An interesting and potentially useful approach to biomarker identification was recently reported by Reddy et al in the journal Cell (1). Using a combinatorial library of synthetic “shape” molecules they screened for ligands that bind antibodies in the serum of subjects with the target condition but do not bind antibodies in the serum of individuals without the target condition. When this approach was used with two mice immunized with myelin oligodendrocyte glycoprotein (MOG) (that, as a result, developed a syndrome resembling human multiple sclerosis) and two mice not immunized with MOG, three peptoids were identified that subsequently discriminated perfectly between new sets of mice, 7 immunized and 7 not immunized with MOG. This was an elegant and convincing proof of principle experiment.
The authors then used this same protocol to determine if the approach “is capable of identifying potentially useful diagnostic antibody-peptoid pairs for a human disease state”. They compared antibody responses to 15,000 peptoids of 6 patients with Alzheimer disease (AD) to antibody responses of 6 age-matched non-demented control individuals and 6 patients with Parkinson’s disease. The authors report sensitivities ≥ 93.7%, specificities ≥ 93.7% and areas under the ROC curve of 0.99±0.01 for the 3 most discriminatory peptoids when those peptoids were tested on serum from 16 different patients with AD and 16 non-demented controls.
The purpose of this Perspective is to highlight the strengths of the study pointed out above and, by contrast, the weaknesses of the study design used to develop and evaluate the biomarkers for Alzheimer’s disease. To the authors’ credit, some of these weaknesses were acknowledged in their Cell paper. However, weak study designs are pervasive in the field of biomarker identification and may be in part responsible for the slow pace of real progress in development of clinically useful biomarkers. A common consequence of poor study design is that biomarkers with seemingly superb performance in early-phase studies are subsequently shown to have mediocre performance in rigorous validation studies. Here, beyond pointing out weaknesses that lead to such false positive findings, we suggest some better general strategies for designing early-phase studies aimed at identifying candidate biomarkers for clinical use.
Consider first the clinical application for which a biomarker of AD is sought. The purpose is to test individuals who have mild cognitive impairment and identify those who are likely to develop AD, at least in the absence of intervention. Reddy et al tested individuals with advanced AD and compared them with non-demented individuals. However, a biomarker that distinguishes between the extremes of advanced AD and normal cognitive function may not distinguish well between individuals with mild cognitive impairment destined to develop AD in the future versus those who will not develop AD. A biomarker common to individuals with cognitive impairment for example, may work for the comparison of patients with and without AD (as described in the Cell paper) but not for the clinical application. Likewise, a biomarker present only when AD is sufficiently advanced may work for the comparison examined in the Cell paper but not for the clinical application. A better strategy, both from discovery and evaluation points of view, would have been to test serum samples from individuals with mild cognitive impairment who subsequently were and were not diagnosed with Alzheimer’s disease. Large prospective cohort studies of ageing individuals such as the Cardiovascular Health Study (http://www.chs-nhlbi.org/) or the Women’s Health Initiative (http://www.nhlbi.nih.gov/whi/) or the Ginkgo Evaluation of Memory Study (http://www.nccam-ginkgo.org/) could provide the specimens for this sort of design and therefore may provide a better basis for identification and evaluation of a biomarker for the intended clinical use.
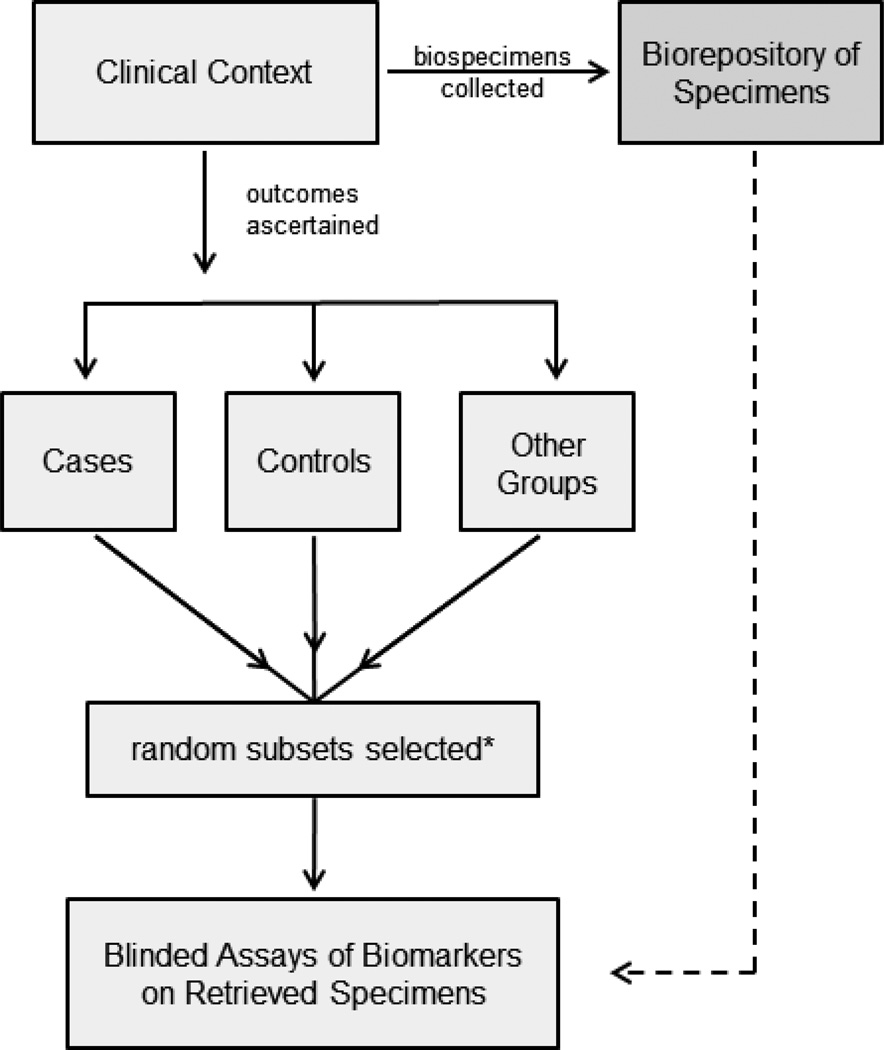
Reddy et al provided no detail on the enrollment of subjects included in their study. Individuals with AD often are under institutional care whereas the non-demented individuals live independently, for example. Institutionalized subjects are likely to differ in many respects from those still living independently. Levels of depression, anxiety, inactivity or medication use are higher in institutionalized individuals. Such factors could give rise to molecular markers that distinguish between AD and non-demented subjects as described in the Cell paper but that would not be useful in testing individuals for future risk of developing AD. Biased comparison groups are a notorious source of false positive findings in early phase biomarker studies. A better strategy is to identify the target population and to select the cases and controls randomly from that population (Figure 1). In other words, individuals who subsequently developed AD (cases) and those who did not (controls) should be representative of those groups in the clinical population of interest and this is made possible by selecting them randomly after classifying all subjects in the relevant population according to the outcome. Again, this is best done in the context of a prospective cohort study of ageing individuals.
Figure 1.
PRoBE design for biomarker discovery and evaluation.
*Selected case, control and other groups may be matched on factors related to diseases and outcomes.
Another advantage of a well-designed prospective collection of specimens is that all specimens are collected, processed and stored in a uniform manner. In the Reddy et al study there are no details about the collection of specimens from the human subjects. It may be that collections from non-demented individuals were relatively recent whereas those from AD subjects occurred further in the past (given the statement that several were autopsy confirmed cases) and this would give rise to spurious differences between the groups for biomarkers that degrade over time, for example. False positive findings could also result from differences between the groups in protocols for specimen collection, processing or storage. Some differences are likely if collections were done by different clinical staff or for different purposes.
Although there is substantial detail in the Cell paper in regard to the technology for measuring the biomarkers and in regard to the mice used in the experiments, scant information is provided in regard to the human studies. The lack of information about sources of subjects and specimens for human subjects as seen in the Reddy et al article is not uncommon in the literature on biomarker identification. To address this general concern, improvements in the reporting standards for biomarker studies are needed to both elevate the science and move it forward to clinical application. Interestingly, journals currently seem to apply much more rigorous standards of reporting to therapeutic studies than to biomarker studies. In particular, more attention to complete reporting of the clinical aspects of study design for biomarker studies is needed. For example, detailed descriptions should be provided for enrollment procedures, eligibility criteria, outcome assessments, selection of cases and controls and protocols for specimen ascertainment as well as techniques for biomarker measurement. Guidelines for reporting of diagnostic test and prognostic marker studies already exist (2–3) and could be adapted for reporting of biomarker discovery/evaluation studies.
The design of biomarker studies would be much improved by using long standing principles of good study design borrowed from population science. The basic concept is straightforward: prospective collection of specimens and outcome ascertainment in the clinical context of interest with biomarker assay on random subsets of cases and controls (Figure 1). We have described the details of this PRoBE design (4) for cancer biomarker studies but the same design is appropriate for early detection, diagnostic and prognostic studies of biomarkers for other diseases. For prognostic biomarkers the clinical study population is patients with disease whereas for early detection biomarkers the study population is healthy individuals. Basic science investigators rely on clinical collaborators to provide specimens. Therefore clinical collaborators must be encouraged to construct specimen repositories according to PRoBE principles and to make available specimen sets for testing. The Early Detection Research Network (http://edrn.nci.nih.gov/) is an organization that facilitates construction of biorepositories and collaboration between basic and clinical scientists. In addition they take advantage of some excellent existing biorepositories such as those of the Women’s Health Initiative and the Prostate Lung Colorectal and Ovarian Cancer Screening Trial (http://prevention.cancer.gov/plco) for early detection biomarkers.
Quality specimen sets should ideally be made available for biomarker identification (discovery) (5). Unfortunately, gatekeepers of biospecimen banks are often inclined to save quality specimen sets for validation of biomarkers and are reluctant to allow their use in discovery research. However, if for discovery we continue to use poorly designed specimen sets that give rise to biased comparison groups, we risk continuing to produce many false leads from discovery studies and remaining stuck in the frustrating wasteful cycle of discovering biomarkers that don’t validate when subjected to more rigorous PRoBE evaluation.
Of course there are circumstances where the availability of specimens is extremely limited, such as for rare diseases, so that practical considerations dictate that discovery research is done with study designs that do not follow the PRoBE criteria. In that event, we must wait for subsequent PRoBE designed validation studies before drawing conclusions about biomarker performance, and acknowledge the limitations of conclusions based on non-PRoBE designed studies. The most serious concern about the Reddy et al paper is the implication that their study evaluations “constitute a fair and critical test of the peptoid-antibody complexes as biomarkers”. Since AD is not rare, it is unfortunate that an unbiased PRoBE set of specimens was not used.
In closing, we congratulate Reddy and colleagues on some elegant experiments that prove the principle that a molecular “shape library” can be useful in finding markers to distinguish between two groups. However, we will not be surprised if the markers identified in his study of AD patients do not validate well in future studies. If that occurs we would encourage the investigators to redo their discovery and evaluation work with a better clinical study design so that the true potential of their technology for identifying useful diagnostic antibody-peptoid pairs for a human disease state can be realized.
Nonstandard abbreviations
- MOG
myelin oligodendrocyte glycoprotein
- ROC
Receiver Operating Characteristic
References
- 1.Reddy MM, Wilson R, Wilson J, Connell S, Gocke A, Hynan L, et al. Identification of candidate lgG biomarkers for Alzheimer’s disease via combinatorial library screening. Cell. 2011;144:132–142. doi: 10.1016/j.cell.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, et al. The STARD statement for reporting studies of diagnostic accuracy: Explanation and elaboration. Clin Chem. 2003;49:1–6. doi: 10.1373/49.1.7. [DOI] [PubMed] [Google Scholar]
- 3.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- 4.Pepe MS, Feng Z, Janes H, Bossuyt P, Potter J. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: Standards for study design. J Nat Cancer Inst. 2008;100(20):1432–1438. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Z, Prentice R, Srivastava S. Research issues and strategies for genomic and proteomic biomarker discovery and validation. Pharmacogenomics. 2004;5(6):709–719. doi: 10.1517/14622416.5.6.709. [DOI] [PubMed] [Google Scholar]