Abstract
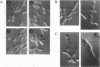
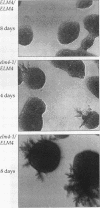
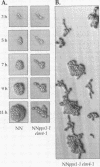
Saccharomyces cerevisiae mutant E124 was selected in a visual screen based on elongated cell shape. Genetic analysis showed that E124 contains two separate mutations, pps1-1 and elm4-1, each causing a distinct phenotype inherited as a single-gene trait. In rich medium, pps1-1 by itself causes increased doubling time but does not affect cell shape, whereas elm4-1 results in a moderate cell elongation phenotype but does not affect growth rate. Reconstructed elm4-1 pps1-1 double mutants display a synthetic phenotype in rich medium including extreme cell elongation and delayed cell separation, both characteristics of pseudohyphal differentiation. The elm4-1 mutation was shown to act as a dominant factor that potentiates pseudohyphal differentiation in response to general nitrogen starvation in a genetic background in which pseudohyphal growth normally does not occur. Thus, elm4-1 allows recognition of, or response to, a pseudohyphal differentiation signal that results from nitrogen limitation. PPS1 was isolated and shown to be a previously undescribed gene coding for a protein similar in amino acid sequence to phosphoribosylpyrophosphate synthase, a rate-limiting enzyme in the biosynthesis of nucleotides, histidine, and tryptophan. Thus, the pps1-1 mutation may generate a nitrogen limitation signal, which when coupled with elm4-1 results in pseudohyphal growth even in rich medium.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker M. A., Raivio K. O., Seegmiller J. E. Synthesis of phosphoribosylpyrophosphate in mammalian cells. Adv Enzymol Relat Areas Mol Biol. 1979;49:281–306. doi: 10.1002/9780470122945.ch7. [DOI] [PubMed] [Google Scholar]
- Blacketer M. J., Koehler C. M., Coats S. G., Myers A. M., Madaule P. Regulation of dimorphism in Saccharomyces cerevisiae: involvement of the novel protein kinase homolog Elm1p and protein phosphatase 2A. Mol Cell Biol. 1993 Sep;13(9):5567–5581. doi: 10.1128/mcb.13.9.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Brown C. M., Hough J. S. Elongation of yeast cells in continuous culture. Nature. 1965 May 15;206(985):676–678. doi: 10.1038/206676a0. [DOI] [PubMed] [Google Scholar]
- Gimeno C. J., Fink G. R. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994 Mar;14(3):2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992 Mar 20;68(6):1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Healy A. M., Zolnierowicz S., Stapleton A. E., Goebl M., DePaoli-Roach A. A., Pringle J. R. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol Cell Biol. 1991 Nov;11(11):5767–5780. doi: 10.1128/mcb.11.11.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Jones M. E. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Annu Rev Biochem. 1980;49:253–279. doi: 10.1146/annurev.bi.49.070180.001345. [DOI] [PubMed] [Google Scholar]
- KORNBERG A., LIEBERMAN I., SIMMS E. S. Enzymatic synthesis and properties of 5-phosphoribosylpyrophosphate. J Biol Chem. 1955 Jul;215(1):389–402. [PubMed] [Google Scholar]
- Köttig H., Rottner G., Beck K. F., Schweizer M., Schweizer E. The pentafunctional FAS1 genes of Saccharomyces cerevisiae and Yarrowia lipolytica are co-linear and considerably longer than previously estimated. Mol Gen Genet. 1991 Apr;226(1-2):310–314. doi: 10.1007/BF00273618. [DOI] [PubMed] [Google Scholar]
- Liu H., Styles C. A., Fink G. R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993 Dec 10;262(5140):1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Ljungdahl P. O., Gimeno C. J., Styles C. A., Fink G. R. SHR3: a novel component of the secretory pathway specifically required for localization of amino acid permeases in yeast. Cell. 1992 Oct 30;71(3):463–478. doi: 10.1016/0092-8674(92)90515-e. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D., Contopoulou C. R., Kans J. A. Genetic map of Saccharomyces cerevisiae, edition 10. Yeast. 1989 Sep-Oct;5(5):321–403. doi: 10.1002/yea.320050503. [DOI] [PubMed] [Google Scholar]
- Olson M. V., Dutchik J. E., Graham M. Y., Brodeur G. M., Helms C., Frank M., MacCollin M., Scheinman R., Frank T. Random-clone strategy for genomic restriction mapping in yeast. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7826–7830. doi: 10.1073/pnas.83.20.7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riles L., Dutchik J. E., Baktha A., McCauley B. K., Thayer E. C., Leckie M. P., Braden V. V., Depke J. E., Olson M. V. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993 May;134(1):81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOWNSEND G. F., LINDEGREN C. C. Characteristic growth patterns of the different members of a polyploid series of Saccharomyces. J Bacteriol. 1954 Apr;67(4):480–483. doi: 10.1128/jb.67.4.480-483.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Foury F. Assembly of the mitochondrial membrane system XVI. Modified form of the ATPase proteolipid in oligomycin-resistant mutants of Saccharomyces cerevisiae. FEBS Lett. 1976 Jun 15;65(3):391–395. doi: 10.1016/0014-5793(76)80154-8. [DOI] [PubMed] [Google Scholar]
- Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989 Jul 28;58(2):409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]