Abstract
Ancient ubiquitous protein 1 (AUP1) is a multifunctional protein, which acts on both lipid droplets (LDs) and the endoplasmic reticulum (ER) membrane. Double localization to these two organelles, featuring very different membrane characteristics, was observed also for several other integral proteins, but little is known about the signals and mechanisms behind dual protein targeting to ER and LDs. Here we dissect the AUP1 targeting signals by analyses of localization and topology of several deletion and point mutants. We found that AUP1 is inserted into the membrane of the ER in a monotopic hairpin fashion, and subsequently transported to the hemi-membrane of LDs. A single domain localized in the N-terminal part of AUP1 enables its ER residence, the monotopic insertion, and the LD localization. Different specific residues within this multifunctional domain are responsible for achieving the complex spatial distribution pattern. A mutation of three amino acids, which changes AUP1 topology from hairpin to transmembrane, abolishes LD localization. These findings suggest that the cell is able to target a protein to multiple intracellular locations using a single domain.
Keywords: acyltransferase, endoplasmic reticulum, triglyceride
The intracellular localization of proteins is achieved by the complex interplay of multiple transport processes using combinations of targeting and retention signals on the cargo proteins that are recognized by the transport machinery. For several localizations, e.g., to the nucleus, mitochondrium, peroxisome, or lysosome, both signals and machinery have been identified and extensively studied (1), but for others, such as the lipid droplet (LD), the situation is less clear. LDs have a unique structure with a triglyceride/sterol ester core surrounded by a phospholipid monolayer (2). They do not exchange material with other organelles by vesicular trafficking, which implies that integral proteins of the LD surface need unconventional targeting mechanisms (3). Previous studies on targeting of proteins to LDs have pointed to the importance of hydrophobic stretches or amphipatic domains in soluble proteins (4–12). For integral proteins, hydrophobic domains, often located at the end of the polypeptide chain (13–19), and charged residues (20) have been found to be essential for droplet targeting. On theoretical grounds, we have proposed that targeting of integral proteins to LDs requires a hairpin-like monotopic topology (21), but experimental evidence for a direct connection between topology and targeting is lacking so far. Additional complexity comes from the fact that many integral proteins of LDs exhibit additional localizations, sometimes to the plasma membrane (22–27), but mostly to the endoplasmic reticulum (ER) (28–34). One such protein is ancient ubiquitous protein 1 (AUP1) that specifically localizes to ER and LDs (35–37). AUP1 is a 410 amino acids long, integral protein with a single predicted membrane domain (36, 37). The membrane domain of AUP1 is located internally and both the N and C termini of AUP1 are facing the cytoplasm, resulting in monotopic/hairpin topology (36).
AUP1 has multiple functional domains and is reported to facilitate ER-associated protein degradation, inside-out signaling in platelets, and neutral lipid storage (35–39). Domains important for a function of AUP1 in ER-associated degradation are found in its C-terminal region. The CUE (coupling of ubiquitin to ER degradation) domain binds dislocation substrates and components of the ER quality control machinery (37), and the G2 binding region (G2BR) domain recruits a specific E2 ubiquitin conjugase, Ube2g2 (36). The function of the N-terminal region of AUP1 is less-well understood. It contains the membrane domain and a predicted acyltransferase domain, which was proposed to function in neutral lipid storage (37). Domains or motifs involved in AUP1 intracellular targeting are not known.
Here we dissect the localization signals of AUP1 and find that the N-terminal region including the membrane domain mediates its ER targeting, LD localization, and monotopic membrane insertion.
MATERIALS AND METHODS
Antibodies
The following antibodies were used: rabbit polyclonal anti-long-chain acyl-CoA synthetase 3 (ACSL3) (32), rabbit polyclonal anti-calnexin, and rabbit polyclonal anti-protein-disulfide isomerase (PDI) (Enzo Life Sciences; Loerrach, Germany), mouse monoclonal anti-GAPDH (Novus Biologicals; Littleton, CO), mouse monoclonal anti-hemagglutinin (clone F-7; Santa Cruz Biotechnology, Santa Cruz, CA), Alexa555- and Alexa488-conjugated secondary antibodies (Invitrogen; Darmstadt, Germany), and HRP-coupled secondary antibodies (Jackson Immuno Research; West Grove, PA).
DNA constructs
Truncation mutants of AUP1 were generated by PCR amplification and inserted into pCDNA3.1 vector with inserted C-terminal 3xHA tag. Site-directed mutagenesis was performed either using the QuikChange® site-directed mutagenesis kit (Stratagene; Amsterdam, The Netherlands) according to the manufacturer's instructions, or using a PCR-based site-directed mutagenesis approach with overlap extensions (40). DNA fragments encoding AUP1 point mutants were inserted into pCDNA3.1 vector with inserted C-terminal 3xHA tag or pN3HA vector with N-terminal 3xHA tag. Supplementary Tables I–III contain detailed information about expression constructs, including primers and restriction sites used. All constructs were verified by sequencing.
Cell culture
The monkey fibroblast cell line, COS-7, was grown at 37°C, 5% CO2 in Dulbecco's modified medium supplemented with 10% fetal calf serum (FCS) (GIBCO; Darmstadt, Germany). When indicated, cells were incubated in medium supplemented with 150 μM oleic acid (Sigma-Aldrich; Taufkirchen, Germany).
DNA transfection
DNA transfection was performed using Lipofectamine2000 transfection reagent (Invitrogen) according to the manufacturer's instructions or the transfection reagent (N4, N9-dioleoyl spermine) (41) using the same protocol but with 20% higher concentration of the transfection reagent.
Fluorescence microscopy
Cells were fixed with 3% (v/v) formaldehyde in PBS for 30 min, washed with PBS, blocked, and permeabilized for 30 min in PBS containing 0.5% BSA and 0.1% saponin (blocking buffer, BB). If indicated, saponin in the BB was replaced by 0.004% digitonin. Cells were incubated with primary antibodies in BB for 1 h, washed three times with BB, incubated with secondary antibody in BB, washed three times in BB and three times in PBS, and counterstained with LD540 (42) in PBS, followed by three washes in PBS. After rinsing in water, coverslips were mounted in Mowiol 4-88 (Merck; Darmstadt, Germany) containing 2.5% DABCO (1,4-diazabicyclo[2.2.2]octane) (Carl Roth; Karlsruhe, Germany). Images were acquired with a ZeissAxio Observer.Z1 microscope (Carl Zeiss; Oberkochen, Germany) equipped with a 63×/NA1.4 objective and a Photometrics Coolsnap K4 camera. Light source was a Polychrome V 150 W xenon lamp (TillPhotonics; Graeffeling, Germany). Images were processed using ImageJ (National Institutes of Health) or Adobe Illustrator software (Adobe).
Live-cell imaging was performed in HEPES-buffered medium (Invitrogen) supplemented with 10% FCS, at 37°C and 5% CO2. Cycloheximide was from Applichem (Darmstadt, Germany).
Purification of LDs
Cells were grown in 10-cm dishes in Dulbecco's modified medium supplemented with 10% FCS and 150 μM oleic acid for 36 h. In experiments with three fractions, four dishes of cells were used per gradient. Cells were washed once in PBS and once with Buffer A (0.2 M sucrose, 20 mM HEPES/NaOH, pH 7.5, protease inhibitor cocktail Complete (Roche; Grenzach, Germany) 1 tablet/50 ml). Subsequently, cells were scraped in Buffer A, passed through a 0.7 × 30 mm needle five times, and homogenized in a European Molecular Biology Laboratory cell cracker (HGM; Heidelberg, Germany) (inner diameter 8.020 mm, ball diameter 8.004 mm, with nine strokes). Nuclei and cell debris were pelleted by centrifugation (1,000 g, 4°C). Postnuclear supernatant was adjusted to 2 ml volume with Buffer A and gently mixed with 1 ml Buffer B (2 M sucrose, 20 mM HEPES/NaOH, pH 7.5, protease inhibitor cocktail 1 tablet/50 ml). The lysate was loaded to the bottom of the tube and overlaid with Buffer A. LDs were floated by ultracentrifugation (SW40Ti rotor at 100,000 g, at 4°C for 3 h). Fractions were collected as follows: LDs (upper 2 ml), intermediate fraction (2 ml), bottom fraction (remaining volume). In the figures, fraction LDs correspond to the LD fraction, fraction II to the intermediate fraction, and fraction III to the bottom fraction. Fractions were generally stored at −20°C prior to further analysis (SDS-PAGE and immunoblotting).
In experiments with four fractions, ultracentrifugation was performed using a different rotor (SW55Ti rotor; 100,000 g, 4°C, 3 h), and fractions were collected as follows: LDs (upper 2 ml), intermediate fraction (1 ml), upper bottom fraction (2 ml), and bottom fraction (remaining volume). In the figure panels, fraction LDs correspond to the LD fraction, fraction II to the intermediate fraction, fraction III to the upper bottom fraction, and fraction IV to the bottom fraction.
N-glycosidase F treatment
For N-glycosidase F treatment, COS-7 cells were grown in 10-cm dishes and transfected with appropriate constructs. After 24 h of expression, cells were washed with PBS and harvested by scraping in cold PBS. Cell lysis was performed using ball bearing EMBL cell cracker (HGM; 10 strokes, inner diameter 8.020 mm, ball diameter 8.004 mm). The lysate was centrifuged to remove nuclei and cellular debris (1,431 g, 10 min, 4°C). The supernatant (containing 800 μg total protein) was subjected to chloroform-methanol protein precipitation. Protein precipitate was resuspended in 20 μl solubilization buffer (20 mM sodium phosphate, pH 7.4, 2% SDS) and 380 μl reaction buffer [20 mM sodium phosphate, pH 7.4, 1% n-octyl-β-d-glucopyranosid (Applichem)]. The sample was divided into two equal parts and either treated with 4–16 U of N-glycosidase F (Roche; Grenzach, Germany) or left untreated. Samples were incubated at 20°C overnight. The reaction was stopped by addition of 5× Laemmli buffer and heating to 95°C for 10 min. Proteins were separated by SDS-PAGE and analyzed by immunoblotting.
RESULTS
Identification of the ER-targeting domain
To reveal the pathway of AUP1 targeting, we transfected COS-7 cells with green fluorescent protein-tagged AUP1 and grew them under lipid-depleted conditions to deprive them of LDs. After recording an initial micrograph, cells were supplemented with oleate and cycloheximide to induce LD formation and to stop protein biosynthesis, and microscopic imaging was continued to observe LD formation and GFP-AUP1 redistribution. As shown in the supplementary movie, GFP-tagged AUP1 initially localized to a reticular structure, presumably the ER. After 24 min, a progressive redistribution to punctate structures became visible. After 72 min, the punctate structures were identified as LDs by in situ staining with the LD-specific dye LD540 (42), demonstrating sequential targeting of the protein from the ER to LDs (see the supplementary Movie and supplementary Fig. I). Therefore, in the following, we first study the ER localization of AUP1, followed by analysis of its LD localization.
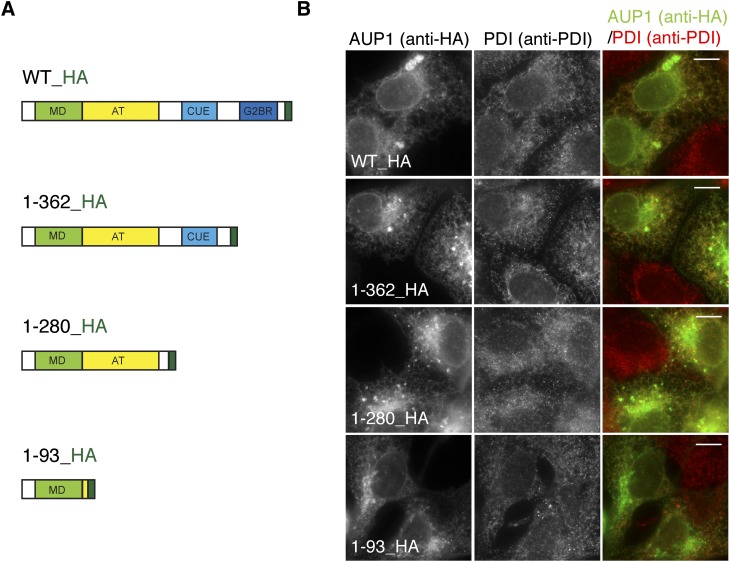
To identify the ER-targeting domain of AUP1, COS-7 cells were transfected with HA-tagged C-terminal truncation constructs (Fig. 1A), and their localization was examined by fluorescence microscopy (Fig. 1B). Full-length AUP1 partially localized to the ER, identified by a marker protein, PDI, as described previously for endogenous AUP1 (35), indicating no apparent influence of the HA tag on the ER localization of AUP1. AUP1 protein lacking either the G2BR domain or both the CUE and the G2BR domains colocalized with the ER marker (Fig. 1B). The shortest truncation construct also colocalized with the ER marker, although an additional perinuclear distribution pattern became visible (Fig. 1B). We conclude that the N-terminal 93 amino acids of AUP1 are sufficient for ER localization of AUP1.
Fig. 1.
Identification of the ER-targeting domain of AUP1. A: Schematic representation of constructs. Full-length protein is shown at the top, truncation mutants are shown below. Colored boxes labeled with capital letters represent domains. MD, membrane domain; AT, predicted acyltransferase domain; CUE, coupling of ubiquitin to ER degradation domain; G2BR, G2 binding region; dark green, HA tag. B: COS-7 cells expressing fusion proteins indicated in panel A were fixed, processed for immunofluorescence microscopy using anti-HA antibody to detect the transfected AUP1 and anti-PDI antibody to detect endoplasmic reticulum protein, and subjected to fluorescence microscopy. Scale bars, 10 μm.
Identification of the LD-targeting domain
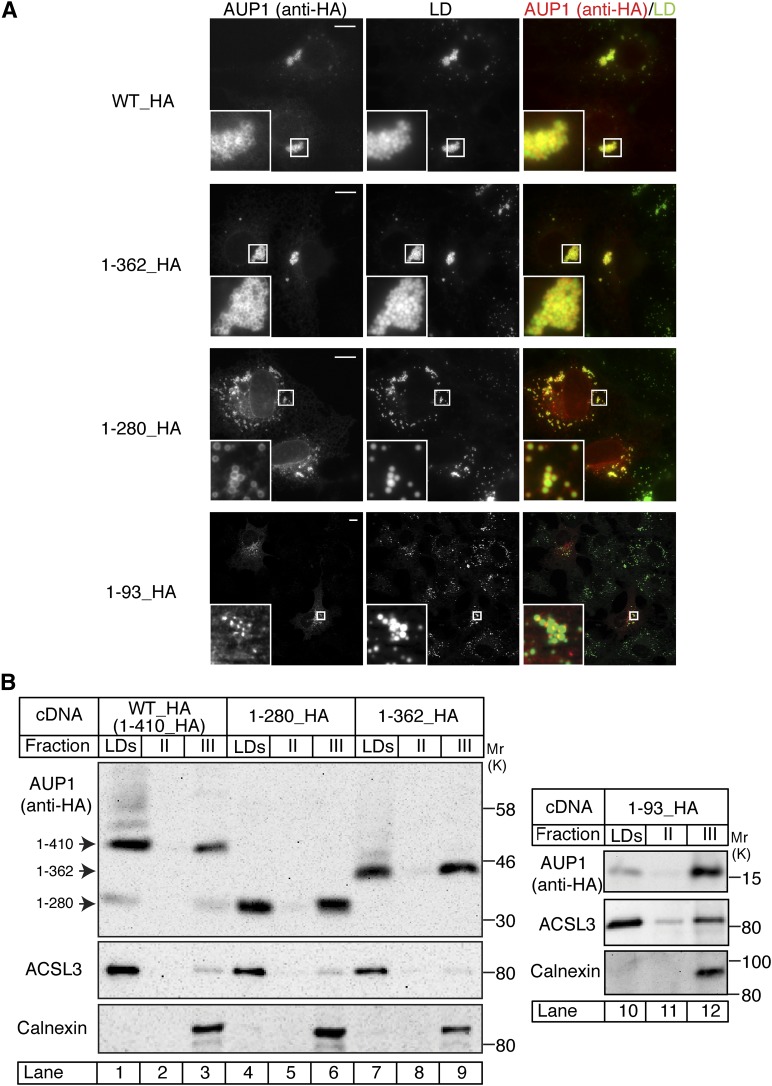
To determine which part of the AUP1 sequence is required for its localization to LDs, we expressed the constructs described above and examined their localization to LDs by fluorescence microscopy (Fig. 2A). Although full-length AUP1 and the two longer truncations displayed continuous rings around LDs, the shortest truncation, containing only 93 N-terminal amino acids of AUP1, formed discrete patches around LDs. The observed staining pattern could represent localization to either LDs or to neighboring organelles, especially ER structures that frequently surround LDs (32, 43). To address this question, localization of full-length AUP1 and the truncation mutants was examined by sucrose density gradient centrifugation and subsequent analysis of the organelle fractions by immunoblotting. A significant fraction of the full-length AUP1 construct and the truncated forms floated with the LD fraction (Fig. 2B), as identified by the LD marker protein, ACSL3 (34). The LD fraction was devoid of ER contamination, as demonstrated by the absence of calnexin, an ER marker protein. We conclude that the N-terminal 93 amino acids of AUP1 are also sufficient for LD localization.
Fig. 2.
Identification of the lipid droplet targeting domain of AUP1. A: COS-7 cells expressing fusion proteins as indicated in Fig. 1A were fixed and processed for immunofluorescence microscopy using anti-HA antibody to detect the transfected AUP1 and the LD-specific dye LD540 to detect LDs. Scale bars, 10 μm. B: COS-7 cells expressing fusion proteins as indicated were grown in medium supplemented with 150 μM oleic acid to promote LD formation. LDs were isolated by floatation in a sucrose gradient. Proteins of the floating LD fraction (LDs) and the two lower fractions (II, III) were analyzed by SDS-PAGE/immunoblotting using antibodies against the transfected AUP1 (anti-HA), LD marker (ACSL3), and ER marker (calnexin).
Key arginines for LD localization
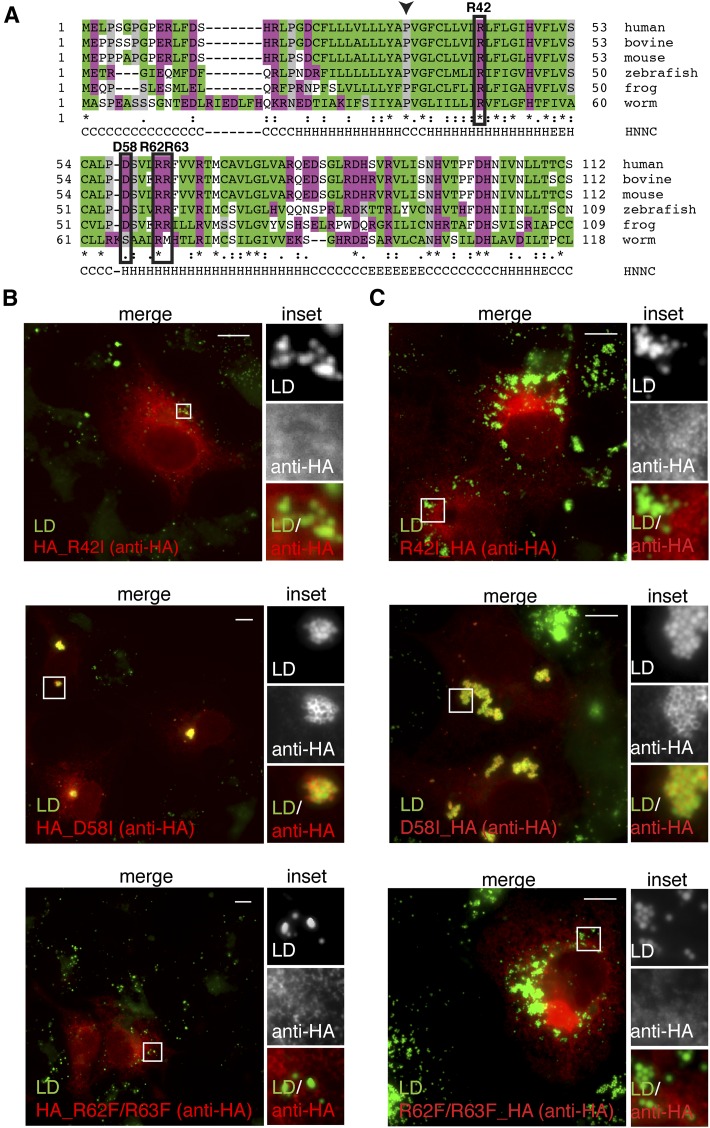
The LD-targeting domain contains a continuous hydrophobic region disrupted by a single arginine residue at position 42 (Fig. 3A). Mutation of this arginine to isoleucine (R42I), within the full-length AUP1, abolished LD targeting of both the N-terminally (Fig. 3B, top panel) and C-terminally (Fig. 3C, top panel) HA-tagged constructs, which was confirmed by subcellular fractionation analysis (see supplementary Fig. II, lanes 7–9). To examine whether charged residues downstream of the hydrophobic domain are also important for LD localization, we generated the C-terminally HA-tagged double mutant of full-length AUP1 (R62F/R63F_HA). This mutant did not accumulate on LDs (Fig. 3C, bottom panel), which was confirmed by the N-terminal tagging of the protein (Fig. 3B, bottom panel) and biochemical examination of the localization of both the N- and C-terminally-tagged constructs (see supplementary Fig. II, lanes 13–15 and 28–31). In contrast, substitution of aspartic acid at position 58 by isoleucine, within the full-length AUP1, had no effect on LD targeting, as assessed by fluorescence microscopy (Fig. 3B, middle panel; Fig. 3C, middle panel), and confirmed by subcellular fractionation (see supplementary Fig. II, lanes 10–12 and 24–27). We conclude that arginines R42 and R62/R63 are necessary for LD localization.
Fig. 3.
Point mutations within the LD-targeting domain that disrupt lipid droplet localization. A: Identification of conserved residues in the LD-targeting domain of AUP1. AUP1 protein sequences of different species were aligned using ClustalW. Graphical display of the sequence alignment is shown. Alignment covering the first 112 amino acids of human AUP1 is shown. Hydrophobic residues are shown in green, charged residues in purple. The boxes show the positions of the conserved charged residues mutated in this study. The arrowhead marks the proline within the PVG motif. The secondary structure prediction obtained using the HNNC prediction method (55) is shown bellow (C, random coil; H, α helix; E, extended strand). B, C: Arginine residues that are essential for LD targeting were revealed by transfecting COS-7 cells with plasmids encoding point-mutated full-length AUP1, either N-terminally (B) or C-terminally (C) HA-tagged. Twenty-four hours after transfection, cells were fixed and processed for immunofluorescence microscopy using anti-HA antibody to detect the transfected AUP1 (anti-HA, red) and LD540 to detect neutral lipids (LD, green) and visualized by fluorescence microscopy. In each panel, the full merged image is shown on the left (merge) and the magnification with the two channels separated and their merge on the right (inset). Scale bars, 10 μm.
Identification of the topogenic domain
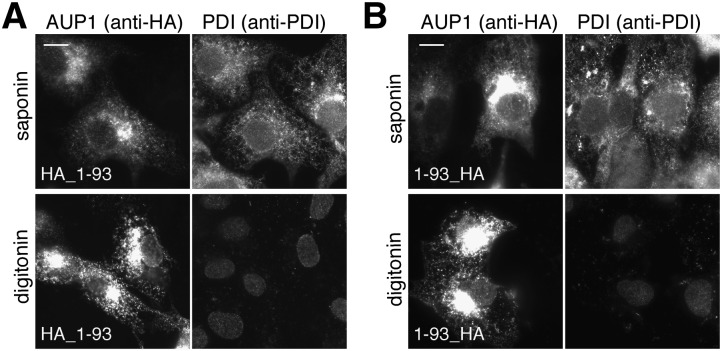
Because the N-terminal 93 amino acids of AUP1 were sufficient for both ER and LD localization of AUP1, we hypothesized that this domain enables the monotopic insertion of AUP1 into the membranes. To test this hypothesis, we expressed N- or C-terminally HA-tagged AUP1 1-93 and subsequently examined its topology using a microscopy-based assay. In this assay, cells were fixed and permeabilized either with saponin (permeabilizes all membranes) or digitonin (selectively permeabilizes plasma membrane, leaving the ER membrane intact), and cells were further processed for immunofluorescence microscopy using anti-HA antibody to detect AUP1 and anti-PDI antibody to detect the soluble luminal ER protein (for monitoring selective permeabilization). Detection of an epitope upon both saponin and digitonin permeabilization conditions indicates its cytoplasmic orientation. In contrast, detection of an epitope only in saponin-permeabilized cells indicates its luminal orientation. Both the N and C termini of the N-terminal domain were detectable upon selective permeabilization with digitonin (Fig. 4A, B), indicating their cytoplasmic orientation. Thus, the N-terminal domain of AUP1 is sufficient for establishing its monotopic topology.
Fig. 4.
Identification of the topogenic domain of AUP1. COS-7 cells were transfected with plasmids encoding either N-terminally (A) or C-terminally (B) HA-tagged AUP1 constructs encoding the first 93 amino acids of AUP1. Cells were fixed and permeabilized with either 0.1% saponin (permeabilizes all membranes) or 0.004% digitonin (selectively permeabilizes plasma membrane) and subsequently processed for immunofluorescence microscopy using anti-HA antibody to stain the transfected AUP1 (anti-HA) and anti-PDI antibody to stain the ER luminal marker PDI (anti-PDI). Scale bars, 10 μm.
Topogenic amino acids
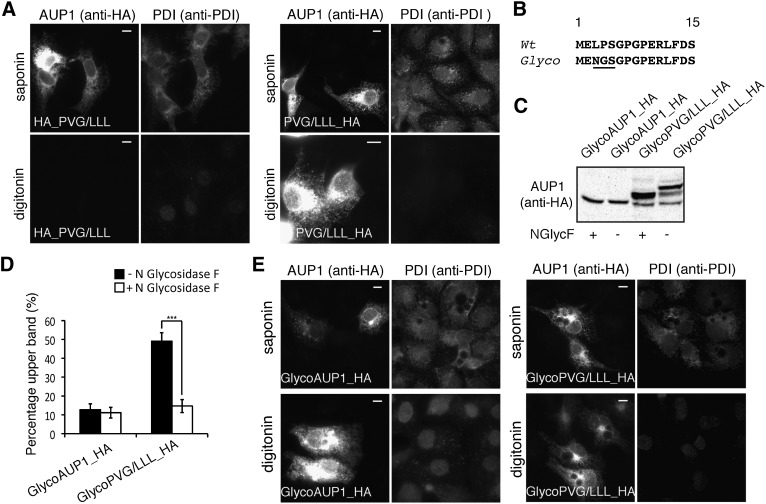
Alignment of AUP1 sequences from several species suggests a continuous helical hydrophobic domain disrupted by a region containing a conserved proline valine glycine (PVG) motif (Fig. 3A, arrowhead). When embedded in a continuous hydrophobic stretch, helix-breaking amino acids, such as proline and glycine, are known to determine protein topology (44, 45). To study whether this PVG motif is a topogenic determinant, we generated a construct substituting three leucines (PVG/LLL) for the PVG motif of full-length AUP1. In the microscopy-based topology assay, the N-terminally HA-tagged construct could be detected only in cells permeabilized with saponin, not in digitonin-permeabilized cells (Fig. 5A, left panel), indicating the luminal orientation of the N-terminus. The C-terminally HA-tagged construct was detectable irrespective of the permeabilization mode (Fig. 5A, right panel), indicating the cytoplasmic orientation of the C terminus.
Fig. 5.
The PVG motif is essential for monotopic insertion. A: COS-7 cells were transfected with plasmids encoding either N-terminally (left panel) or C-terminally (right panel) HA-tagged PVG/LLL mutant of full-length AUP1. Twenty-four hours after transfection, cells were fixed and permeabilized with either 0.1% saponin (permeabilizes all membranes) or 0.004% digitonin (selectively permeabilizes plasma membrane). Cells were subsequently processed for immunofluorescence microscopy using anti-HA antibody to stain the transfected AUP1 (anti-HA) and anti-PDI antibody to stain the ER luminal marker PDI (anti-PDI). Note the lack of staining in the bottom left image, which indicates luminal orientation of the N-terminus. Scale bars, 10 μm. B–E: Topology of wild-type and PVG/LLL constructs of full-length AUP1 bearing an N-terminal glycosylation site and a C-terminal HA tag (GlycoAUP1_HA and GlycoPVG/LLL_HA, respectively). B: Schematic representation of the first 15 amino acids of wild-type AUP1 (Wt) and the first 15 amino acids of glycosylation tag-bearing mutants (Glyco). The consensus glycosylation site is underlined. C, D: Biochemical analysis of AUP1 topology using the N-terminal glycosylation tag. COS-7 cells were transfected with constructs encoding indicated proteins. After 24 h, cellular proteins were incubated either with (+) or without (−) N-glycosidase F (NGlycF), separated by SDS-PAGE, and immunoblotted using anti-HA antibody to detect the transfected AUP1 (C). Note that GlycoPVG/LLL_HA is glycosylated, indicating luminal orientation of its N-terminus. In contrast, GlycoAUP1_HA is not glycosylated, suggesting its cytoplasmic orientation. The quantification of deglycosylation reaction is shown in panel D (Mean ± s.e.m., n = 4 or more independent experiments, t-test,*** designates P = 0.0001). E: Orientation of the C terminus of the glycosylation tag bearing constructs. COS-7 cells were transfected with plasmid encoding C-terminally HA-tagged either full-length GlycoAUP1 (left panel) or full-length GlycoPVG/LLL (right panel). Twenty-four hours after transfection, cells were fixed and processed for immunofluorescence microscopy as described in A. Note the presence of staining in bottom left image of both panels, which indicates the cytoplasmic orientation of the C terminus. Scale bars, 10 μm.
To confirm this result, we used a glycosylation assay. In this assay, an N-glycosylation site within a protein is used as topological marker, because it can only be glycosylated if it is translocated into the lumen of the ER. Because AUP1 does not have a consensus glycosylation site, we introduced an engineered glycosylation site close to the N-terminus of wild-type full-length AUP1 and full-length PVG/LLL (Fig. 5B), generating GlycoAUP1_HA and GlycoPVG/LLL_HA, respectively. To examine potential glycosylation, the constructs were expressed in COS-7 cells and analyzed by immunoblotting that detected the HA tag. GlycoAUP1_HA migrated as a single unglycosylated band (Fig. 5C, left lanes). In contrast, the GlycoPVG/LLL_HA displayed two additional bands with higher apparent molecular mass, representing N-glycosidase F-resistant and -sensitive glycosylated forms (Fig. 5C, right lanes). These results show that the glycosylation tag and the N-terminus of GlycoPVG/LLL_HA face the ER lumen. To determine the orientation of the C terminus, we examined the above-described glyco mutants in the microscopy-based assay. As expected for both proteins (GlycoAUP1_HA and GlycoPVG/LLL_HA), the C-terminal HA tag was detectable irrespective of the permeabilization mode (Fig. 5E), indicating its cytoplasmic orientation. These results suggested that AUP1 containing the PVG/LLL mutation adopts a bitopic topology with luminal orientation of the N-terminus.
The PVG motif is required for droplet localization
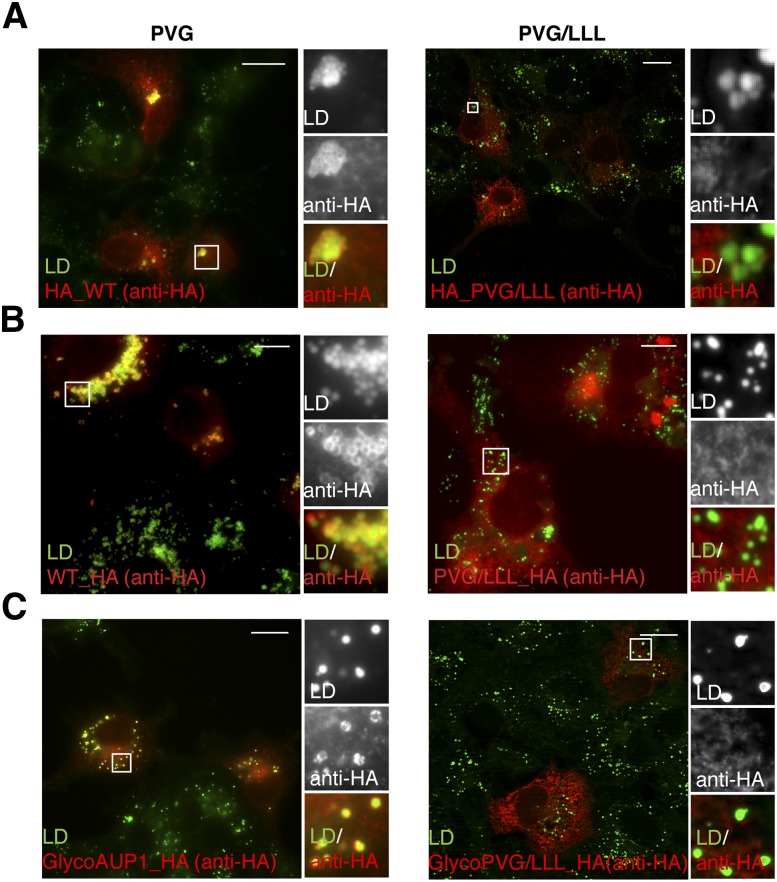
To examine whether the PVG motif also affects the distribution of AUP1 between the ER and LDs, we used fluorescence microscopy and examined the localization of tagged AUP1 PVG/LLL mutants in COS-7 cells. In contrast to full-length AUP1 with intact PVG motif (Fig. 6, left panels), the corresponding full-length PVG/LLL mutants were virtually absent from LDs (Fig. 6, right panels) but present on the ER (colocalization data not shown, but were reviewed). These results were confirmed by subcellular fractionation analysis (see supplementary Fig. II, lanes 1–6, 16–23, 32–37).
Fig. 6.
The PVG motif is required for lipid droplet localization. COS-7 cells were transfected with plasmids encoding point-mutated full-length AUP1, either N-terminally (A) or C-terminally (B, C) HA-tagged. Twenty-four hours after transfection, cells were fixed and processed for immunofluorescence microscopy using anti-HA antibody to detect the transfected AUP1 (anti-HA, red) and LD540 to detect neutral lipids (LD, green) and visualized by fluorescence microscopy. In each panel, the full merged image is shown on the left and the magnification with the two channels separated and their merge on the right. Scale bars, 20 μm (A, C) or 10 μm (B).
We further examined whether the point mutants of the charged residues (R42I, R62F/R63F) that disrupt LD localization also influence the topology of AUP1. In both the microscopy-based and glycosylation assay, the mutants retained their monotopic topology and localization to the ER (data not shown, but were reviewed). We conclude that the PVG motif is required for monotopic topology and for LD localization, whereas the specific arginine residues are required for LD localization but are dispensable for a monotopic topology.
Topogenic determinant of AUP1 prevails over the signal peptide
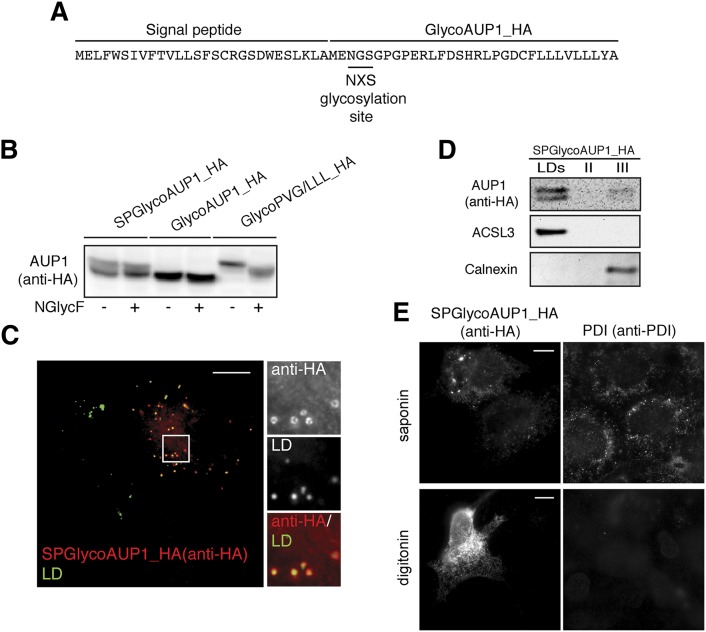
To examine the hierarchical relationship of the topogenic determinants of AUP1 and a classical topogenic determinant, the membrane translocating signal peptide, we generated a full-length AUP1 variant fused to the signal peptide of rabbit lactase-phlorizin hydrolase (Fig. 7A). This signal peptide had previously been used to translocate GFP into the lumen of the ER (46). Upon Western blotting, the protein displayed two bands, which were insensitive to glycosidase treatment (Fig. 7B) but likely to represent the intact protein and the form after cleavage of the signal peptide. Apparently, the signal peptide initially allows partial translocation and subsequent signal peptide cleavage, but the N-terminus is then retranslocated, as indicated by the lack of glycosylation. By fluorescence microscopy the protein localized to LDs (Fig. 7C). Subcellular fractionation showed that actually both forms of the protein, with and without the signal peptide, localized to LDs (Fig. 7D). Also, the cytoplasmic localization of the C terminus was unaffected by addition of the signal peptide (Fig. 7E). We conclude that the hydrophobic stretch of AUP1 is a strong topogenic determinant that prevents permanent translocation of the N-terminus by a fused signal peptide.
Fig. 7.
The topogenic determinant of full-length AUP1 can dominate a signal peptide. A: A plasmid was prepared that encodes full-length GlycoAUP1_HA with the N-terminally attached cleavable signal sequence of rabbit lactase-phlorizin hydrolase (SPGlycoAUP1_HA). B: Biochemical analysis of topology using N-terminal glycosylation tag. On day 0, COS-7 cells were transfected with constructs encoding indicated proteins (SPGlycoAUP1_HA, GlycoAUP1_HA, or GlycoPVG/LLL_HA). On day 1, total cellular proteins were precipitated, resuspended in a reaction buffer, and incubated either with (+) or without (−) N-glycosidase F (NGlycF). Proteins were separated by SDS-PAGE and immunoblotted using anti-HA antibody to detect the transfected AUP1. SPGlycoAUP1_HA was not N-glycosylated, in contrast to the positive control (GlycoPVG/LLL_HA). This result indicates cytoplasmic orientation of SPGlycoAUP1_HA N-terminus. C: COS-7 cells expressing SPGlycoAUP1_HA were fixed and processed for immunofluorescence microscopy using anti-HA antibody to detect the transfected AUP1 (anti-HA, red) and LD540 to detect neutral lipids (LD, green) and visualized by fluorescence microscopy. Fluorescence microscopy image is shown. Scale bar, 10 μm. D: COS-7 cells expressing SPGlycoAUP1_HA were grown in medium supplemented with 150 μM oleic acid to promote LD formation. LDs were isolated by floatation in a sucrose gradient. Proteins of the floating LD fraction (LDs) and the two lower fractions (II, III) were analyzed by SDS-PAGE/immunoblotting using antibodies against the transfected AUP1 (anti-HA), LD marker (ACSL3), and ER marker (calnexin). E: Orientation of the C terminus of the SPGlycoAUP1_HA. COS-7 cells were transfected with plasmid encoding C-terminally HA-tagged SPGlycoAUP1_HA. Twenty-four hours after transfection, cells were fixed and permeabilized with either 0.1% saponin (permeabilizes all membranes) or 0.004% digitonin (selectively permeabilizes plasma membrane). Cells were subsequently processed for immunofluorescence microscopy using anti-HA antibody to stain the transfected AUP1 (anti-HA) and anti-PDI antibody to stain the ER luminal marker PDI (anti-PDI). Fluorescence microscopy images are shown. The staining in the bottom left image indicates cytoplasmic orientation of the C terminus of SPGlycoAUP1_HA. Scale bars, 10 μm.
DISCUSSION
Intracellular targeting of AUP1 is mediated by a single domain at the N-terminal region of the protein. In contrast, the acyltransferase, CUE, and G2BR domains are dispensable for AUP1 targeting. This indicates that targeting signals of AUP1 are overlapping, i.e., sorting to multiple locations using a single domain.
Combining our results with analysis of hydrophobicity (Fig. 3A, green residues), the following model emerges. The targeting domain of AUP1 contains an internal continuous hydrophobic stretch that probably integrates into the membrane (Fig. 8A), and thus we refer to it as a membrane domain. This membrane domain presumably binds to the signal recognition particle, resulting in insertion of AUP1 into the ER membrane. The leucine-rich stretch at the start of the AUP1 membrane domain that resembles a signal recognition particle binding site (21, 47) would drive this process. Also, insertion into microsomes in a signal recognition particle-dependent manner was reported for the structurally similar hairpin-monotopic protein caveolin 1 (48) and the plant LD protein oleosin (49). This initial targeting to the ER resembles that of ALDI/AAM-B, an LD protein with an N-terminal hydrophobic domain (3, 50).
Fig. 8.
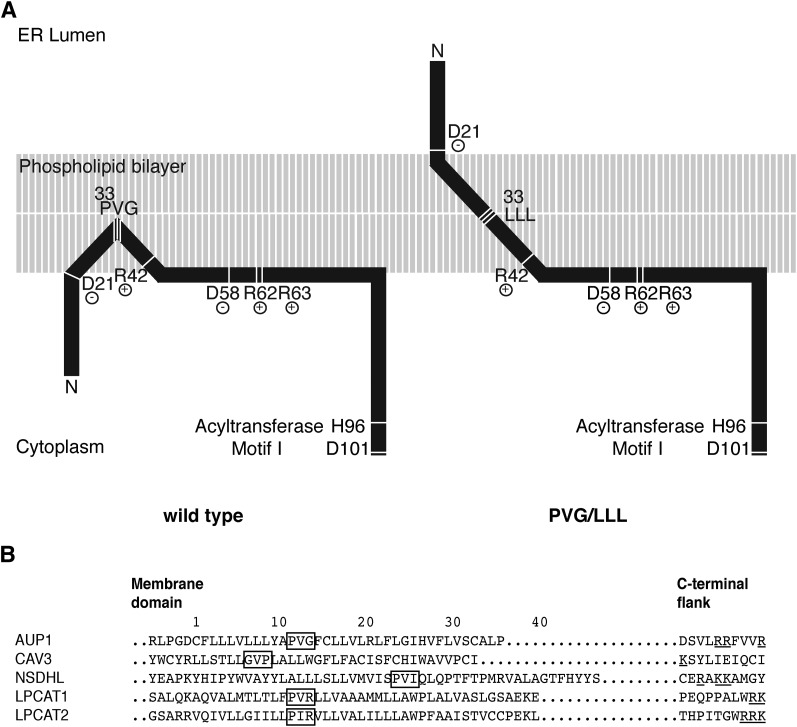
Model of the AUP1 targeting domain. A: The N-terminal region of AUP1 is represented schematically. The phospholipid bilayer is in gray, protein is in black. The targeting domain of AUP1 contains a continuous hydrophobic stretch surrounded by two hydrophilic flanks. The hydrophobic stretch of AUP1 is probably integrated into the membrane, and contains the PVG motif and the accessory motifs. The PVG motif is required for establishing monotopic/hairpin topology and LD localization. The accessory motifs (R42 and R62+R63) are required for LD targeting. B: Sequence comparison between AUP1 and the LD proteins CAV3, NSDHL, LPCAT1, and LPCAT2 (21, 32). Position 1 indicates the first residue of the hydrophobic/membrane domain. To compensate for the different length of the hydrophobic domains, we aligned the C-terminal flanks. PVG motif and similar motifs are highlighted with a black frame. Positively charged residues (R,K) in the C-terminal flank are underlined. Sequences are taken from Swissprot with the following accession numbers: AUP1 Q9Y679, CAV3 P56539, NSDHL Q15738, LPCAT1 Q8NF37, LPCAT2 Q7L5N7.
Important features of the AUP1-targeting domain are the PVG motif and the accessory charge motifs (Fig. 8A, left). When the PVG motif is intact, AUP1 adopts a monotopic/hairpin topology. In contrast, when the PVG motif is mutated, AUP1 becomes a transmembrane protein (Fig. 8A, right). The PVG motif is located in the center of the hydrophobic stretch and contains two amino acids (proline and glycine) with a helix-breaking propensity. These residues would force the hydrophobic domain to adopt a loop shape, resulting in a monotopic/hairpin topology. The stability of this hairpin conformation is illustrated by the fact that a fused signal peptide preceding the AUP1 sequence is unable to permanently invert the topology of AUP1. The PVG motif is also a prerequisite for targeting of AUP1 to LDs. With the PVG motif present, AUP1 localizes to LDs, whereas lack of the PVG motif results in the absence of AUP1 from LDs, indicating the importance of the hydrophobic stretch conformation for sorting AUP1 to LDs. Although theories have been proposed that could explain incorporation of transmembrane proteins into LDs (51, 52), the bitopic AUP1 mutant is virtually absent from LDs, supporting the conclusion that monotopic topology is required for LD targeting. Monotopic topology is, however, not sufficient to drive a protein to LDs, as illustrated by AUP1 arginine R42 and R62/63 point mutants.
These arginines form an accessory motif that supports targeting of AUP1 to LDs (Fig. 8A). When they are mutated, AUP1 is virtually absent from LDs, but retains the ER localization and the monotopic topology. In contrast, mutation of a conserved negatively charged residue within the targeting domain (aspartic acid at position 58) did not alter LD localization. This suggests that the positively charged residues specifically support the LD association of AUP1, e.g., by electrostatic interactions. Due to its proximity to the membrane, the arginine motif may bind to negatively charged phospholipids or other components that contribute to the negative potential of the LD surface (53). In this case, posttranslational modifications influencing the protein charge, such as phosphorylation and acetylation, could regulate AUP1 targeting to LDs. An analogous observation has been made for the MARCKS protein (54), whose translocation to the membrane depends on N-terminal myristoylation and neighboring positively charged amino acids. Phosphorylation of amino acids adjacent to these positively charged amino acids reduces membrane targeting of MARCKS (54).
The membrane domains of several other LD proteins with monotopic topology display motifs similar to the PVG motif of AUP1 (Fig. 8B), suggesting that introduction of turns into long hydrophobic stretches is a more general requirement in lipid droplet protein targeting. Also, the C-terminal flanks of NSDHL, LPCAT1, and LPCAT2 contain positively charged amino acids similar to the accessory arginines of AUP1 (Fig. 8B), further suggesting the existence of a common targeting strategy.
In conclusion, here we show that a single domain of AUP1 enables its ER residence, monotopic membrane insertion, and LD localization. This overlap of targeting signals is probably a consequence of the distinct structure of the LD that lacks a luminal aqueous phase and thereby imposes topological restrictions on protein targeting to LDs. This mechanism, the selective targeting of monotopic proteins to LDs using the overlapping targeting signals, may be a common mechanism for localization of several other integral LD proteins.
Supplementary Material
Acknowledgments
The authors thank Dr. Lars Kuerschner, Dr. Johanna Spandl, Dr. Anke Penno, and Daniel Lohmann for proofreading the manuscript. They would like to thank Prof. Kai Simons and Dr. Daniel Lingwood for kindly providing the GFP-GPI construct.
Footnotes
Abbreviations:
- ACSL-3
- long-chain acyl-CoA synthetase 3
- AUP1
- ancient ubiquitous protein 1
- BB
- blocking buffer
- CUE
- coupling of ubiquitin to ER degradation
- ER
- endoplasmic reticulum
- FCS
- fetal calf serum
- G2BR
- G2 binding region
- HA
- hemagglutinin
- LD
- lipid droplet
- PDI
- protein-disulfide isomerase
The authors gratefully acknowledge the financial support of Deutsche Forschungsgemeinschaft Collaborative Research Center SFB 645 (C.T., A.S.) and the EU Framework 7 project “LipidomicNet” (C.T.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one movie, three tables, and two figures.
REFERENCES
- 1.Matlin K. S. 2011. Spatial expression of the genome: the signal hypothesis at forty. Nat. Rev. Mol. Cell Biol. 12: 333–340 [DOI] [PubMed] [Google Scholar]
- 2.Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277: 44507–44512 [DOI] [PubMed] [Google Scholar]
- 3.Zehmer J. K., Bartz R., Bisel B., Liu P., Seemann J., Anderson R. G. 2009. Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J. Cell Sci. 122: 3694–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole N. B., Murphy D. D., Grider T., Rueter S., Brasaemle D., Nussbaum R. L. 2002. Lipid droplet binding and oligomerization properties of the Parkinson's disease protein α-synuclein. J. Biol. Chem. 277: 6344–6352 [DOI] [PubMed] [Google Scholar]
- 5.McManaman J. L., Zabaronick W., Schaack J., Orlicky D. J. 2003. Lipid droplet targeting domains of adipophilin. J. Lipid Res. 44: 668–673 [DOI] [PubMed] [Google Scholar]
- 6.Nakamura N., Fujimoto T. 2003. Adipose differentiation-related protein has two independent domains for targeting to lipid droplets. Biochem. Biophys. Res. Commun. 306: 333–338 [DOI] [PubMed] [Google Scholar]
- 7.Targett-Adams P., Chambers D., Gledhill S., Hope R. G., Coy J. F., Girod A., McLauchlan J. 2003. Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J. Biol. Chem. 278: 15998–16007 [DOI] [PubMed] [Google Scholar]
- 8.Ohsaki Y., Maeda T., Maeda M., Tauchi-Sato K., Fujimoto T. 2006. Recruitment of TIP47 to lipid droplets is controlled by the putative hydrophobic cleft. Biochem. Biophys. Res. Commun. 347: 279–287 [DOI] [PubMed] [Google Scholar]
- 9.Bulankina A., Deggerich A., Wenzel D., Mutenda K., Wittmann J. G., Rudolph M. G., Burger K. N., Hoening S. 2009. TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 185: 641–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruber A., Cornaciu I., Lass A., Schweiger M., Poeschl M., Eder C., Kumari M., Schoiswohl G., Wolinski H., Kohlwein S. D., et al. 2010. The N-terminal region of Comparative Gene Identification-58 (CGI-58) is important for lipid droplet binding and activation of adipose triglyceride lipase. J. Biol. Chem. 285: 12289–12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Zhang J., Qiu W., Han G. S., Carman G. M., Adeli K. 2011. Lipin-1γ isoform is a novel lipid droplet-associated protein highly expressed in the brain. FEBS Lett. 585: 1979–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H. W., Schmidt-Supprian M., Vance D. E., Mann M., et al. 2011. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14: 504–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia A., Sekowski A., Subramanian V., Brasaemle D. L. 2003. The central domain is required to target and anchor perilipin A to lipid droplets. J. Biol. Chem. 278: 625–635 [DOI] [PubMed] [Google Scholar]
- 14.Subramanian V., Garcia A., Sekowski A., Brasaemle D. L. 2004. Hydrophobic sequences target and anchor perilipin A to lipid droplets. J. Lipid Res. 45: 1983–1991 [DOI] [PubMed] [Google Scholar]
- 15.Muellner H., Zweytick D., Leber R., Turnowsky F., Daum G. 2004. Targeting of proteins involved in sterol biosynthesis to lipid particles of the yeast Sacharomyces cerevisiae. Biochim. Biophys. Acta. 1663: 9–13 [DOI] [PubMed] [Google Scholar]
- 16.Zehmer J. K., Bartz R., Liu P., Anderson R. G. 2008. Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets. J. Cell Sci. 121: 1852–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horiguchi Y., Araki M., Motojima K. 2008. Identification and characterization of the ER/lipid droplet-targeting sequence in 17β-hydroxysteroid dehydrogenase type 11. Arch. Biochem. Biophys. 479: 121–130 [DOI] [PubMed] [Google Scholar]
- 18.Schweiger M., Schoiswohl G., Lass A., Radner F. P., Haemmerle G., Malli R., Graier W., Cornaciu I., Oberer M., Salvayre R., et al. 2008. The C-terminal region of human adipose triglyceride lipase affects enzyme activity and lipid droplet binding. J. Biol. Chem. 283: 17211–17220 [DOI] [PubMed] [Google Scholar]
- 19.Lu X., Yang X., Liu J. 2010. Differential control of ATGL-mediated lipid droplet degradation by CGI-58 and G0S2. Cell Cycle. 9: 2719–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingelmo-Torres M., Gonzalez-Moreno E., Kassan A., Hanzal-Bayer M., Tebar F., Herms A., Grewal T., Hancock J. F., Enrich C., Bosch M., et al. 2009. Hydrophobic and basic domains target proteins to lipid droplets. Traffic. 10: 1785–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiele C., Spandl J. 2008. Cell biology of lipid droplets. Curr. Opin. Cell Biol. 20: 378–385 [DOI] [PubMed] [Google Scholar]
- 22.Fujimoto T., Kogo H., Ishiguro K., Tauchi K., Nomura R. 2001. Caveolin-2 is targeted to lipid droplets, a new “membrane domain” in the cell. J. Cell Biol. 152: 1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostermeyer A. G., Paci J. M., Zeng Y., Lublin D. M., Munro S., Brown D. A. 2001. Accumulation of caveolin in the endoplasmic reticulum redirects the protein to lipid storage droplets. J. Cell Biol. 152: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pol A., Luetterforst R., Lindsay M., Heino S., Ikonen E., Parton R. G. 2001. A caveolin dominant negative mutant associates with lipid bodies and induces intracellular cholesterol imbalance. J. Cell Biol. 152: 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umlauf E., Csaszar E., Moertelmaier M., Schuetz G. J., Parton R. G., Prohaska R. 2004. Association of stomatin with lipid bodies. J. Biol. Chem. 279: 23699–23709 [DOI] [PubMed] [Google Scholar]
- 26.Le Lay S., Hajduch E., Lindsay M. R., Le Liepvre X., Thiele C., Ferre P., Parton R. G., Kurzchalia T., Simons K., Dugail I. 2006. Cholesterol-induced caveolin targeting to lipid droplets in adipocytes: a role for caveolar endocytosis. Traffic. 7: 549–561 [DOI] [PubMed] [Google Scholar]
- 27.Rajendran L., Le Lay S., Illges H. 2007. Raft association and lipid droplet targeting of flotillins are independent of caveolin. Biol. Chem. 388: 307–314 [DOI] [PubMed] [Google Scholar]
- 28.Leber R., Landl K., Zinser E., Ahorn H., Spoek A., Kohlwein S. D., Turnowsky F., Daum G. 1998. Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol. Biol. Cell. 9: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohashi M., Mizushima N., Kabeya Y., Yoshimori T. 2003. Localization of mammalian NAD(P)H steroid dehydrogenase-like protein on lipid droplets. J. Biol. Chem. 278: 36819–36829 [DOI] [PubMed] [Google Scholar]
- 30.Caldas H., Herman G. E. 2003. NSDHL, an enzyme involved in cholesterol biosynthesis, traffics through the Golgi and accumulates on ER membranes and on the surface of lipid droplets. Hum. Mol. Genet. 12: 2981–2991 [DOI] [PubMed] [Google Scholar]
- 31.Kuerschner L., Moessinger C., Thiele C. 2008. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 9: 338–352 [DOI] [PubMed] [Google Scholar]
- 32.Moessinger C., Kuerschner L., Spandl J., Shevchenko A., Thiele C. 2011. Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J. Biol. Chem. 286: 21330–21339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McFie P. J., Banman S. L., Kary S., Stone S. J. 2011. Murine diacylglycerol acyltransferase-2 (DGAT2) can catalyze triacylglycerol synthesis and promote lipid droplet formation independent of its localization to the endoplasmic reticulum. J. Biol. Chem. 286: 28235–28246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poppelreuther M., Rudolph B., Du C., Grossmann R., Becker M., Thiele C., Ehehalt R., Fuellekrug J. 2012. The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J. Lipid Res. 53: 888–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller B., Klemm E. J., Spooner E., Claessen J. H., Ploegh H. L. 2008. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. USA. 105: 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spandl J., Lohmann D., Kuerschner L., Moessinger C., Thiele C. 2011. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J. Biol. Chem. 286: 5599–5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klemm E. J., Spooner E., Ploegh H. L. 2011. Dual role of ancient ubiquitous protein 1 (AUP1) in lipid droplet accumulation and endoplasmic reticulum (ER) protein quality control. J. Biol. Chem. 286: 37602–37614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kato A., Kawamata N., Tamayose K., Egashira M., Miura R., Fujimura T., Murayama K., Oshimi K. 2002. Ancient ubiquitous protein 1 binds to the conserved membrane-proximal sequence of the cytoplasmic tail of the integrin alpha subunits that plays a crucial role in the inside-out signaling of alpha IIbbeta 3. J. Biol. Chem. 277: 28934–28941 [DOI] [PubMed] [Google Scholar]
- 39.Kato A., Oshimi K. 2009. Ancient ubiquitous protein 1 and Syk link cytoplasmic tails of the integrin alpha(IIb)beta(3). Platelets. 20: 105–110 [DOI] [PubMed] [Google Scholar]
- 40.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 77: 51–59 [DOI] [PubMed] [Google Scholar]
- 41.Ahmed O. A., Adjimatera N., Pourzand C., Blagbrough I. S. 2005. N4,N9-dioleoyl spermine is a novel nonviral lipopolyamine vector for plasmid DNA formulation. Pharm. Res. 22: 972–980 [DOI] [PubMed] [Google Scholar]
- 42.Spandl J., White D. J., Peychl J., Thiele C. 2009. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic. 10: 1579–1584 [DOI] [PubMed] [Google Scholar]
- 43.Blanchette-Mackie E. J., Dwyer N. K., Barber T., Coxey R. A., Takeda T., Rondinone C. M., Theodorakis J. L., Greenberg A. S., Londos C. 1995. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J. Lipid Res. 36: 1211–1226 [PubMed] [Google Scholar]
- 44.Nilsson I., von Heijne G. 1998. Breaking the camel's back: proline-induced turns in a model transmembrane helix. J. Mol. Biol. 284: 1185–1189 [DOI] [PubMed] [Google Scholar]
- 45.Monne M., Hermansson M., von Heijne G. 1999. A turn propensity scale for transmembrane helices. J. Mol. Biol. 288: 141–145 [DOI] [PubMed] [Google Scholar]
- 46.Keller P., Toomre D., Diaz E., White J., Simons K. 2001. Multicolour imaging of post-Golgi sorting and trafficking in live cells. Nat. Cell Biol. 3: 140–149 [DOI] [PubMed] [Google Scholar]
- 47.von Heijne G. 1990. The signal peptide. J. Membr. Biol. 115: 195–201 [DOI] [PubMed] [Google Scholar]
- 48.Monier S., Parton R. G., Vogel F., Behlke J., Henske A., Kurzchalia T. V. 1995. VIP21-Caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol. Biol. Cell. 6: 911–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beaudoin F., Wilkinson B. M., Stirling C. J., Napier J. A. 2000. In vivo targeting of sunflower oil body protein in yeast secretory (sec) mutants. Plant J. 23: 159–170 [DOI] [PubMed] [Google Scholar]
- 50.Turro S., Ingelmo-Torres M., Estanyol J. M., Tebar F., Fernandez M. A., Albor C. V., Gaus K., Grewal T., Enrich C., Pol A. 2006. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 7: 1254–1269 [DOI] [PubMed] [Google Scholar]
- 51.Ploegh H. L. 2007. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 448: 435–438 [DOI] [PubMed] [Google Scholar]
- 52.Walther T. C., Farese R. V., Jr 2009. The life of lipid droplets. Biochim. Biophys. Acta. 1791: 459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carvalho F. A., Carneiro F. A., Martins I. C., Assuncao-Miranda I., Faustino A. F., Pereira R. M., Bozza P. T., Castanho M. A., Mohana-Borges R., Da Poian A. T., et al. 2012. Dengue virus capsid protein binding to hepatic lipid droplets (LD) is potassium ion dependent and is mediated by LD surface proteins. J. Virol. 86: 2096–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLaughlin S., Aderem A. 1995. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem. Sci. 20: 272–276 [DOI] [PubMed] [Google Scholar]
- 55.Combet C., Blanchet C., Geourjon C., Deleage G. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25: 147–150 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.