harris, S. E. & Orpen, A. G. (2004). J. Chem. Inf. Comput. Sci. 44, 2133–2144. [DOI] [PubMed]
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Kirillov, N. F., Melekhin, V. S. & Aliev, Z. G. (2010). J. Struct. Chem, 51, 996–997.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Palatinus, L. & Chapuis, G. (2007). J. Appl. Cryst. 40, 786–790.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Souza, L. C. de, Imbroisi, D. de O., De Simone, C. A., Pereira, M. A. & Malta, V. R. S. (2009). Acta Cryst. E65, o250. [DOI] [PMC free article] [PubMed]
- Souza, L. C., Soares de Araujo, A., Sant’Ana, A. E. G. & Oliveira Imbroisi, D. (2004). Bioorg. Med. Chem. 12, 865–869.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Tait, B. D., Hagen, S., Domagala, J., Ellsworth, E. L., Gajda, C., Hamilton, H. W., Vara Prasad, J. V. N., Ferguson, D., Graham, N., Hupe, D., Nouhan, C., Tummino, P. J., Humblet, C., Lunney, E. A., Pavlovsky, A., Rubin, J., Gracheck, S. J., Baldwin, E. T., Bhat, T. N., Erickson, J. W., Gulnik, S. V. & Liu, B. (1997). J. Med. Chem. 40, 3782–3791. [DOI] [PubMed]
- Wang, Y., Li, Z., Li, J., Li, S. & Zhang, S. (1999). Gaodeng Xuexiao Huaxue Xuebao, 20, 1559–1563.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812049781/bx2433sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812049781/bx2433Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812049781/bx2433Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



