Abstract
The lipid compositions of different breast tumor microenvironments are largely unknown due to limitations in lipid imaging techniques. Imaging lipid distributions would enhance our understanding of processes occurring inside growing tumors, such as cancer cell proliferation, invasion, and metastasis. Recent developments in MALDI mass spectrometry imaging (MSI) enable rapid and specific detection of lipids directly from thin tissue sections. In this study, we performed multimodal imaging of acylcarnitines, phosphatidylcholines (PC), a lysophosphatidylcholine (LPC), and a sphingomyelin (SM) from different microenvironments of breast tumor xenograft models, which carried tdTomato red fluorescent protein as a hypoxia-response element-driven reporter gene. The MSI molecular lipid images revealed spatially heterogeneous lipid distributions within tumor tissue. Four of the most-abundant lipid species, namely PC(16:0/16:0), PC(16:0/18:1), PC(18:1/18:1), and PC(18:0/18:1), were localized in viable tumor regions, whereas LPC(16:0/0:0) was detected in necrotic tumor regions. We identified a heterogeneous distribution of palmitoylcarnitine, stearoylcarnitine, PC(16:0/22:1), and SM(d18:1/16:0) sodium adduct, which colocalized primarily with hypoxic tumor regions. For the first time, we have applied a multimodal imaging approach that has combined optical imaging and MALDI-MSI with ion mobility separation to spatially localize and structurally identify acylcarnitines and a variety of lipid species present in breast tumor xenograft models.
Keywords: multimodal imaging, phospholipids, matrix-assisted laser desorption/ionization, mass spectrometry
MALDI mass spectrometry imaging (MSI) can detect, localize, and identify multiple biologically relevant molecules directly from thin tissue sections without the necessity for any labeling (1). The sample preparation of thin tissue sections for MALDI-MSI requires the application of laser absorbing matrix crystals on the tissue surface. During an MSI experiment, the laser beam ablates the matrix coated tissue sample surface while the mass spectrometer acquires a collection of spectra that contain comprehensive information about the local biomolecular composition of the sample. Biomolecules present in the tissue are desorbed and separated by a mass spectrometer according to their m/z ratios. Each m/z value present in this spectral collection can be converted to an image by using dedicated imaging software. The chemical structure of each ion detected from the tissue surface can be identified after its isolation and fragmentation inside a mass spectrometer.
An additional advantage of MSI is its compatibility with other imaging techniques such as optical bright field and fluorescence microscopy. Such a multimodal approach is particularly useful for analyzing the molecular composition of complex, heterogeneous tumor tissue, which comprises several distinct tumor microenvironments. One example of such heterogeneity is tissue hypoxia, a state of low oxygen tension (pO2 values ≤2.5 mmHg) observed in many solid tumors (2). Hypoxia increases overall tumor aggressiveness, induces radio and drug resistance, triggers resistance to apoptosis, and enhances cellular migration and invasiveness, eventually leading to cancer metastasis (3). At the molecular level, hypoxia triggers many known and unknown metabolome, lipidome, and proteome changes that modulate tumor vascular expansion and provide an overall advantage for malignant growth, causing rapid disease progression (4). Severe, prolonged hypoxia can lead to cell death and tissue necrosis (4). Tumor expansion results in the formation of metabolically diverse, viable and necrotic, normoxic and hypoxic, vascularized and avascular tissue regions, which, to date, have not been imaged in a single experiment or by a single imaging technique. We hypothesize that these different tumor tissue regions have different distributions of metabolites and lipids. MSI has the capability of parallel detection and visualization of biomolecules present in these specific tissue microenvironments, which allow for a more-comprehensive analysis of tumor biology.
Unlike cancer cell cultures, human tumor xenograft models in mice provide a three-dimensional, native-like environment for studying different aspects of tumor growth (5). Additionally, tumor xenograft models allow for genetic modifications such as incorporation of genes encoding fluorescent proteins for visualizing and monitoring selected processes such as the influence of tissue hypoxia on tumor extracellular matrix (6) or cancer metastasis (7).
MALDI-MSI is well-suited for ex vivo detection and localization of different lipid categories and classes from samples of various origins and different sizes. MALDI-MSI has recently been applied in lipidome studies of human skin (8), veins (9), and ovary (10); mouse embryo implantation sites (11), tongue (12), and leg muscle (13, 14); and rat spinal cord (15), cardiac tissue (16), and liver (17). Mammalian brain tissue, due to its high lipid content, has also been extensively studied by MSI. Ion images of lipid distributions have been obtained for rat (17–22), mouse (23, 24), and human (25, 26) brain sections. Lipid MSI has also been performed for various types of tumor tissue, such as human brain tumors (27), seminoma (28), ovarian cancer (29, 30), myxoid liposarcomas (31), human astrocytoma (32), human prostate cancer tissue (33), colon cancer liver metastasis (34), and human liver adenocarcinoma (35).
Lipid-rich tumors have been associated with increased tumor aggressiveness and metastasis (36–38). To develop effective cancer treatment strategies, it is crucial not only to identify but also to localize the lipid molecules involved in cancer progression. To date, lipid distributions in breast tumors remain largely unknown due to limitations in lipid imaging methods, which often require incorporation of fluorescent tags and provide limited specificity (39–41). Here, we have adopted a novel multimodal imaging approach, which integrates microscopy and MSI, for a label-free, ex vivo visualization of different phospholipids directly from breast tumor xenograft tissue sections. We present spatial distributions of sphingomyelin (SM) and multiple phosphatidylcholine (PC) species, which were colocalized with normoxic, hypoxic, or necrotic tumor regions. MSI also localized the distribution of two acylcarnitine molecular ions, which were identified from tumor tissue by ion mobility separation (IMS) followed by tandem mass spectrometry. We have demonstrated that the incorporation of MSI into a multimodal imaging approach revealed the molecular complexity of tumor tissue at an unprecedented level.
MATERIALS AND METHODS
Chemicals and materials
The matrix α-cyano-4-hydroxycinnamic acid (CHCA) was purchased from Sigma-Aldrich (Germany). Water, acetonitrile (ACN), and trifluoroacetic acid (TFA) were purchased from Biosolve (The Netherlands). Cresyl violet acetate, Ponceau S, and paraformaldehyde powder were purchased from Sigma-Aldrich. Gelatin Type A was purchased from Sigma. Mayer's hematoxylin was purchased from Sigma, and aqueous Eosin Y from EMD Chemicals, Inc. Cytoseal 60 Mounting Medium, Richard-Allan Scientific, was purchased from Thermo Scientific.
Breast tumor xenograft model
The MDA-MB-231 human breast cancer cell line was purchased from the American Type Culture Collection (ATCC) and used within 6 months of obtaining from ATCC. This cell line was tested and authenticated by ATCC using two independent methods: the ATCC cytochrome C oxidase I PCR assay, and short tandem-repeat profiling using multiplex PCR. The MDA-MB-231 cell line was genetically modified to express tdTomato red fluorescent protein under the control of hypoxia response elements (MDA-MB-231-HRE-tdTomato) as previously described (42). Cells were injected into the upper thoracic mammary fat pad of athymic nude mice (2 × 106 cells/injection), and tumor growth was monitored using standard calipers. The research was conducted in conformity with the Public Health Service policy on Humane Care and Use of Laboratory Animals. All experimental animal protocols were approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine.
Tumor dissection and optical imaging
When tumors reached a volume of approximately 500 mm3, mice were euthanized and tumors were removed. Three individual tumors were embedded into 10% gelatin blocks prepared using 15 mm × 15 mm × 5 mm cryomolds (Sakura Finetek). Cresyl violet acetate and Ponceau S were prepared in 10% warm (37°C) gelatin at a concentration of 10 mg/ml, mixed in a 1:1 ratio, and injected into 10% gelatin blocks next to the tumor tissue. Each block was sectioned into serial 2 mm-thick fresh tumor sections using an acrylic adjustable tissue slicer (12 mm depth up to 25 mm width, Braintree Scientific, Inc.; Braintree, MA), and tissue slicer blades (Braintree Scientific, Inc.) as previously described (43, 44). These serial fresh tumor xenograft sections were each placed on individual microscope slides (Fisherbrand catalog number 12-550-34, Fisher Scientific; Pittsburgh, PA), and stored in an ice box containing ice on the bottom, with the slides located on a perforated plate at approximately 1-cm above the ice to minimize tissue degradation (43, 44). These fresh sections were imaged by bright-field and fluorescence microscopy with a 1× objective attached to a Nikon inverted microscope, equipped with a filter set for 528 to 553 nm excitation and 600 to 660 nm emission, and a Nikon Coolpix digital camera (Nikon Instruments, Inc.; Melville, NY). Bright-field imaging captured the position of the fiducial markers present inside the gelatin block, as well as the shape of the tumor tissue. The fluorescence from tdTomato expression in hypoxic regions of these tumor sections was detected by fluorescence microscopy. The GNU Image Manipulation Program (GIMP 2.6) was used for two-dimensional (2D) coregistration and overlay of bright-field and fluorescence images of 2 mm-thick tumor sections. All 2 mm-thick sections were snap-frozen immediately after microscopic imaging and stored at −80°C until further analysis.
Tumor sectioning
From each 2 mm-thick section, 10 µm-thick sections were cut at −16°C for MSI using a Microm HM550 cryo-microtome (Microm International GmbH; Walldorf, Germany) along with adjacent 10 µm-thick sections for histological staining. Tissue sections for MSI analysis were mounted onto 25 mm × 50 mm × 1.1 mm, Rs = 4–8 Ω Indium Tin Oxide-coated slides (Delta Technologies) and for histological staining onto Superfrost Slides (VWR International; Cat. 48311-600).
H and E staining method
Tissue sections were stained using a modified H and E staining protocol as previously described (43–45). Briefly, 10 µm sections attached to Superfrost Slides (VWR International, Cat. 48311-600) were washed with PBS, fixed with freshly depolymerized 3% paraformaldehyde solution for 30 min, washed with distilled water (dH2O), and treated with Mayer's hematoxylin for 30 min at room temperature, followed by five washes with dH2O. Sections were immediately immersed in aqueous Eosin Y for 30 min, followed by five washes with dH2O, mounting with aqueous mounting medium, and attaching of a coverslip. Bright-field images of H and E-stained sections were acquired using 1× or 20× objectives attached to a Nikon microscope equipped with a Nikon Coolpix digital camera (Nikon Instruments, Inc.; Melville, NY).
MALDI-IMS-MSI
Six tissue sections obtained from three individual tumors were subjected to MSI analysis. Before MSI analysis, tissue sections were placed inside a vacuum desiccator for 30 min. CHCA matrix was prepared at a concentration of 10 mg/ml in 1:1 ACN:H2O/0.1% TFA and was applied by an ImagePrep (Bruker; Germany) application system. Tissue sections were analyzed on a MALDI-Q-TOF (Synapt HDMS; Waters, UK) instrument in IMS mode detecting positive ions. Images were acquired with 150 μm × 150 μm spatial resolution and a 100 μm laser beam spot size. For better detection of masses above m/z 700, the instrument was set to suppress all masses below m/z 700. For 2D MSI analysis and overlay of images, data were visualized using BioMap software (Novartis; Basel, Switzerland). Ion mobility data analysis and visualization were performed by using DriftScope software (version 2.1; Waters, UK) as well as high-definition imaging (HDI) software (version 31; Waters, UK). The LIPID Metabolites And Pathways Strategy database (LIPID MAPS, www.lipidmaps.org) was used to search for possible lipid structures (46, 47).
Lipid identification by MS/MS analysis
The identification of selected lipid species was performed on tissue in positive-ion mode using a MALDI-Q-TOF instrument (Synapt; Waters, UK) after completing MSI experiments. Lipid ion precursors were selected within a 2 Da selection window and fragmented using 20–30 V collision energy applied in the trap cell (in TOF mode) or 30–40 V applied in the transfer cell (in IMS mode). Spectra were analyzed using MassLynx software (Waters, UK). Ion mobility-separated ions were visualized by Driftscope software (Waters, UK), and selected drift time windows were exported to MassLynx for spectral analysis. FA chains of lipids of interest were identified from a combination of the mass of the lipid and the MS/MS fragmentation pattern obtained with a mass accuracy of 0.1 Da at m/z 1,000. The LIPID MAPS, Human Metabolome Database version 2.5 (www.hmdb.ca) and the METLIN Metabolite Database (metlin.scripps.edu) were used to search for possible lipid structures and MS/MS fragmentation spectra.
Overlap analysis
We analyzed three MDA-MB-231-HRE-tdTomato breast tumor xenografts that were imaged by the multimodal approach described above. Necrotic tumor regions were outlined in H and E-stained 10 µm-thick sections. Hypoxic tumor regions were detected from the fluorescence images of tdTomato protein expression in 2 mm-thick sections. The remaining tumor regions were normoxic. Ion images of each individual m/z were overlaid with the corresponding optical images and coregistered by using the positions of fiducial markers and tumor boundaries as previously described (43, 44). The tumor region was masked by applying an active-shape model to the registered optical bright-field images (48). The highest 5% of the area under the histogram of images was automatically thresholded as the ionic or the hypoxic region inside the tumor, respectively. The volume percentage of ion overlap with normoxic, hypoxic, and necrotic regions inside the tumor was calculated to quantify their colocalizations as previously described (43, 44).Volume percentage of hypoxia was defined as the number of hypoxic voxels that overlap with a given ion normalized to total hypoxic voxels in the tumor. Similarly, volume percentage of ions with normoxic voxels and necrotic voxels was measured. The image registration and colocalization analysis program was written in Matlab 2012b (Mathworks, Inc.; Natick, MA). Mean and standard error of overlap volume percentage from tumor center sections (n = 3) were calculated for each m/z and tumor region using MS Excel 2003.
RESULTS
Multimodal imaging approach
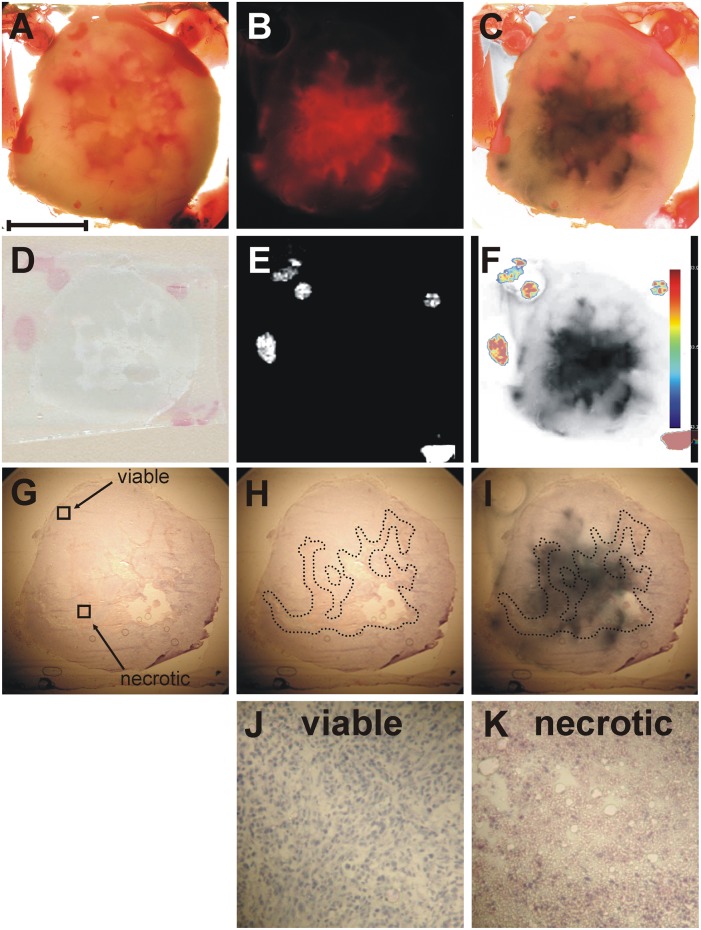
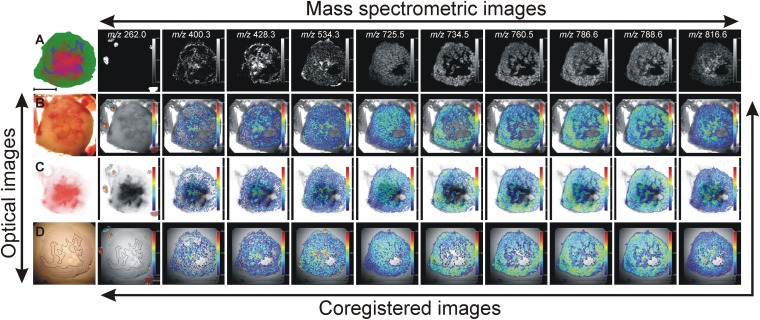
We have employed a multimodal imaging approach for investigating the effects of tumor hypoxia and tumor necrosis on the regionally distinct, spatially heterogeneous lipid composition in a breast tumor model. In this approach, we have integrated optical microscopy with MSI. Bright-field microscopy visualized the shape of the tumor, fluorescence microscopy outlined the position of hypoxic tumor regions, and histochemical staining revealed the localization of necrotic tumor tissue. MSI employing MALDI-MSI visualized the distribution of intact phospholipid molecular ions. All modalities were combined through the presence of fiducial markers visible in all imaging modalities (45). Figure 1 presents results obtained from a representative breast tumor xenograft using our multimodal imaging approach. Figure 1A shows a bright-field image of a 2 mm-thick tumor section obtained from the tumor center, which was embedded in a gelatin block containing the fiducial markers, as described in Materials and Methods. Figure 1B presents the fluorescence image of tdTomato red fluorescent protein exclusively expressed in the hypoxic tissue regions of this genetically modified tumor model (42). The coregistration of the bright-field image (Fig. 1A) with the tdTomato fluorescence image (Fig. 1B) was performed based on the position of the fiducial markers, which were visible in both images. The red color of the tdTomato signal was changed to black for better visualization of the hypoxic tumor regions and subsequently overlaid on the bright-field image of the tumor tissue as shown in Fig. 1C. Figure 1D presents the 10 µm-thick section obtained from this tumor for MSI. The position of the fiducial markers as visualized by MSI (shown in Fig. 1E) was used for coregistration of the MSI dataset with the fluorescence image of tdTomato as shown in Fig. 1F. Figure 1G shows an adjacent 10 µm-thick section stained with hematoxylin and eosin (H and E) for visualization of the necrotic tumor regions. Figure 1H presents the shape of the necrotic regions inside the tumor, outlined by a dotted line. Figure 1I shows the coregistration of the necrotic tumor regions obtained from the H and E image (Fig. 1H) with the hypoxic tumor regions from the fluorescence image (Fig. 1B). The fiducial markers were not clearly visible in some H and E images, in which case, the coregistration was performed based on the tumor shape by using the ImageJ-TurboReg plug-in (http://rsb.info.nih.gov/ij). Magnified images of viable and necrotic tumor regions, as indicated in Fig. 1G, are shown in Fig. 1J, K, respectively.
Fig. 1.
Co-registration of optical microscopy and mass spectrometric imaging. A: Representative bright-field image of a 2 mm-thick fresh breast tumor tissue section embedded in gelatin containing fiducial markers. B: Corresponding fluorescence image of tdTomato expression in the hypoxic tumor regions. C: Coregistration of the bright-field image shown in A and the fluorescence image shown in B based on the position of the fiducial markers present in both images. The red color of the tdTomato signal was changed to black prior to generating this overlay for better visualization of the hypoxic tumor regions. D: Corresponding optical image of a thin tissue section used for MSI. E: Corresponding MSI image of the fiducial markers. F: Coregistration of fiducial markers imaged by MSI and the hypoxic regions imaged by fluorescence microscopy. G: Image of the adjacent H and E-stained tissue section. H: Image of the outlined necrotic tissue present inside the tumor. I: Coregistration of the necrotic region visualized by H and E staining and the hypoxic region imaged by fluorescence microscopy. J: Magnified image of viable tumor region as indicated in G. K: Magnified image of necrotic tumor region as indicated in G. Scale bar, 5 mm A–I; 250 µm J–K.
Mass spectrometric analysis of lipids
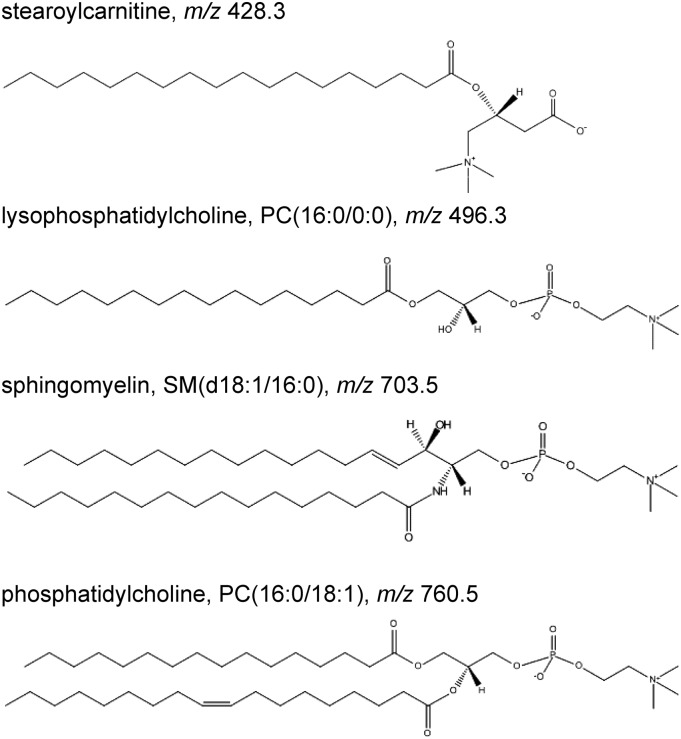
Mass spectrometric imaging was employed directly on thin tumor tissue sections for detection, visualization, and identification of a number of lipid biomolecules. Figure 2 presents selected molecular structures of acylcarnitine and lipids detected by MSI. These biomolecules all have a quaternary amine group, which can attain a positive charge, resulting in good ionization properties.
Fig. 2.
Representative examples of acylcarnitine and lipid molecular species such as stearoylcarnitine, LPC, SM, and PC detected from tumor tissue by MSI. The molecular structures were obtained from the LipidMaps web site.
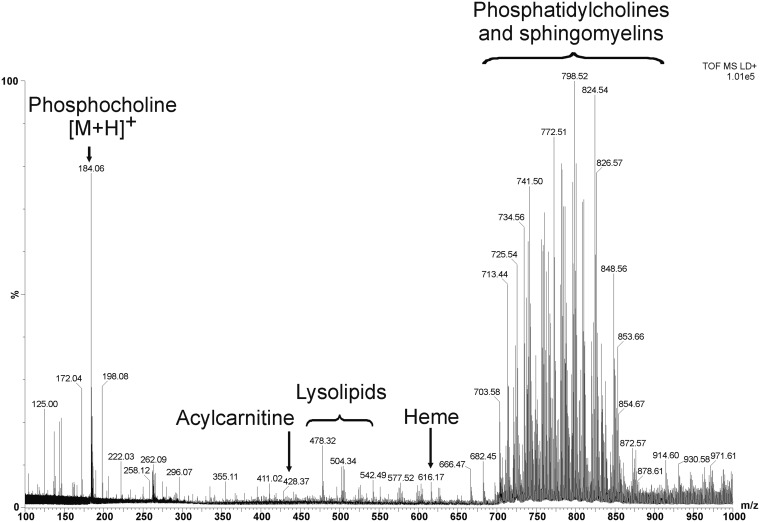
Figure 3 shows a representative combined mass spectrum of positively charged ions detected by MALDI-MSI from MDA-MB-231-HRE-tdTomato breast tumor xenograft tissue. In the low-mass region, we observed phosphate-related metabolite ions at m/z 125.0 [M+H]+, m/z 146.9 [M+Na]+, and m/z 162.9 [M+K]+, the phosphocholine at m/z 184.0 [M+H]+ as well as a phosphocholine-related ion at m/z 198.0 [M+H]+. Matrix-related ions were detected at m/z 172.0 [M-H2O]+, m/z 190.0 [M+H]+, and m/z 379.0 [2M+H]+. The fiducial marker cresyl violet was observed at m/z 262.0. Two acylcarnitines, palmitoylcarnitine and stearoylcarnitine, were observed at m/z 400.3 [M+H]+ and at m/z 428.3 [M+H]+, respectively. Lysolipid species were detected in the mass range from m/z 450 to m/z 550. Heme was observed at m/z 616.1 [M]+ as a singly charged ion. Intact lipid molecular ions were detected as protonated ions as well as sodium and potassium salt adducts in the mass range from m/z 700 to m/z 860.
Fig. 3.
Representative combined mass spectrum detected by MALDI-MSI from an MDA-MB-231-HRE-tdTomato breast tumor xenograft. In the low-mass range, we detected phosphate-containing ions at m/z 125.0, m/z 146.9, and m/z 162.9, the phosphocholine head group at m/z 184.0 and a related ion at m/z 198.0, matrix-related ions at m/z 172.0, m/z 190.0, and m/z 379.0, as well as the fiducial marker cresyl violet at m/z 262.0. Stearoylcarnitine was observed at m/z 428.3, and lysolipids were detected in the mass range from m/z 450 to m/z 550. The heme ion was present at m/z 616.1. Abundant intact phospholipid molecular ions were observed in the mass range from m/z 700 to m/z 860.
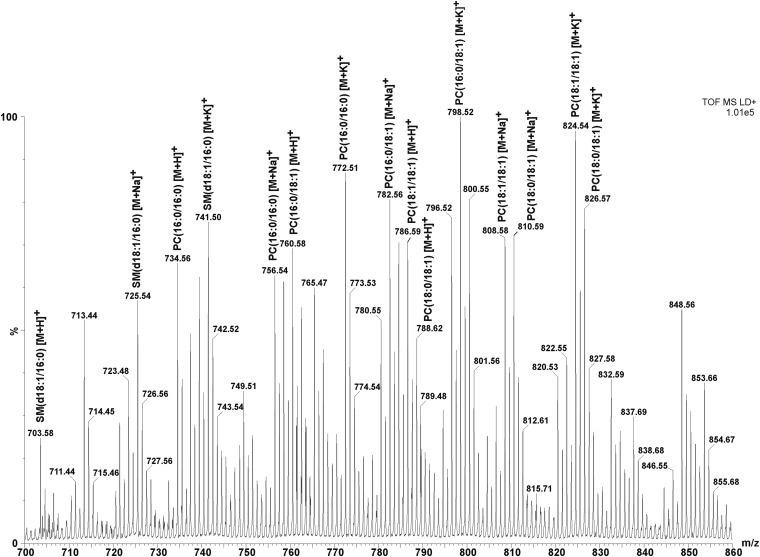
Figure 4 presents an expanded view of the spectrum shown in Fig. 3 showing multiple sphingomyelin and phosphatidylcholine species detected as proton, sodium, and potassium adducts. We detected and identified protonated species of SM(d18:1/16:0) at m/z 703.5, PC(16:0/16:0) at m/z 734.5, PC(16:0/18:1) at m/z 760.5, PC(18:1/18:1) at m/z 786.6, and PC(18:0/18:1) at m/z 788.6. The most-abundant SM and PC lipid peaks detected from MDA-MB-231-HRE-tdTomato breast tumor xenograft sections by MALDI-MS and MSI acquisitions as sodium and potassium adducts of lipids are listed in Table 1.
Fig. 4.
Expanded spectrum of proton, sodium, and potassium adducts of PC and SM lipid species identified in the mass range from m/z 700 to m/z 860 from an MDA-MB-231-HRE-tdTomato breast tumor xenograft section.
TABLE 1.
Most-abundant PC and SM species with their observed m/z values, maximum peak intensities per pixel, and ion drift times (bins) detected by MSI
| Measured m/z values |
Max peak-intensity/pixel |
Drift time (bins) |
|||||||
| Phospholipid | [M+H]+ | [M+Na]+ | [M+K]+ | [M+H]+ | [M+Na]+ | [M+K]+ | [M+H]+ | [M+Na]+ | [M+K]+ |
| SM(d18:1/16:0) | 703.58 | 725.54 | 741.50 | 81 | 112 | 136 | 89.81 | 90.74 | 91.59 |
| PC(16:0/16:0) | 734.56 | 756.54 | 772.51 | 131 | 108 | 153 | 90.84 | 91.70 | 92.28 |
| PC(16:0/18:1) | 760.58 | 782.56 | 798.52 | 140 | 139 | 173 | 92.22 | 92.55 | 93.91 |
| PC(18:1/18:1) | 786.59 | 808.58 | 824.54 | 121 | 118 | 160 | 94.09 | 94.68 | 95.19 |
| PC(18:0/18:1) | 788.62 | 810.59 | 826.57 | 95 | 135 | 122 | 95.59 | 95.99 | 96.73 |
Values are listed for protonated ions [M+H]+, sodium adducts [M+Na]+, and potassium adducts [M+K]+ of lipids.
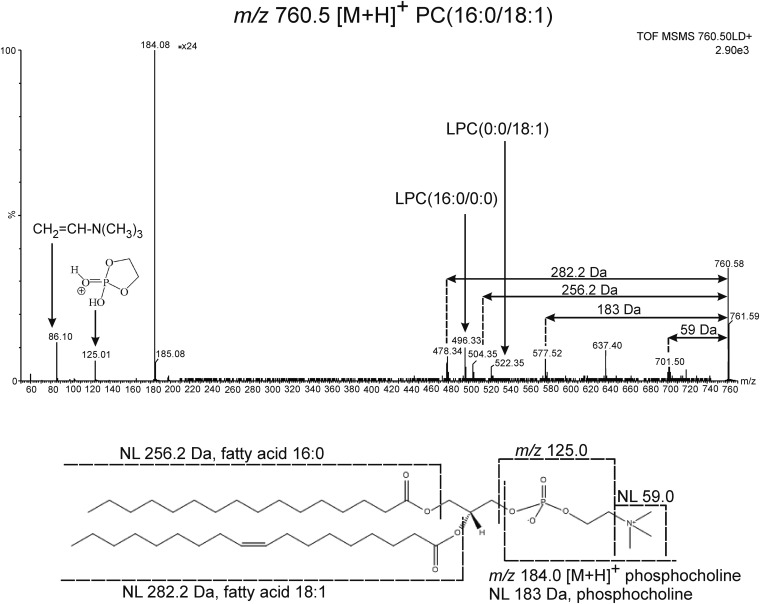
The structures of all described lipid species were confirmed by MALDI-MS/MS profiling experiments performed directly on tumor tissue sections. Figure 5 shows a typical MS/MS spectrum of protonated PC(16:0/18:1), which has a highly abundant fragment signal at m/z 184.0 arising from the protonated phosphocholine ion, a signature peak in the MS/MS of protonated lysophosphatidylcholine (LPC), PC, and SM species. A less-abundant signal at m/z 86 is characteristic of the choline head group. The loss of trimethylamine from the phosphocholine head group leads to the fragment ion [M+H-59]+ at m/z 701.5, whereas the loss of the entire phosphocholine head group leads to the formation of the fragment ion [M+H-183]+ at m/z 577.5. Additionally, the peak at m/z 125.0 is a phosphate-related ion, which forms from the protonated phosphocholine head group after neutral loss (NL) of the quaternary amine (NL 59). Other structurally relevant fragment ions are of low abundance. The fragment at m/z 478.3 is generated by loss of the 18:1 fatty acyl chain, whereas the fragment at m/z 504.3 forms from loss of the 16:0 fatty acyl chain from the PC precursor. The fragment at m/z 496.3 corresponds to the protonated LPC(16:0/0:0). Similarly, the fragment at m/z 522.3 corresponds to the protonated LPC(0:0/18:1). These four distinct fragments confirm the identity of m/z 760.5 as PC(16:0/18:1).
Fig. 5.
MS/MS spectrum and structure of PC(16:0/18:1). The ion at m/z 86 is a characteristic fragment of the choline head group, which ionizes as an intact molecular ion at m/z 184.0. The fragment ion at m/z 478.3 is generated from loss of the 18:1 fatty acyl chain from the intact lipid molecule, whereas the m/z 504.3 fragment forms from loss of the 16:0 fatty acyl chain from the PC precursor. The protonated LPC(16:0/0:0) is formed as a fragment at m/z 496.3. Similarly, the fragment at m/z 522.3 corresponds to the protonated LPC(0:0/18:1). NL of the entire phosphocholine head group leads to the formation of the ion [M+H-183]+ at m/z 577.5, whereas the loss of trimethylamine from the phosphocholine head group generates the fragment ion [M+H-59]+ at m/z 701.5.
Mass spectrometric imaging of lipids
MALDI imaging.
Images obtained from optical imaging and MALDI-MSI were coregistered as previously described based on the position of the fiducial markers detected by both optical microscopy and MALDI-MSI (45). During MALDI-MS analysis, both PC and SM species ionized as singly charged molecular ions due to the choline head group. Figure 6 shows the distributions of protonated [M+H]+ ions derived from PC(16:0/16:0) at m/z 734.5, PC(16:0/18:1) at m/z 760.5, PC(18:1/18:1) at m/z 786.6, and PC(18:0/18:1) at m/z 788.6. These highly abundant phospholipid species were detected predominantly from viable (both normoxic as well as hypoxic) tumor regions. We detected the [M+H]+ ion of palmitoylcarnitine at m/z 400.3, the [M+H]+ ion of stearoylcarnitine at m/z 428.3, the [M+K]+ ion of LPC(16:0/0:0) at m/z 534.3, the sodium adduct [M+Na]+ of sphingomyelin SM(d18:1/16:0) at m/z 725.5, and the [M+H]+ ion of PC(16:0/22:1) at m/z 816.6, mostly in hypoxic and necrotic tumor regions. The fragmentation spectra of these lipid species were detected by MALDI-MS/MS directly from the breast tumor xenograft sections and are provided in the supplementary materials.
Fig. 6.
Multimodal imaging combining optical microscopy to visualize different tumor microenvironments and mass spectrometric imaging to localize multiple lipid molecular ions. Optical images: A: Diagram showing the localization of viable (in green), necrotic (in blue) and hypoxic regions (in red) in a representative MDA-MB-231-HRE-tdTomato breast tumor section. B: Bright-field image of the corresponding fresh tumor section and fiducial markers visible as red points. C: Image of the hypoxic region obtained by fluorescence microscopy of tdTomato. The black background has been removed for better coregistration with MSI. D: H and E-stained image of a tumor section obtained from the fresh section shown in B. The necrotic region present in the tumor center has been outlined. The empty hole within this section originated from loss of tissue integrity in a small part of the necrotic tumor region. Mass spectrometric images from left to right: [M-CH3COO−]+ of fiducial markers at m/z 262.0, [M+H]+ of palmitoylcarnitine at m/z 400.3, [M+H]+ of stearoylcarnitine at m/z 428.3, [M+K]+ of LPC(16:0/0:0) at m/z 534.3, [M+Na]+ of SM(d18:1/16:0) at m/z 725.5, [M+H]+ of PC(16:0/16:0) at m/z 734.5, [M+H]+ of PC(16:0/18:1) at m/z 760.5, [M+H]+ of PC(18:1/18:1) at m/z 786.6, [M+H]+ of PC(18:0/18:1) at m/z 788.6, and [M+H]+ of PC(16:0/22:1) at m/z 816.6. Coregistered images: The mass spectrometric images in the top row were coregistered with the optical images in the left column based on the position of the fiducial markers. Scale bar, 5 mm.
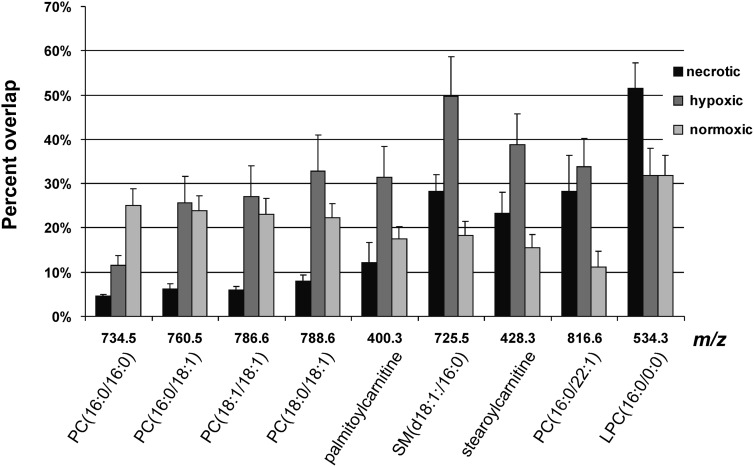
To quantify the ion signals in different breast tumor regions, we performed an overlap analysis of each ion distribution with each of the three distinct regions, namely the normoxic, hypoxic, and necrotic tumor regions, which were visualized by our multimodal imaging approach from three MDA-MB-231-HRE-tdTomato tumor xenografts. Figure 7 present the results of the overlay analysis. PC(16:0/16:0) overlapped predominantly with normoxic tumor regions, whereas LPC(16:0/0:0) was highly enriched in necrotic tumor regions. The remaining PCs, SM, and acylcarnitines displayed the highest percentage of overlap with hypoxic tumor regions.
Fig. 7.
Overlap analysis between the MSI-detected molecules and different tumor regions. The necrotic region was separated from the viable region. The latter was further divided into hypoxic and normoxic tumor regions. The necrotic, hypoxic, and normoxic regions together make up the entire tumor tissue. PC(16:0/16:0) was primarily detected from normoxic tumor regions, whereas LPC(16:0/0:0) was characteristic of necrotic tissue. The remaining PCs, SM, and acylcarnitines were colocalizing primarily with hypoxic tumor regions and, to a lesser extent, with normoxic tumor regions. Data are shown as mean ± standard error.
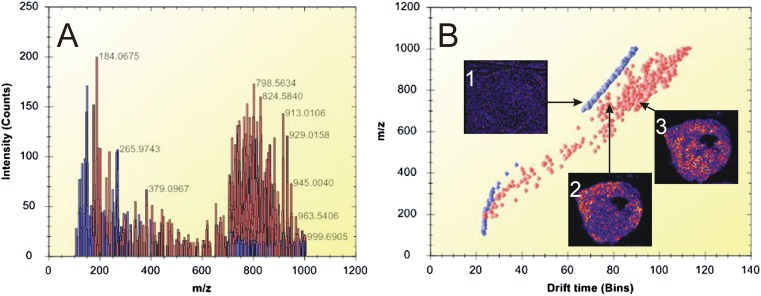
Imaging of isobaric species in IMS mode.
IMS enables the separation of ions on the basis of their collisional cross-section, which depends on their shape and charge. In our study, IMS separated the background ions (chemical noise, matrix clusters, etc.) from the biomolecular lipid species. Figure 8A presents a spectrum of peak-picked data acquired during an IMS imaging experiment, and Fig. 8B shows a drift plot of the separated ions. In Figs. 8A and background ions are highlighted in blue, whereas biomolecular lipid ions are highlighted in red. Ion image 1 in Fig. 8B shows the distribution of a background matrix ion, which was separated from two lipid ions, shown in ion images 2 and 3.
Fig. 8.
IMS of biomolecular ions detected from thin tumor tissue sections. A: Representative spectrum of peak-picked data acquired during an IMS imaging experiment. The lipid peaks are highlighted in red, whereas matrix-related ions are shown in blue. B: Drift plot of the separated ions. Ion image 1 shows the distribution of a background matrix ion. Ion images 2 (m/z 746.5) and 3 (m/z 746.6) show the distribution of different lipids in the tumor tissue. Lipid-related ions (highlighted in red) were separated from background ions (blue).
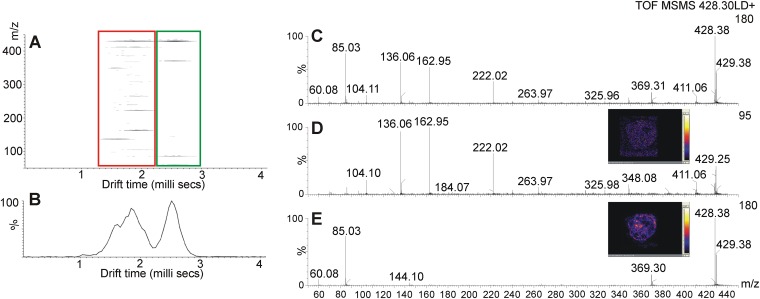
IMS not only improved the MSI data obtained from complex tumor tissue, but also enabled the identification of several biomolecules, which otherwise could not have been separated from other ions of similar mass. Figure 9 shows an example of two ions at m/z 428.25 and m/z 428.38, which were analyzed in IMS mode. Separated precursor ions were fragmented, which resulted in the identification of m/z 428.3 as stearoylcarnitine [M+H]+. Figure 9A presents the ion mobility graph from two fragmented ions. The left side, circled by the red box, shows the separated faster ion and its fragments, while the green box encircles the slower ion and its fragments. Figure 9B presents the corresponding drift time plot showing the intensity of the slow and fast ions and their drift times, which are 1.7 milliseconds and 2.5 milliseconds, respectively. Figure 9C presents a full spectrum of detected ions and their fragments, obtained without IMS. IMS performed prior to fragmentation resulted in two spectra of fast (Fig. 9D) and slow (Fig. 9E) ions. Subsequent collision-induced dissociation allowed for the identification of the slower ion at m/z 428.3 as protonated stearoylcarnitine.
Fig. 9.
Imaging ion mobility mass spectrometry of molecules present in the MDA-MB-231-HRE-tdTomato breast tumor xenograft model. A: 2D map (m/z vs. drift time) showing ions at m/z 428.2 (red) and m/z 428.3 (green) separated and fragmented during an MS/MS experiment. B: Drift time plot showing drift times of the two clearly separated ions shown in A. C: MS/MS spectrum of fragments detected from both ions without IMS. D: MS/MS spectrum of fragments detected only from the ion at m/z 428.2, whose separation was achieved by IMS. E: MS/MS spectrum of fragments detected only from the stearoylcarnitine ion [M+H]+ at m/z 428.3, whose separation was achieved by IMS.
DISCUSSION
Advantages of MSI-IMS over other techniques used in lipidomics studies
In the present study, we analyzed human breast tumor xenograft tissue using MSI-IMS, and localized multiple metabolites and phospholipid species in different tumor regions. Unlike classical MALDI MS, which is widely used to identify lipids, MSI does not require extraction of the lipids prior to analysis, but rather visualizes their localization directly in the analyzed tissue. MALDI-MSI does not require any labeling of the lipids. Lipid identification is achieved on the basis of their molecular mass and characteristic fragmentation patterns. Such an imaging approach requires very little sample modification prior to imaging, and introduces minimal alterations in lipid localizations and structures. Phospholipid ion formation by MALDI-MSI, when performed directly on a tissue section, generates abundant alkali metal adduct ions, a phenomenon not observed when MALDI is performed on pure lipid extracts. Here, we have employed IMS, because tissue washing removes certain metabolites and lipids from the tissue or leads to their diffusion. Hence, we decided to follow a sample preparation procedure that introduces minimal changes to the composition of the tissue. IMS provided a separation step for our imaging experiments that is usually performed on lipid extracts by HPLC or TLC prior to mass analysis (49, 50). IMS, in which separation occurs in milliseconds, can be easily coupled with MSI and helps obtain ion images of individual lipid species without requiring labor-intensive sample preparation procedures or long acquisition times. The development of new HDI software (Waters, UK) incorporating drift plots and ion drift time made our data analysis fast and simple. The effects of acyl chain length, degree of unsaturation, lipid class, head group, and cation formation on an ion's collision cross-section in the gas phase and on its ionization efficiency have been studied for direct tissue profiling (51). Such information greatly improves our understanding of ion behaviors detected from a complex tissue sample during MSI-IMS. To date, IMS has been applied to map the distribution of PC and cerebroside species from coronal rat brain sections (52) and for imaging of glucosylceramide in spleen sections from a mouse model of Gaucher disease (53).
Identification of lipids and their biological role
Most of the lipids identified in this study were SM and PC species detected in the mass range from m/z 700 to m/z 850. The presence of a positively charged quaternary ammonium group in both PC and SM species aids in the detection of these two classes of phospholipids in positive-ion mode (51). Previous MALDI-MS studies of phospholipid mixtures have demonstrated that in positive-ion mode, PC and SM species can inhibit the detection of other classes of phospholipids, such as phosphatidylethanolamines, present in the tissue at similar concentrations (54, 55). In this study, we focused on identification and imaging of phosphocholine-containing lipids present in MDA-MB-231-HRE-tdTomato breast tumor xenograft models.
Analysis of MALDI mass spectra directly obtained from MDA-MB-231-HRE-tdTomato breast tumor sections revealed multiple PC species with FA chains ranging between 16 and 22 carbons and containing 0 to 2 unsaturated bonds (Fig. 4). These PC species were detected primarily in viable regions of MDA-MB-231-HRE-tdTomato breast tumors, with PC(16:0/16:0) predominantly localized to normoxic tumor regions and all other PC species localized primarily to hypoxic tumor regions (Fig. 7). The identification of all lipid species involved collisional activation (see Fig. 5 and supplementary Fig. I–V). The protonated LPCs and PCs yielded a major product ion at m/z 184.0, corresponding to the phosphocholine headgroup, as well as a phosphate-related ion at m/z 125.0 [M+H]+, whereas the alkali-cationized molecular species produced characteristic ions at m/z 146.9 [M+Na]+ and m/z 162.9 [M+K]+ (24) instead of m/z 125.0.
Our MSI and MS/MS analyses of MDA-MB-231-HRE-tdTomato breast tumor tissue revealed an abundant presence of LPC(16:0/0:0) in necrotic tumor regions, which could be due to breakdown of intact PC species. LPC species are formed during inflammation, in cases of chemical exposure (56), and following heart (16) and brain (19, 57) injury. Multiple different bioactive lysophospholipids exhibit pleomorphic effects on multiple cell lineages, including breast and ovarian cancer cells (58, 59). LPC activates the ovarian cancer G protein-coupled receptor 1 family of G protein-coupled receptors , known to regulate cellular motility, growth, differentiation, and gene transcription, all of which are factors central to the biology of cancer (60). The effects of LPC on intracellular signaling in breast cancer cells is translated to functional changes such as increased production of multiple growth factors from breast cancer cells, including interleukin-6 and -8, which are potent regulators of neovascularization (61). The mechanisms regulating the production and degradation of lysophospholipids are just beginning to be elucidated.
MSI analysis of MDA-MB-231-HRE-tdTomato breast tumor tissue also revealed the presence of SM(d18:1/16:0), which was detected primarily in hypoxic tissue regions. SM(d18:1/16:0) is known to be involved in tissue inflammation and acts as a precursor of sphingolipid mediators (62, 63). Sphingolipids such as SM(d18:1/16:0) are detected by MSI in the same mass range as PC species. However, the protonated ions of PC species have even m/z values, while those of SM species have odd m/z values; therefore, although they occupy similar mass ranges, the peaks do not overlap, as evident in Fig. 4 (51). Sphingolipids contribute to the pathogenesis of cancer as well as neurological and autoimmune disorders (64). Several sphingolipid-regulated functions are implicated in tumor initiation, promotion, progression, and response to chemotherapy due to their importance in regulating growth, survival, adhesion, and migration of cancer cells (65). Sphingolipids have been proposed as cancer markers (66), and tumors often display alterations in sphingolipid composition (66). Sphingolipid intermediates may trigger proliferation and impair the ability of damaged cells to undergo apoptosis (65). Signaling-induced hydrolysis of sphingolipids produces a series of messenger lipids, such as ceramides and ceramide-1-phosphate, which, as part of signaling cascades, act by recruiting cytosolic proteins. Sphingosine, sphingosine-1-phosphate, ceramide, and ceramide-1-phosphate have pivotal roles in apoptosis, inflammation, and arachidonic acid signaling, all of which are pathways that are impaired in many cancers (67, 68).
Carnitine and acylcarnitines are essential compounds in FA metabolism (69). Carnitine assists with transport of FAs into mitochondria, where FAs are oxidized by β-oxidation as a major source of energy. We have detected and imaged two acylcarnitines directly from MDA-MB-231-HRE-tdTomato breast tumor tissue sections by applying MSI together with on-tissue IMS-MS/MS analysis combined with a Human Metabolome Database search for their identification. IMS was successfully employed for removing background ion fragments. We have identified ions at m/z 400.3 and m/z 428.3 as protonated ions of palmitoylcarnitine and stearoylcarnitine, respectively, which both localized to hypoxic tumor regions. This accumulation of acylcarnitines in hypoxic tumor regions probably occurred because of a blockage of oxidative processes in these hypoxic tumor regions (70). The inhibition of β-oxidation during hypoxia leads to an accumulation of acylcarnitines in the cytoplasm of hypoxic cells (69). The concentration of long-chain acylcarnitines increased rapidly within minutes after the onset of ischemia in vivo or hypoxia in vitro (71), as shown in plasma and hearts of hypoxic rats (72–74), a newborn rat model of hypoxia-ischemia (75), adult canine myocytes (76), rabbit fetal lungs (70), and blood samples from neonates (77). Acylcarnitines are highly polar, zwitterionic molecules and therefore tend to elute together with phospholipids such as phosphatidylcholine in many chromatographic systems. Here, we have demonstrated that due to the presence of their positively charged quaternary ammonium group, acylcarnitines can also be easily detected and localized by MALDI-MSI.
CONCLUSIONS
We detected, identified, and visualized several important lipid species in a human breast tumor xenograft model using MALDI-IMS-MSI. The detected lipids belonged to the lipid classes of PC and SM. For the first time, different lipids were visualized in different microenvironmental tumor regions. Most PC species were found in viable normoxic and hypoxic breast tumor regions, whereas LPC was localized to necrotic regions, which could be attributed to PC breakdown triggered by tissue injury. Two acylcarnitines were localized in hypoxic tumor regions and identified by using IMS. Their presence in the hypoxic regions suggests blockage of the β-oxidation process of FAs inside mitochondria. In addition, SM(d18:1/16:0), known to be involved in cellular signaling processes, was enriched in hypoxic tumor regions. Our study demonstrated the first visualization of phospholipids in a human breast tumor xenograft model and shed more light on processes occurring during tumor growth.
Supplementary Material
Acknowledgments
The authors thank Drs. Paul T. Winnard, Jr., Venu Raman, and Zaver M. Bhujwalla for providing the MDA-MB-231-HRE-tdTomato breast cancer cell line.
Footnotes
Abbreviations:
- ACN
- acetonitrile
- ATCC
- American Type Culture Collection
- 2D
- two-dimensional
- H and E
- hematoxylin and eosin
- HDI
- high-definition imaging
- IMS
- ion mobility separation
- LPC
- lysophosphatidylcholine
- MSI
- mass spectrometry imaging
- NL
- neutral loss
- PC
- phosphatidylcholine
- SM
- sphingomyelin
- TFA
- trifluoroacetic acid
This work is part of the research program of the Foundation for Fundamental Research on Matter, which is financially supported by the The Netherlands Organization for Scientific Research. The authors gratefully acknowledge financial support from National Institutes of Health Grant R01 CA-134695.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five figures.
REFERENCES
- 1.Chughtai K., Heeren R. M. 2010. Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 110: 3237–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaupel P., Mayer A., Briest S., Hockel M. 2003. Oxygenation gain factor: a novel parameter characterizing the association between hemoglobin level and the oxygenation status of breast cancers. Cancer Res. 63: 7634–7637 [PubMed] [Google Scholar]
- 3.Hoogsteen I. J., Marres H. A., van den Hoogen F. J., Rijken P. F., Lok J., Bussink J., Kaanders J. H. Expression of EGFR under tumor hypoxia: identification of a subpopulation of tumor cells responsible for aggressiveness and treatment resistance. Int. J. Radiat. Oncol. Biol. Phys. Epub ahead of print. March 13, 2012; doi: 10.1016/j.ijrobp.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 4.Vaupel P., Mayer A. 2007. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 26: 225–239 [DOI] [PubMed] [Google Scholar]
- 5.Morton C. L., Houghton P. J. 2007. Establishment of human tumor xenografts in immunodeficient mice. Nat. Protoc. 2: 247–250 [DOI] [PubMed] [Google Scholar]
- 6.Kakkad S. M., Solaiyappan M., O'Rourke B., Stasinopoulos I., Ackerstaff E., Raman V., Bhujwalla Z. M., Glunde K. 2010. Hypoxic tumor microenvironments reduce collagen I fiber density. Neoplasia. 12: 608–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penet M. F., Pathak A. P., Raman V., Ballesteros P., Artemov D., Bhujwalla Z. M. 2009. Noninvasive multiparametric imaging of metastasis-permissive microenvironments in a human prostate cancer xenograft. Cancer Res. 69: 8822–8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hart P. J., Francese S., Claude E., Woodroofe M. N., Clench M. R. 2011. MALDI-MS imaging of lipids in ex vivo human skin. Anal. Bioanal. Chem. 401: 115–125 [DOI] [PubMed] [Google Scholar]
- 9.Tanaka H., Zaima N., Yamamoto N., Sagara D., Suzuki M., Nishiyama M., Mano Y., Sano M., Hayasaka T., Goto-Inoue N., et al. 2010. Imaging mass spectrometry reveals unique lipid distribution in primary varicose veins. Eur. J. Vasc. Endovasc. Surg. 40: 657–663 [DOI] [PubMed] [Google Scholar]
- 10.Meriaux C., Franck J., Wisztorski M., Salzet M., Fournier I. 2010. Liquid ionic matrixes for MALDI mass spectrometry imaging of lipids. J. Proteomics. 73: 1204–1218 [DOI] [PubMed] [Google Scholar]
- 11.Burnum K. E., Cornett D. S., Puolitaival S. M., Milne S. B., Myers D. S., Tranguch S., Brown H. A., Dey S. K., Caprioli R. M. 2009. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J. Lipid Res. 50: 2290–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enomoto H., Sugiura Y., Setou M., Zaima N. 2011. Visualization of phosphatidylcholine, lysophosphatidylcholine and sphingomyelin in mouse tongue body by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal. Bioanal. Chem. 400: 1913–1921 [DOI] [PubMed] [Google Scholar]
- 13.Touboul D., Piednoel H., Voisin V., De La Porte S., Brunelle A., Halgand F., Laprevote O. 2004. Changes of phospholipid composition within the dystrophic muscle by matrix-assisted laser desorption/ionization mass spectrometry and mass spectrometry imaging. Eur. J. Mass Spectrom. (Chichester, Eng.). 10: 657–664 [DOI] [PubMed] [Google Scholar]
- 14.Chughtai S., Chughtai K., Cillero-Pastor B., Kiss A., Agrawal P., MacAleese L., Heeren R. M. A. 2012. A multimodal mass spectrometry imaging approach for the study of musculoskeletal tissues. Int. J. Mass Spectrom. 325–327: 150–160 [Google Scholar]
- 15.Landgraf R. R., Prieto Conaway M. C., Garrett T. J., Stacpoole P. W., Yost R. A. 2009. Imaging of lipids in spinal cord using intermediate pressure matrix-assisted laser desorption-linear ion trap/Orbitrap MS. Anal. Chem. 81: 8488–8495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menger R. F., Stutts W. L., Anbukumar D. S., Bowden J. A., Ford D. A., Yost R. A. 2012. MALDI mass spectrometric imaging of cardiac tissue following myocardial infarction in a rat coronary artery ligation model. Anal. Chem. 84: 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Astigarraga E., Barreda-Gomez G., Lombardero L., Fresnedo O., Castano F., Giralt M. T., Ochoa B., Rodriguez-Puertas R., Fernandez J. A. 2008. Profiling and imaging of lipids on brain and liver tissue by matrix-assisted laser desorption/ ionization mass spectrometry using 2-mercaptobenzothiazole as a matrix. Anal. Chem. 80: 9105–9114 [DOI] [PubMed] [Google Scholar]
- 18.Cha S., Yeung E. S. 2007. Colloidal graphite-assisted laser desorption/ionization mass spectrometry and MSn of small molecules. 1. Imaging of cerebrosides directly from rat brain tissue. Anal. Chem. 79: 2373–2385 [DOI] [PubMed] [Google Scholar]
- 19.Koizumi S., Yamamoto S., Hayasaka T., Konishi Y., Yamaguchi-Okada M., Goto-Inoue N., Sugiura Y., Setou M., Namba H. 2010. Imaging mass spectrometry revealed the production of lyso-phosphatidylcholine in the injured ischemic rat brain. Neuroscience. 168: 219–225 [DOI] [PubMed] [Google Scholar]
- 20.Puolitaival S. M., Burnum K. E., Cornett D. S., Caprioli R. M. 2008. Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J. Am. Soc. Mass Spectrom. 19: 882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trim P. J., Atkinson S. J., Princivalle A. P., Marshall P. S., West A., Clench M. R. 2008. Matrix-assisted laser desorption/ionisation mass spectrometry imaging of lipids in rat brain tissue with integrated unsupervised and supervised multivariant statistical analysis. Rapid Commun. Mass Spectrom. 22: 1503–1509 [DOI] [PubMed] [Google Scholar]
- 22.Wang H. Y., Jackson S. N., Post J., Woods A. S. 2008. A minimalist approach to MALDI imaging of glycerophospholipids and sphingolipids in rat brain sections. Int. J. Mass Spectrom. 278: 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Allegood J., Liu Y., Wang E., Cachon-Gonzalez B., Cox T. M., Merrill A. H., Jr, Sullards M. C. 2008. Imaging MALDI mass spectrometry using an oscillating capillary nebulizer matrix coating system and its application to analysis of lipids in brain from a mouse model of Tay-Sachs/Sandhoff disease. Anal. Chem. 80: 2780–2788 [DOI] [PubMed] [Google Scholar]
- 24.Murphy R. C., Hankin J. A., Barkley R. M. 2009. Imaging of lipid species by MALDI mass spectrometry. J. Lipid Res. 50 (Suppl.): 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veloso A., Astigarraga E., Barreda-Gomez G., Manuel I., Ferrer I., Giralt M. T., Ochoa B., Fresnedo O., Rodriguez-Puertas R., Fernandez J. A. 2011. Anatomical distribution of lipids in human brain cortex by imaging mass spectrometry. J. Am. Soc. Mass Spectrom. 22: 329–338 [DOI] [PubMed] [Google Scholar]
- 26.Veloso A., Fernandez R., Astigarraga E., Barreda-Gomez G., Manuel I., Giralt M. T., Ferrer I., Ochoa B., Rodriguez-Puertas R., Fernandez J. A. 2011. Distribution of lipids in human brain. Anal. Bioanal. Chem. 401: 89–101 [DOI] [PubMed] [Google Scholar]
- 27.Eberlin L. S., Norton I., Dill A. L., Golby A. J., Ligon K. L., Santagata S., Cooks R. G., Agar N. Y. 2012. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 72: 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masterson T. A., Dill A. L., Eberlin L. S., Mattarozzi M., Cheng L., Beck S. D., Bianchi F., Cooks R. G. 2011. Distinctive glycerophospholipid profiles of human seminoma and adjacent normal tissues by desorption electrospray ionization imaging mass spectrometry. J. Am. Soc. Mass Spectrom. 22: 1326–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gustafsson J. O., Oehler M. K., Ruszkiewicz A., McColl S. R., Hoffmann P. 2011. MALDI imaging mass spectrometry (MALDI-IMS)—application of spatial proteomics for ovarian cancer classification and diagnosis. Int. J. Mol. Sci. 12: 773–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Chen Y., Momin A., Shaner R., Wang E., Bowen N. J., Matyunina L. V., Walker L. D., McDonald J. F., Sullards M. C., et al. 2010. Elevation of sulfatides in ovarian cancer: an integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol. Cancer. 9: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willems S. M., van Remoortere A., van Zeijl R., Deelder A. M., McDonnell L. A., Hogendoorn P. C. 2010. Imaging mass spectrometry of myxoid sarcomas identifies proteins and lipids specific to tumour type and grade, and reveals biochemical intratumour heterogeneity. J. Pathol. 222: 400–409 [DOI] [PubMed] [Google Scholar]
- 32.Eberlin L. S., Dill A. L., Golby A. J., Ligon K. L., Wiseman J. M., Cooks R. G., Agar N. Y. 2010. Discrimination of human astrocytoma subtypes by lipid analysis using desorption electrospray ionization imaging mass spectrometry. Angew. Chem. Int. Ed. Engl. 49: 5953–5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eberlin L. S., Dill A. L., Costa A. B., Ifa D. R., Cheng L., Masterson T., Koch M., Ratliff T. L., Cooks R. G. 2010. Cholesterol sulfate imaging in human prostate cancer tissue by desorption electrospray ionization mass spectrometry. Anal. Chem. 82: 3430–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimma S., Sugiura Y., Hayasaka T., Hoshikawa Y., Noda T., Setou M. 2007. MALDI-based imaging mass spectrometry revealed abnormal distribution of phospholipids in colon cancer liver metastasis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 855: 98–103 [DOI] [PubMed] [Google Scholar]
- 35.Dill A. L., Ifa D. R., Manicke N. E., Ouyang Z., Cooks R. G. 2009. Mass spectrometric imaging of lipids using desorption electrospray ionization. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2883–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le T. T., Huff T. B., Cheng J. X. 2009. Coherent anti-Stokes Raman scattering imaging of lipids in cancer metastasis. BMC Cancer. 9: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramos C. V., Taylor H. B. 1974. Lipid-rich carcinoma of the breast. A clinicopathologic analysis of 13 examples. Cancer. 33: 812–819 [DOI] [PubMed] [Google Scholar]
- 38.Sijens P. E., Levendag P. C., Vecht C. J., van Dijk P., Oudkerk M. 1996. 1H MR spectroscopy detection of lipids and lactate in metastatic brain tumors. NMR Biomed. 9: 65–71 [DOI] [PubMed] [Google Scholar]
- 39.Folick A., Min W., Wang M. C. 2011. Label-free imaging of lipid dynamics using coherent anti-Stokes Raman scattering (CARS) and stimulated Raman scattering (SRS) microscopy. Curr. Opin. Genet. Dev. 21: 585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le T. T., Yue S., Cheng J. X. 2010. Shedding new light on lipid biology with coherent anti-Stokes Raman scattering microscopy. J. Lipid Res. 51: 3091–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maier O., Oberle V., Hoekstra D. 2002. Fluorescent lipid probes: some properties and applications (a review). Chem. Phys. Lipids. 116: 3–18 [DOI] [PubMed] [Google Scholar]
- 42.Krishnamachary B., Penet M. F., Nimmagadda S., Mironchik Y., Raman V., Solaiyappan M., Semenza G. L., Pomper M. G., Bhujwalla Z. M. Hypoxia regulates CD44 and its variant isoforms through HIF-1α in triple negative breast cancer. PLoS ONE. Epub August 28, 2012; doi: 10.1371/journal.pone.0044078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang L., Greenwood T. R., Artemov D., Raman V., Winnard P. T., Jr, Heeren R. M. A., Bhujwalla Z. M., Glunde K. 2012.. Localized hypoxia results in spatially heterogeneous metabolic signatures in breast tumor models. Neoplasia. 14: 732–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang L., Greenwood T. R., Amstalden van Hove E. R., Chughtai K., Raman V., Winnard P. T., Jr, Heeren R. M. A., Artemov D., Glunde K. 2012. Combined MR, fluorescence and histology imaging strategy in a human breast tumor xenograft model. NMR Biomed. Epub ahead of print. September 4, 2012. doi: 10.1002/nbm.2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chughtai K., Jiang L., Greenwood T. R., Klinkert I., Amstalden van Hove E. R., Heeren R. M., Glunde K. 2012. Fiducial markers for combined 3-dimensional mass spectrometric and optical tissue imaging. Anal. Chem. 84: 1817–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fahy E., Subramaniam S., Murphy R. C., Nishijima M., Raetz C. R., Shimizu T., Spener F., van Meer G., Wakelam M. J., Dennis E. A. 2009. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50 (Suppl.): 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fahy E., Sud M., Cotter D., Subramaniam S. 2007. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 35: W606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cootes T. F., Taylor C. J., Cooper D. H., Graham J. 1995. Active shape models—their training and application. Comput. Vis. Image Underst. 61: 38–59 [Google Scholar]
- 49.Peterson B. L., Cummings B. S. 2006. A review of chromatographic methods for the assessment of phospholipids in biological samples. Biomed. Chromatogr. 20: 227–243 [DOI] [PubMed] [Google Scholar]
- 50.Watson A. D. 2006. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J. Lipid Res. 47: 2101–2111 [DOI] [PubMed] [Google Scholar]
- 51.Jackson S. N., Ugarov M., Post J. D., Egan T., Langlais D., Schultz J. A., Woods A. S. 2008. A study of phospholipids by ion mobility TOFMS. J. Am. Soc. Mass Spectrom. 19: 1655–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson S. N., Ugarov M., Egan T., Post J. D., Langlais D., Albert Schultz J., Woods A. S. 2007. MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J. Mass Spectrom. 42: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snel M. F., Fuller M. 2010. High-spatial resolution matrix-assisted laser desorption ionization imaging analysis of glucosylceramide in spleen sections from a mouse model of Gaucher disease. Anal. Chem. 82: 3664–3670 [DOI] [PubMed] [Google Scholar]
- 54.Petkovic M., Schiller J., Muller M., Benard S., Reichl S., Arnold K., Arnhold J. 2001. Detection of individual phospholipids in lipid mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: phosphatidylcholine prevents the detection of further species. Anal. Biochem. 289: 202–216 [DOI] [PubMed] [Google Scholar]
- 55.Estrada R., Yappert M. C. 2004. Alternative approaches for the detection of various phospholipid classes by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Mass Spectrom. 39: 412–422 [DOI] [PubMed] [Google Scholar]
- 56.Arnhold J., Osipov A. N., Spalteholz H., Panasenko O. M., Schiller J. 2002. Formation of lysophospholipids from unsaturated phosphatidylcholines under the influence of hypochlorous acid. Biochim. Biophys. Acta. 1572: 91–100 [DOI] [PubMed] [Google Scholar]
- 57.Wang H. Y., Liu C. B., Wu H. W., Kuo J. S. 2010. Direct profiling of phospholipids and lysophospholipids in rat brain sections after ischemic stroke. Rapid Commun. Mass Spectrom. 24: 2057–2064 [DOI] [PubMed] [Google Scholar]
- 58.Goetzl E. J., Dolezalova H., Kong Y., Zeng L. 1999. Dual mechanisms for lysophospholipid induction of proliferation of human breast carcinoma cells. Cancer Res. 59: 4732–4737 [PubMed] [Google Scholar]
- 59.Imai A., Furui T., Tamaya T., Mills G. B. 2000. A gonadotropin-releasing hormone-responsive phosphatase hydrolyses lysophosphatidic acid within the plasma membrane of ovarian cancer cells. J. Clin. Endocrinol. Metab. 85: 3370–3375 [DOI] [PubMed] [Google Scholar]
- 60.Spiegelberg B. D., Hamm H. E. 2007. Roles of G-protein-coupled receptor signaling in cancer biology and gene transcription. Curr. Opin. Genet. Dev. 17: 40–44 [DOI] [PubMed] [Google Scholar]
- 61.Umezu-Goto M., Tanyi J., Lahad J., Liu S., Yu S., Lapushin R., Hasegawa Y., Lu Y., Trost R., Bevers T., et al. 2004. Lysophosphatidic acid production and action: validated targets in cancer? J. Cell. Biochem. 92: 1115–1140 [DOI] [PubMed] [Google Scholar]
- 62.Modrak D. E., Gold D. V., Goldenberg D. M. 2006. Sphingolipid targets in cancer therapy. Mol. Cancer Ther. 5: 200–208 [DOI] [PubMed] [Google Scholar]
- 63.Augé N., Maupas-Schwalm F., Elbaz M., Thiers J. C., Waysbort A., Itohara S., Krell H. W., Salvayre R., Negre-Salvayre A. 2004. Role for matrix metalloproteinase-2 in oxidized low-density lipoprotein-induced activation of the sphingomyelin/ceramide pathway and smooth muscle cell proliferation. Circulation. 110: 571–578 [DOI] [PubMed] [Google Scholar]
- 64.Zeidan Y. H., Hannun Y. A. 2010. The acid sphingomyelinase/ceramide pathway: biomedical significance and mechanisms of regulation. Curr. Mol. Med. 10: 454–466 [DOI] [PubMed] [Google Scholar]
- 65.Signorelli P., Ghidoni R. 2005. Breast cancer and sphingolipid signalling. J. Dairy Res. 72: 5–13 [DOI] [PubMed] [Google Scholar]
- 66.Momin A. A., Park H., Portz B. J., Haynes C. A., Shaner R. L., Kelly S. L., Jordan I. K., Merrill A. H., Jr 2011. A method for visualization of “omic” datasets for sphingolipid metabolism to predict potentially interesting differences. J. Lipid Res. 52: 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chalfant C. E., Spiegel S. 2005. Sphingosine 1-phosphate and ceramide 1-phosphate: expanding roles in cell signaling. J. Cell Sci. 118: 4605–4612 [DOI] [PubMed] [Google Scholar]
- 68.Pettus B. J., Bielawska A., Subramanian P., Wijesinghe D. S., Maceyka M., Leslie C. C., Evans J. H., Freiberg J., Roddy P., Hannun Y. A., et al. 2004. Ceramide 1-phosphate is a direct activator of cytosolic phospholipase A2. J. Biol. Chem. 279: 11320–11326 [DOI] [PubMed] [Google Scholar]
- 69.Kerner J., Hoppel C. 2000. Fatty acid import into mitochondria. Biochim. Biophys. Acta. 1486: 1–17 [DOI] [PubMed] [Google Scholar]
- 70.Das D. K., Ayromlooi J., Neogi A. 1983. Effect of ischemia on fatty acid metabolism in fetal lung. Life Sci. 33: 569–576 [DOI] [PubMed] [Google Scholar]
- 71.Yamada K. A., McHowat J., Yan G. X., Donahue K., Peirick J., Kleber A. G., Corr P. B. 1994. Cellular uncoupling induced by accumulation of long-chain acylcarnitine during ischemia. Circ. Res. 74: 83–95 [DOI] [PubMed] [Google Scholar]
- 72.Bruder E. D., Raff H. 2010. Cardiac and plasma lipid profiles in response to acute hypoxia in neonatal and young adult rats. Lipids Health Dis. 9: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hütter J. F., Alves C., Soboll S. 1990. Effects of hypoxia and fatty acids on the distribution of metabolites in rat heart. Biochim. Biophys. Acta. 1016: 244–252 [DOI] [PubMed] [Google Scholar]
- 74.Whitmer J. T., Idell-Wenger J. A., Rovetto M. J., Neely J. R. 1978. Control of fatty acid metabolism in ischemic and hypoxic hearts. J. Biol. Chem. 253: 4305–4309 [PubMed] [Google Scholar]
- 75.Wainwright M. S., Kohli R., Whitington P. F., Chace D. H. 2006. Carnitine treatment inhibits increases in cerebral carnitine esters and glutamate detected by mass spectrometry after hypoxia-ischemia in newborn rats. Stroke. 37: 524–530 [DOI] [PubMed] [Google Scholar]
- 76.McHowat J., Yamada K. A., Saffitz J. E., Corr P. B. 1993. Subcellular distribution of endogenous long chain acylcarnitines during hypoxia in adult canine myocytes. Cardiovasc. Res. 27: 1237–1243 [DOI] [PubMed] [Google Scholar]
- 77.Meyburg J., Schulze A., Kohlmueller D., Linderkamp O., Mayatepek E. 2001. Postnatal changes in neonatal acylcarnitine profile. Pediatr. Res. 49: 125–129 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.