Abstract
In the title compound, [Cu(C4H2O4)(C11H9N3O)2(H2O)]n, CuII ions on crystallographic twofold rotation axes are coordinated in a square pyramidal environment by two trans O atoms belonging to two monodentate fumarate anions, two trans isonicotinamide pyridyl N-donor atoms from monodentate, pendant 3-pyridylisonicotinamide (3-pina) ligands, and one apical aqua ligand, also sited on the crystallographic twofold rotation axis. The exobidentate fumarate ligands form [Cu(fumarate)(3-pina)2(H2O)]n coordination polymer chains that are arranged parallel to [001]. In the crystal, these polymeric chains are anchored into supramolecular layers parallel to (100) by O—H⋯O hydrogen bonds between aqua ligands and unligating fumarate O atoms, and N—H⋯O(=C) hydrogen bonds between 3-pina ligands. In turn, the layers aggregate by weak C—H⋯N and C—H⋯O hydrogen bonds, affording a three-dimensional network.
Related literature
For the preparation of 3-pyridylisonicotinamide, see: Gardner et al. (1954 ▶). For the preparation of other dicarboxylate coordination polymers containing 3-pyridylisonicotinamide, see: Kumar (2009 ▶).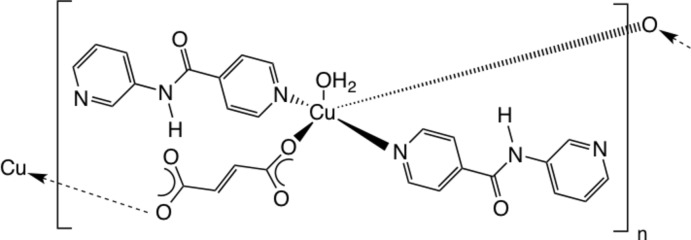
Experimental
Crystal data
[Cu(C4H2O4)(C11H9N3O)2(H2O)]
M r = 594.04
Monoclinic,
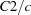
a = 29.854 (4) Å
b = 5.3535 (7) Å
c = 17.353 (2) Å
β = 118.686 (2)°
V = 2433.0 (6) Å3
Z = 4
Mo Kα radiation
μ = 0.96 mm−1
T = 173 K
0.25 × 0.13 × 0.09 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.794, T max = 0.919
9406 measured reflections
2240 independent reflections
1758 reflections with I > 2σ(I)
R int = 0.053
Refinement
R[F 2 > 2σ(F 2)] = 0.036
wR(F 2) = 0.083
S = 1.04
2240 reflections
188 parameters
5 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.34 e Å−3
Δρmin = −0.36 e Å−3
Data collection: APEX2 (Bruker, 2006 ▶); cell refinement: SAINT (Bruker, 2006 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: Crystal Maker (Palmer, 2007 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812047101/lh5556sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812047101/lh5556Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3O⋯O1i | 0.84 (1) | 1.83 (1) | 2.660 (2) | 169 (3) |
| N2—H2N⋯O2i | 0.88 (2) | 2.33 (2) | 3.153 (3) | 155 (3) |
| C2—H2⋯O4ii | 0.95 | 2.48 | 3.360 (4) | 153 |
| C7—H7⋯O1iii | 0.95 | 2.48 | 3.430 (4) | 178 |
| C9—H9⋯N1iv | 0.95 | 2.39 | 3.263 (4) | 153 |
| C12—H12⋯O1v | 0.95 | 2.43 | 3.374 (4) | 171 |
Symmetry codes: (i) 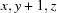 ; (ii)
; (ii) 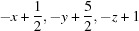 ; (iii)
; (iii) 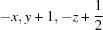 ; (iv)
; (iv) 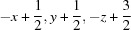 ; (v)
; (v) 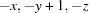 .
.
Acknowledgments
We gratefully acknowledge the donors of the American Chemical Society Petroleum Research Fund for funding this work.
supplementary crystallographic information
Comment
In comparison to divalent metal coordination polymers containing rigid rod dipyridine ligands such as 4,4'-bipyridine, related materials containing the kinked dipodal ligand 3-pyridylisonicotinamide (3-pina) are much less common (Kumar, 2009). The title compound was obtained as blue crystals through the hydrothermal reaction of copper nitrate, fumaric acid, and 3-pina.
The asymmetric unit of the title compound contains a divalent copper atom and an aqua ligand on a crystallographic twofold rotation axis, a 3-pina ligand, and one half of a fumarate ligand whose centroid rests on a crystallographic inversion centre. The copper atom is square pyramidally coordinated (Fig. 1), with the basal plane containing trans isonicotinamide pyridyl N atom donors from two 3-pina ligands and trans O atom donors from monodentate carboxylate groups belonging to two fumarate ligands. The aqua ligand is located in the apical position.
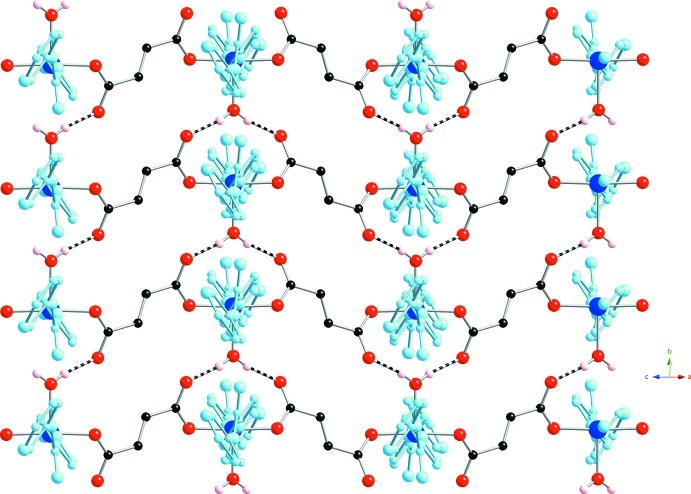
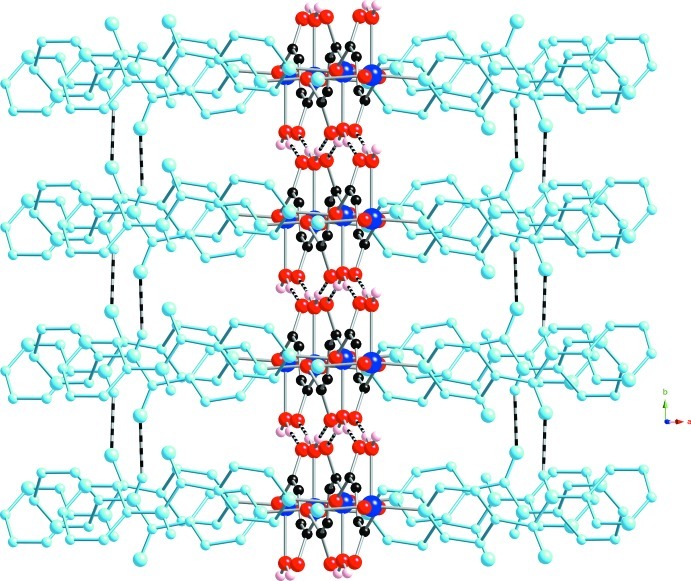
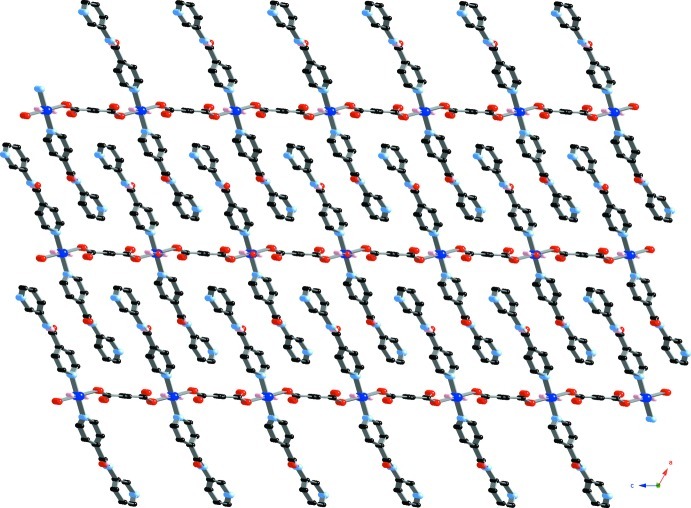
The Cu atoms are linked by exobidentate, bis(monodentate) fumarate ligands to form [Cu(fumarate)(3-pina)2(H2O)]n coordination polymer chains that are oriented parallel to [0 0 1] (Fig. 2). Each individual chain is anchored to two others via O—H···O pairwise hydrogen bonding (Table 1) between aqua ligands and unligated fumarate O atoms, thereby constructing supramolecular two-dimensional layers arranged parallel to the bc crystal planes (Fig. 3). The stability of the layer motifs is enhanced by N—H···O hydrogen bonding between amide groups of adjacent 3-pina ligands (Fig. 4). In turn the layers stack along [1 0 0] in an AAA pattern via C—H···N interactions mediated by unligated 3-pyridyl N atoms belonging to the pendant 3-pina ligands (Fig. 5), thus forming the three-dimensional structure of the title compound which is also stabilized by weak C—H···O interactions.
Experimental
Copper(II) nitrate hydrate and fumaric acid were obtained commercially. 3-Pyridylisonicotinamide (3-pina) was prepared via a published procedure (Gardner et al., 1954). A mixture of copper nitrate hydrate (86 mg, 0.37 mmol), fumaric acid (42 mg, 0.36 mmol), 3-pina (74 mg, 0.37 mmol) and 10.0 g water (550 mmol) was placed into a 23 ml Teflon-lined Parr acid digestion bomb, which was then heated under autogenous pressure at 393 K for 24 h. Blue needles of the title compound were obtained.
Refinement
All H atoms bound to C atoms were placed in calculated positions, with C—H = 0.95 Å, and refined in riding mode with Uiso = 1.2Ueq(C). The H atom within the amide group of the 3-pina ligand was found in a difference Fourier map, restrained with N—H = 0.90 (2) Å and refined with Uiso = 1.2Ueq(N). The H atoms within the aqua ligand were found in a difference Fourier map, restrained with O—H = 0.85 (2) Å and refined with Uiso = 1.2Ueq(O).
Figures
Fig. 1.
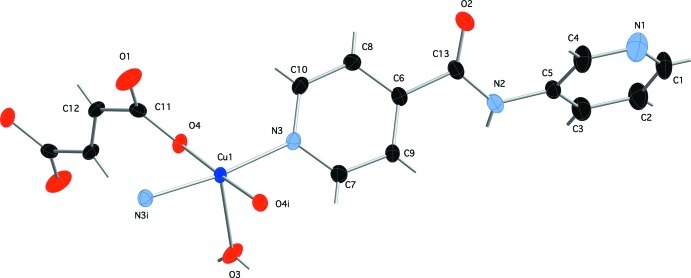
The coordination environment of the title compound, showing 50% probability ellipsoids and atom numbering scheme. Hydrogen atom positions are shown as grey sticks. Color codes: dark blue Cu, red O, light blue N, black C, pink H. Symmetry code: (i) -x, y, -z + 1/2
Fig. 2.
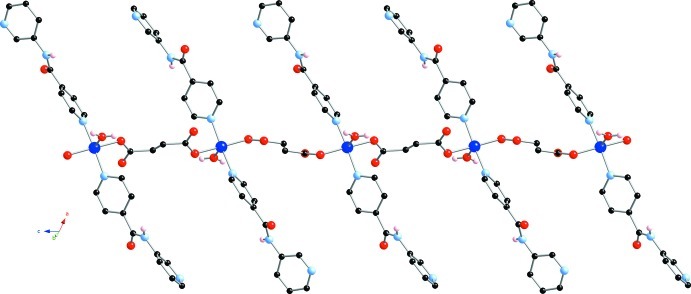
A single [Cu(fumarate)(3-pina)2(H2O)]n coordination polymer chain.
Fig. 3.
Supramolecular layer of [Cu(fumarate)(3-pina)2(H2O)]n coordination polymer chains. O—H···O hydrogen bonding is shown as dashed lines.
Fig. 4.
Side view of the supramolecular layer of [Cu(fumarate)(3-pina)2(H2O)]n coordination polymer chains. N—H···O hydrogen bonding between 3-pina amide groups is shown as dashed lines.
Fig. 5.
Stacking of supramolecular layers within the title compound.
Crystal data
| [Cu(C4H2O4)(C11H9N3O)2(H2O)] | F(000) = 1220 |
| Mr = 594.04 | Dx = 1.622 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 3329 reflections |
| a = 29.854 (4) Å | θ = 2.7–25.3° |
| b = 5.3535 (7) Å | µ = 0.96 mm−1 |
| c = 17.353 (2) Å | T = 173 K |
| β = 118.686 (2)° | Needle, blue |
| V = 2433.0 (6) Å3 | 0.25 × 0.13 × 0.09 mm |
| Z = 4 |
Data collection
| Bruker APEXII CCD diffractometer | 2240 independent reflections |
| Radiation source: fine-focus sealed tube | 1758 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.053 |
| ω–φ scans | θmax = 25.4°, θmin = 2.4° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −35→35 |
| Tmin = 0.794, Tmax = 0.919 | k = −6→6 |
| 9406 measured reflections | l = −20→20 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.036 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.083 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0303P)2 + 4.2344P] where P = (Fo2 + 2Fc2)/3 |
| 2240 reflections | (Δ/σ)max < 0.001 |
| 188 parameters | Δρmax = 0.34 e Å−3 |
| 5 restraints | Δρmin = −0.36 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cu1 | 0.0000 | 0.98462 (9) | 0.2500 | 0.01332 (15) | |
| O1 | −0.01260 (9) | 0.5988 (4) | 0.10232 (13) | 0.0329 (6) | |
| O2 | 0.23784 (7) | 0.6328 (4) | 0.58520 (13) | 0.0277 (5) | |
| O3 | 0.0000 | 1.3863 (5) | 0.2500 | 0.0288 (7) | |
| H3O | −0.0035 (12) | 1.471 (4) | 0.2067 (12) | 0.035* | |
| O4 | 0.01923 (7) | 0.9749 (3) | 0.15746 (11) | 0.0170 (4) | |
| N1 | 0.34390 (10) | 0.8621 (5) | 0.83508 (17) | 0.0384 (7) | |
| N2 | 0.24546 (9) | 1.0496 (5) | 0.61401 (16) | 0.0232 (6) | |
| H2N | 0.2343 (11) | 1.199 (4) | 0.5910 (18) | 0.028* | |
| N3 | 0.07405 (8) | 0.9573 (4) | 0.34350 (14) | 0.0159 (5) | |
| C1 | 0.37744 (12) | 1.0459 (6) | 0.8530 (2) | 0.0344 (8) | |
| H1 | 0.4076 | 1.0448 | 0.9083 | 0.041* | |
| C2 | 0.37059 (12) | 1.2372 (6) | 0.7956 (2) | 0.0339 (8) | |
| H2 | 0.3955 | 1.3652 | 0.8108 | 0.041* | |
| C3 | 0.32648 (12) | 1.2391 (6) | 0.7149 (2) | 0.0295 (8) | |
| H3 | 0.3203 | 1.3698 | 0.6739 | 0.035* | |
| C4 | 0.30186 (11) | 0.8643 (6) | 0.75726 (19) | 0.0292 (7) | |
| H4 | 0.2777 | 0.7335 | 0.7438 | 0.035* | |
| C5 | 0.29173 (10) | 1.0479 (5) | 0.69522 (18) | 0.0201 (6) | |
| C6 | 0.16993 (10) | 0.8946 (5) | 0.48796 (18) | 0.0187 (6) | |
| C7 | 0.09141 (10) | 1.1150 (5) | 0.41137 (18) | 0.0199 (6) | |
| H7 | 0.0702 | 1.2498 | 0.4094 | 0.024* | |
| C8 | 0.15215 (10) | 0.7307 (5) | 0.41788 (18) | 0.0195 (6) | |
| H8 | 0.1725 | 0.5933 | 0.4188 | 0.023* | |
| C9 | 0.13897 (10) | 1.0906 (5) | 0.48445 (18) | 0.0214 (7) | |
| H9 | 0.1502 | 1.2068 | 0.5315 | 0.026* | |
| C10 | 0.10459 (10) | 0.7686 (5) | 0.34665 (18) | 0.0193 (6) | |
| H10 | 0.0930 | 0.6575 | 0.2981 | 0.023* | |
| C11 | 0.00282 (10) | 0.8103 (5) | 0.09670 (18) | 0.0180 (6) | |
| C12 | 0.00079 (10) | 0.8823 (5) | 0.01221 (17) | 0.0169 (6) | |
| H12 | 0.0006 | 0.7540 | −0.0257 | 0.020* | |
| C13 | 0.22077 (10) | 0.8447 (5) | 0.56670 (18) | 0.0205 (6) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu1 | 0.0147 (2) | 0.0154 (3) | 0.0090 (2) | 0.000 | 0.00500 (19) | 0.000 |
| O1 | 0.0667 (16) | 0.0171 (11) | 0.0272 (12) | −0.0040 (11) | 0.0324 (12) | −0.0003 (9) |
| O2 | 0.0219 (11) | 0.0249 (12) | 0.0250 (12) | 0.0040 (9) | 0.0022 (9) | 0.0016 (10) |
| O3 | 0.058 (2) | 0.0146 (16) | 0.0144 (16) | 0.000 | 0.0181 (16) | 0.000 |
| O4 | 0.0193 (10) | 0.0222 (11) | 0.0111 (9) | −0.0034 (8) | 0.0086 (8) | −0.0021 (9) |
| N1 | 0.0373 (16) | 0.0429 (18) | 0.0212 (15) | −0.0081 (14) | 0.0030 (13) | 0.0075 (13) |
| N2 | 0.0191 (13) | 0.0230 (15) | 0.0187 (13) | 0.0024 (11) | 0.0020 (11) | 0.0017 (11) |
| N3 | 0.0172 (12) | 0.0188 (13) | 0.0120 (11) | −0.0005 (10) | 0.0073 (10) | −0.0011 (10) |
| C1 | 0.0261 (17) | 0.043 (2) | 0.0208 (16) | −0.0033 (15) | 0.0005 (14) | −0.0006 (15) |
| C2 | 0.0243 (17) | 0.035 (2) | 0.0302 (19) | −0.0102 (14) | 0.0033 (15) | −0.0011 (15) |
| C3 | 0.0307 (18) | 0.0241 (18) | 0.0255 (18) | −0.0030 (14) | 0.0068 (15) | 0.0037 (14) |
| C4 | 0.0262 (17) | 0.0343 (19) | 0.0206 (16) | −0.0115 (15) | 0.0060 (14) | 0.0012 (14) |
| C5 | 0.0144 (14) | 0.0256 (17) | 0.0171 (15) | 0.0011 (12) | 0.0050 (12) | −0.0024 (12) |
| C6 | 0.0153 (14) | 0.0218 (15) | 0.0169 (15) | 0.0008 (12) | 0.0060 (12) | 0.0019 (12) |
| C7 | 0.0193 (15) | 0.0225 (16) | 0.0172 (15) | 0.0030 (12) | 0.0082 (12) | −0.0008 (13) |
| C8 | 0.0159 (14) | 0.0199 (16) | 0.0214 (16) | 0.0017 (12) | 0.0080 (13) | −0.0020 (12) |
| C9 | 0.0195 (15) | 0.0234 (16) | 0.0156 (15) | −0.0008 (12) | 0.0040 (12) | −0.0076 (12) |
| C10 | 0.0223 (15) | 0.0201 (16) | 0.0158 (15) | −0.0008 (12) | 0.0094 (13) | −0.0035 (12) |
| C11 | 0.0209 (15) | 0.0160 (15) | 0.0168 (15) | 0.0057 (12) | 0.0088 (12) | 0.0027 (12) |
| C12 | 0.0215 (14) | 0.0157 (13) | 0.0154 (15) | −0.0008 (12) | 0.0103 (12) | −0.0035 (12) |
| C13 | 0.0161 (14) | 0.0241 (17) | 0.0176 (15) | 0.0023 (13) | 0.0051 (12) | 0.0020 (13) |
Geometric parameters (Å, º)
| Cu1—O4i | 1.9485 (17) | C2—C3 | 1.388 (4) |
| Cu1—O4 | 1.9485 (17) | C2—H2 | 0.9500 |
| Cu1—N3 | 2.024 (2) | C3—C5 | 1.379 (4) |
| Cu1—N3i | 2.024 (2) | C3—H3 | 0.9500 |
| Cu1—O3 | 2.151 (3) | C4—C5 | 1.380 (4) |
| O1—C11 | 1.244 (3) | C4—H4 | 0.9500 |
| O2—C13 | 1.222 (3) | C6—C9 | 1.380 (4) |
| O3—H3Oi | 0.839 (11) | C6—C8 | 1.382 (4) |
| O3—H3O | 0.839 (11) | C6—C13 | 1.500 (4) |
| O4—C11 | 1.278 (3) | C7—C9 | 1.383 (4) |
| N1—C1 | 1.330 (4) | C7—H7 | 0.9500 |
| N1—C4 | 1.331 (4) | C8—C10 | 1.378 (4) |
| N2—C13 | 1.356 (4) | C8—H8 | 0.9500 |
| N2—C5 | 1.423 (3) | C9—H9 | 0.9500 |
| N2—H2N | 0.883 (17) | C10—H10 | 0.9500 |
| N3—C7 | 1.335 (3) | C11—C12 | 1.489 (4) |
| N3—C10 | 1.344 (3) | C12—C12ii | 1.323 (5) |
| C1—C2 | 1.374 (4) | C12—H12 | 0.9500 |
| C1—H1 | 0.9500 | ||
| O4i—Cu1—O4 | 176.95 (12) | N1—C4—C5 | 123.0 (3) |
| O4i—Cu1—N3 | 88.74 (8) | N1—C4—H4 | 118.5 |
| O4—Cu1—N3 | 91.04 (8) | C5—C4—H4 | 118.5 |
| O4i—Cu1—N3i | 91.04 (8) | C3—C5—C4 | 118.6 (3) |
| O4—Cu1—N3i | 88.74 (8) | C3—C5—N2 | 119.8 (3) |
| N3—Cu1—N3i | 171.71 (13) | C4—C5—N2 | 121.5 (3) |
| O4i—Cu1—O3 | 91.52 (6) | C9—C6—C8 | 118.5 (2) |
| O4—Cu1—O3 | 91.52 (6) | C9—C6—C13 | 122.6 (3) |
| N3—Cu1—O3 | 94.15 (6) | C8—C6—C13 | 118.8 (2) |
| N3i—Cu1—O3 | 94.15 (6) | N3—C7—C9 | 122.8 (3) |
| H3Oi—O3—Cu1 | 122.6 (17) | N3—C7—H7 | 118.6 |
| H3Oi—O3—H3O | 115 (3) | C9—C7—H7 | 118.6 |
| Cu1—O3—H3O | 122.6 (17) | C10—C8—C6 | 119.4 (3) |
| C11—O4—Cu1 | 123.39 (17) | C10—C8—H8 | 120.3 |
| C1—N1—C4 | 117.9 (3) | C6—C8—H8 | 120.3 |
| C13—N2—C5 | 125.5 (2) | C6—C9—C7 | 118.9 (3) |
| C13—N2—H2N | 119 (2) | C6—C9—H9 | 120.5 |
| C5—N2—H2N | 115 (2) | C7—C9—H9 | 120.5 |
| C7—N3—C10 | 118.1 (2) | N3—C10—C8 | 122.3 (3) |
| C7—N3—Cu1 | 118.25 (18) | N3—C10—H10 | 118.9 |
| C10—N3—Cu1 | 123.06 (18) | C8—C10—H10 | 118.9 |
| N1—C1—C2 | 123.3 (3) | O1—C11—O4 | 125.0 (3) |
| N1—C1—H1 | 118.4 | O1—C11—C12 | 118.1 (2) |
| C2—C1—H1 | 118.4 | O4—C11—C12 | 116.9 (2) |
| C1—C2—C3 | 118.4 (3) | C12ii—C12—C11 | 122.7 (3) |
| C1—C2—H2 | 120.8 | C12ii—C12—H12 | 118.6 |
| C3—C2—H2 | 120.8 | C11—C12—H12 | 118.6 |
| C5—C3—C2 | 118.8 (3) | O2—C13—N2 | 123.7 (3) |
| C5—C3—H3 | 120.6 | O2—C13—C6 | 121.1 (3) |
| C2—C3—H3 | 120.6 | N2—C13—C6 | 115.2 (2) |
| O4i—Cu1—O3—H3Oi | 22 (3) | C13—N2—C5—C4 | −36.8 (4) |
| O4—Cu1—O3—H3Oi | −158 (3) | C10—N3—C7—C9 | −0.6 (4) |
| N3—Cu1—O3—H3Oi | −66 (3) | Cu1—N3—C7—C9 | 170.8 (2) |
| N3i—Cu1—O3—H3Oi | 114 (3) | C9—C6—C8—C10 | 0.6 (4) |
| N3—Cu1—O4—C11 | 121.8 (2) | C13—C6—C8—C10 | 177.4 (3) |
| N3i—Cu1—O4—C11 | −49.9 (2) | C8—C6—C9—C7 | 0.3 (4) |
| O3—Cu1—O4—C11 | −143.99 (19) | C13—C6—C9—C7 | −176.4 (3) |
| O4i—Cu1—N3—C7 | −50.8 (2) | N3—C7—C9—C6 | −0.3 (4) |
| O4—Cu1—N3—C7 | 132.2 (2) | C7—N3—C10—C8 | 1.5 (4) |
| O3—Cu1—N3—C7 | 40.6 (2) | Cu1—N3—C10—C8 | −169.4 (2) |
| O4i—Cu1—N3—C10 | 120.2 (2) | C6—C8—C10—N3 | −1.5 (4) |
| O4—Cu1—N3—C10 | −56.8 (2) | Cu1—O4—C11—O1 | −24.5 (4) |
| O3—Cu1—N3—C10 | −148.4 (2) | Cu1—O4—C11—C12 | 153.62 (18) |
| C4—N1—C1—C2 | 0.4 (5) | O1—C11—C12—C12ii | 156.4 (3) |
| N1—C1—C2—C3 | 0.1 (5) | O4—C11—C12—C12ii | −21.9 (5) |
| C1—C2—C3—C5 | −0.8 (5) | C5—N2—C13—O2 | −6.2 (4) |
| C1—N1—C4—C5 | −0.1 (5) | C5—N2—C13—C6 | 174.0 (2) |
| C2—C3—C5—C4 | 1.0 (5) | C9—C6—C13—O2 | 150.1 (3) |
| C2—C3—C5—N2 | 178.0 (3) | C8—C6—C13—O2 | −26.5 (4) |
| N1—C4—C5—C3 | −0.6 (5) | C9—C6—C13—N2 | −30.1 (4) |
| N1—C4—C5—N2 | −177.6 (3) | C8—C6—C13—N2 | 153.3 (3) |
| C13—N2—C5—C3 | 146.3 (3) |
Symmetry codes: (i) −x, y, −z+1/2; (ii) −x, −y+2, −z.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3O···O1iii | 0.84 (1) | 1.83 (1) | 2.660 (2) | 169 (3) |
| N2—H2N···O2iii | 0.88 (2) | 2.33 (2) | 3.153 (3) | 155 (3) |
| C2—H2···O4iv | 0.95 | 2.48 | 3.360 (4) | 153 |
| C7—H7···O1v | 0.95 | 2.48 | 3.430 (4) | 178 |
| C9—H9···N1vi | 0.95 | 2.39 | 3.263 (4) | 153 |
| C12—H12···O1vii | 0.95 | 2.43 | 3.374 (4) | 171 |
Symmetry codes: (iii) x, y+1, z; (iv) −x+1/2, −y+5/2, −z+1; (v) −x, y+1, −z+1/2; (vi) −x+1/2, y+1/2, −z+3/2; (vii) −x, −y+1, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LH5556).
References
- Bruker (2006). APEX2 and SAINT Bruker AXS, Inc., Madison, Wisconsin, USA.
- Gardner, T. S., Wenis, E. & Lee, J. (1954). J. Org. Chem. 19, 753–757.
- Kumar, D. K. (2009). Inorg. Chim. Acta, 362, 1767–1771.
- Palmer, D. (2007). CrystalMaker CrystalMaker Software Ltd, Bicester, England.
- Sheldrick, G. M. (1996). SADABS, University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812047101/lh5556sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812047101/lh5556Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report