Abstract
Bacterial populations have a remarkable capacity to cope with extreme environmental fluctuations in their natural environments. In certain cases, adaptation to one stressful environment provides a fitness advantage when cells are exposed to a second stressor, a phenomenon that has been coined as cross-stress protection. A tantalizing question in bacterial physiology is how the cross-stress behavior emerges during evolutionary adaptation and what the genetic basis of acquired stress resistance is. To address these questions, we evolved Escherichia coli cells over 500 generations in five environments that include four abiotic stressors. Through growth profiling and competition assays, we identified several cases of positive and negative cross-stress behavior that span all strain–stress combinations. Resequencing the genomes of the evolved strains resulted in the identification of several mutations and gene amplifications, whose fitness effect was further assessed by mutation reversal and competition assays. Transcriptional profiling of all strains under a specific stress, NaCl-induced osmotic stress, and integration with resequencing data further elucidated the regulatory responses and genes that are involved in this phenomenon. Our results suggest that cross-stress dependencies are ubiquitous, highly interconnected, and can emerge within short timeframes. The high adaptive potential that we observed argues that bacterial populations occupy a genotypic space that enables a high phenotypic plasticity during adaptation in fluctuating environments.
Keywords: cross-stress protection, evolutionary trade-offs, microbial evolution, stress adaptation
Introduction
Outside the laboratory, microbes rarely live in conditions of optimum growth as environmental parameters constantly fluctuate. ‘Stress’ is the term used when these fluctuations vary severely from the optimum, and the ability of microbes to sense and respond to environmental stress is critical to their survival. Due to their importance to medical, agricultural and industrial fields, the underlying response mechanisms have been studied extensively and as a result we now understand their key components (Storz and Hengge-Aronis, 2000).
Previous work on the adaptive potential of the bacterium Escherichia coli to fluctuations in temperature (Riehle et al, 2003; Bennett and Lenski, 2007; Sleight et al, 2008), UV irradiation (Alcántara-Díaz et al, 2004), high ethanol (Goodarzi et al, 2010), isobutanol (Atsumi et al, 2008), and various carbon sources (Silver and Mateles, 1969; Dekel and Alon, 2005; Blount et al, 2008) has provided initial evidence regarding the genetic basis of microbial long-term adaptation to abiotic environmental factors. However, little is known about how stress selection affects the capacity of an organism to be protected under other stresses. ‘Cross-stress’ protection, that is, acquired stress resistance to a specific stress after pretreatment with another stressor, has been documented for various stress combinations and it has been observed in many species across the tree of life, ranging from microbes (Tesone et al, 1981; Jenkins et al, 1988, 1990; Leyer and Johnson, 1993; Mattimore and Battista, 1996; Lou and Yousef, 1997; Fletcher and Csonka, 1998; Garren et al, 1998; Ryu and Beuchat, 1998; Cánovas et al, 2001; Begley et al, 2002; Sghaier et al, 2007; Berry and Gasch, 2008; Machielsen et al, 2010), to plants (Schenk et al, 2000; Durrant and Dong, 2004) and humans (Raffaghello et al, 2008; Zhao, 2009). For example in E. coli, pre-adaptation to glucose or nitrogen limitation increases survival rates after a heat shock or oxidative stress (Jenkins et al, 1988), and a possible link was suggested to be RpoH, a heat shock regulator, which was found to have an important role in protein synthesis under carbon starvation (Jenkins et al, 1991). Similarly, increased survival was observed after osmotic shock, when cells were pre-treated with a mild increase in osmolarity (Jenkins et al, 1990). Transcriptional profiling in E. coli revealed a high degree of overlap between different stresses (Supplementary Results; Supplementary Figure S1), such as starvation, osmotic and acidic stress (∼140 genes) (Weber et al, 2005), and it was later found that high osmolarity or high temperature induces the oxidative stress regulons (SoxRS and OxyR) that can at least partially explain the cross-stress protection between these stresses (Gunasekera et al, 2008; White-Ziegler et al, 2008). Responses to n-butanol were recently found to share the same high overlap with those in heat shock, oxidative and acidic stress (Rutherford et al, 2010). Interestingly, recent work with gene-deletion libraries in yeast shows that acquired hydrogen peroxide (H2O2) tolerance after three distinct pretreatments occurs by different mechanisms depending on prior cellular experiences (Berry et al, 2011).
Given the importance of cross-stress protection in bacterial physiology, we investigate the extent to which cross-stress protection emerges spontaneously during microbial evolution, the evolutionary trade-offs in stress selection, and the genetic basis of both acquired resistance and evolutionary cross-stress dependencies. Towards this goal, we performed laboratory evolution of E. coli cells over 500 generations in minimal media and four stresses: osmotic, acidic, oxidative, and n-butanol (Figure 1). Whole genome resequencing (Araya et al, 2010; Cooper and Lenski, 2010; Conrad et al, 2011) and transcriptional profiling (RNA-Seq) revealed several genetic and transcriptional differences between the control and evolved strains. To understand the genotypic–phenotypic dependencies and genetic basis of acquired stress resistance during laboratory selection, mutations were individually reversed and the fitness of the corresponding repaired mutants was assessed. Systematic analysis of growth curves and competition assays among the evolved strains reveals a wide spectrum of evolutionary cross-stress protection and evolutionary trade-off cases for various strain–environment combinations.
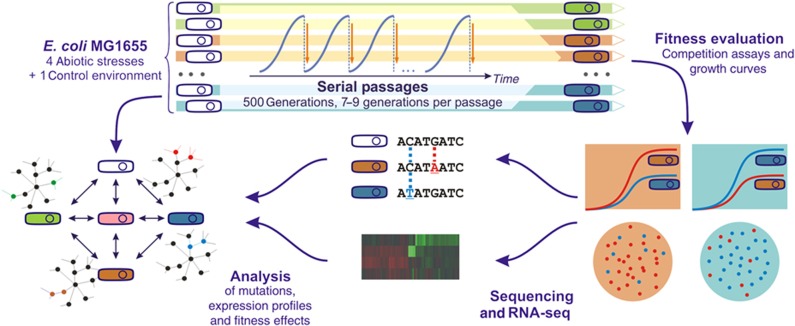
Figure 1.
Overview of the experimental setting. E. coli MG1655 cell lines were evolved for 500 generations in five environments with minimal M9 salt media and glucose as the sole carbon source. These environments included four abiotic stresses (acidic, osmotic, oxidative, and n-butanol stress) and a control medium-only environment. The relative fitness of all evolved strains was measured under all stresses by competition assays and growth curves. Selected clones from adapted populations were sequenced and transcriptional profiles were obtained by RNA-Seq. Individual mutations from the resequenced clones where associated to phenotypic fitness by mutation reversals and competition assays. Identified mutations, expression data and relative fitness of the evolved strains under various stressors were analyzed in a system-level approach.
Results
Evolutionary potential and cross-stress behavior
We adapted E. coli strains for 500 generations in five environments (osmotic, acidic, oxidative, n-butanol, and control; four biological replicates per environment) with minimal M9 salts media and glucose as the sole carbon source (Figure 1). Prior to evolutionary adaptation in these environments we tested the MG1655 and ΔlacZ strains that were used in order to enable direct competition assays of evolved populations, in all growth environments to ensure neutrality of the lacZ deletion marker in all conditions (Supplementary Results; Supplementary Table S-I; Supplementary Figures S2–S4). Growth curves and maximum growth rates of all strain–environment combinations were initially acquired to assess the growth differences in the respective populations (Supplementary Table S-II and S-III; Supplementary Figures S7–S9). Competition assays between adapted strains in all environments were then performed at a population level to obtain a relative fitness profile for each strain (four biological replicates per environment, competition over population averages, Supplementary Tables S-IV and S-V; Supplementary Figures S10–S15). We observed significant phenotypic heterogeneity in individual populations (Supplementary Figure S6), which is in agreement with what has been reported previously in short-term evolution experiments (Wang et al, 2010). Notably, there was only a small evolutionary trade-off between adaptation to the medium and to the respective stress, as all strains had similar fitness (Darwinian fitness w=0.96–1.04) in the control (non-stress) condition (Figure 2). In contrast, medium-only evolved cells (G500) had significantly lower fitness under other stressors (w=0.51–0.75 relative to the stress-adapted strains), which argues that adaptation to the medium and to the specific stress are independent processes.
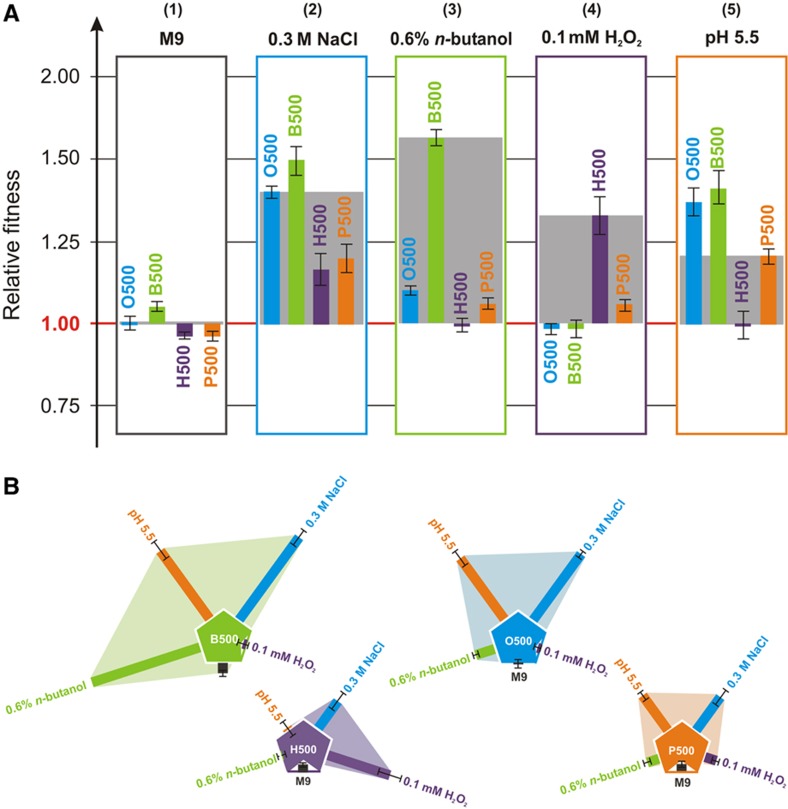
Figure 2.
Relative fitness of the evolved populations under all environmental conditions. The relative fitness of populations evolved in all five environments was assessed through competition assays. Fitness refers to Darwinian fitness as measured through competition assays relative to the medium-only adapted population (G500). (A) Environment-based representation: (1) In the absence of other stressors, all populations showed small differences in fitness; (2) all populations where significantly more protected in osmotic stress in comparison to the G500 population; (3–4) while the B500 and O500 populations have high fitness under n-butanol stress, they showed a small evolutionary trade-off under oxidative stress; (5) Interestingly, O500 and B500 populations outcompeted P500 under acidic stress. Shaded gray areas depict the fitness difference between the strain evolved in the respective environment relative to the G500 population. (B) Population-based representation of the relative fitness data shown in (A). Shaded areas depict the degree of cross-stress protection in each respective environment. Error bars show standard error of the mean for eight independent competitions; counts for each competition were averaged over two plates in each experiment. Competition assays were obtained over 48 h of growth with one transfer (1:500 dilution) at 24 h. Supplementary Table S-V summarizes the competition assay results. Source data for this figure is available on the online supplementary information page.
Evolutionary cross-stress protection was observed in various stress pairs. The n-butanol-adapted strains (B500) had the highest fitness under hyper-osmotic conditions, which was similar to the fitness of the osmotic-adapted strains O500 (w=1.51±0.05 and w=1.39±0.02, respectively). A similar profile was observed under n-butanol stress where the O500 strains had an increased fitness (w=1.10±0.02), although still lesser than the B500 strains (w=1.58±0.03). Remarkably, all B500 and O500 strains had higher fitness in the acidic environment, outcompeting even the acidic-evolved strain (P500), a surprising result that clearly argues that similar or higher fitness neighborhoods can be reached through adaptive evolution under different environmental contexts. This result is in agreement with previous reports that adaptation in pH-related stress has been known to confer fitness effects of low magnitude (Hughes et al, 2007), and lower viability in acidic stress was observed in ethanol-adapted (Goodarzi et al, 2010), but not isobutanol-adapted (Atsumi et al, 2010) cell lines. Evolutionary cross-stress protection was also observed in the case of cells evolved under oxidative stress (H500), which were found to have the same Darwinian fitness as acidic-evolved cells (P500) under osmotic stress. A negative cross-stress relationship of lesser magnitude was observed in the case of butanol-evolved (B500) and osmotic-evolved (O500) cells under oxidative stress. Interestingly, in competition assays between the B500-O500 strains in the oxidative environment, the O500 clearly showed increased fitness relative to the B500 strains (1.436±0.030, see Supplementary Text and Supplementary Table S-V), which further substantiates the negative evolutionary cross-stress dependency between selection in n-butanol and oxidative stress.
To investigate whether the phenotypic similarity of the B500 and O500 strains holds for higher stress levels and for different medium composition, we evolved them for another 500 generations and compared their growth in different levels of osmotic and n-butanol stress (Supplementary Figures S19 and S20). We found that both sets of strains demonstrated almost linear correlation between stress tolerance and number of generations in the stressful environment. Interestingly, osmotic-adapted cells exhibited a striking trade-off (sixfold lower density compared with wild-type) when the medium was switched to LB under the same stress conditions, in contrast to the butanol-adapted cell lines.
Identifying the genetic basis of adaptation
To elucidate the genetic basis of the observed behavior, we used high-throughput sequencing to resequence the genomes of selected clones from the adapted cell lines (Figure 3; Supplementary Tables S-VI–S-VIII; Supplementary Figure S16). Although we did not resequence the ancestral genome, its reconstruction from the sequences of the five adapted cell lines revealed seven novel genetic variations to the MG1655 genome (Hayashi et al, 2006), six of which were confirmed independently (Freddolino et al, 2012) (Supplementary Table S-IX). The overall number of fixed mutations and type of observed genomic changes is consistent with our expectations from previous laboratory evolution experiments (see Supplementary Results and Supplementary Table S-X).
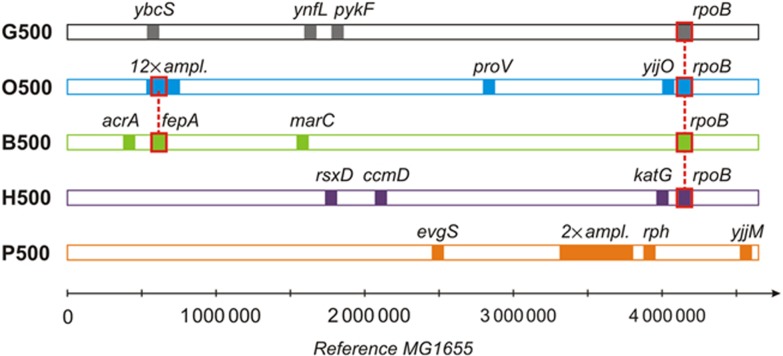
Figure 3.
Mutation map of fixed genetic changes in the evolved strains. The map of mutations identified in E. coli strains evolved for 500 generations. Coordinates are relative to the reference MG1655 genome. The seven identified mutations in the ancestral genome are not shown here (Supplementary Table S-IX).
The glucose-adapted strain G500 showed four SNP mutations in the ynfL, ybcS, pykF and rpoB genes (Figure 3; Table 1). From these, the ynfL mutation is outside the coding region, and the mutation in the ybcS gene may be neutral or inactivating as the latter encodes a prophage lysozyme that is implicated in cell lysis (Srividhya and Krishnaswamy, 2007). Mutations in the pykF gene are known to be beneficial for M9-glucose minimal media adaptation, as a previous study has shown that an IS insertion into the pykF gene has a significant positive effect in glucose-limited media (Barrick et al, 2010). Similarly, mutations on the β subunit of RNA polymerase (rpoB gene) have been shown to confer higher growth rates and efficiency in carbon source metabolism than the wild-type strain (Conrad et al, 2010). In fact, we identified rpoB mutations in five out of the six strains (with P500 being the exception). Previous results highlight the importance of RNA polymerase subunits as targets during microbial laboratory selection experiments. For example, the ratio of the housekeeping sigma factor σ70 and the stress-related sigma factor σS is crucial for shifting the balance from nutritional competence to stress protection (Patten et al, 2004; Ferenci, 2008; Klein-Marcuschamer et al, 2009). In the current study, we identified only mutations in the rpoB gene and no other RNA polymerase subunits. This may indeed indicate that selection towards increased performance in a selected growth medium and an additional stressor can be independent from each other. Sequence analysis in the osmotic-adapted strain O500 revealed a 12-fold amplification of a 8.5-kbp region that includes full copies of the enterobactin-related proteins fepA, entD, fes, hokE, and ybdZ genes (Supplementary Figure S17A). The three first genes are involved in enterobactin-iron transport and catalysis (Sansom, 1999) and there is evidence that fepA is upregulated under adversarial environments (Wachi et al, 1999; Muela et al, 2008), although its implication in stress protection is not clear. Additionally, we identified a premature stop codon mutation in proV, a subunit of the proline ABC-transporter which is essential for the uptake of potent osmoprotectants, such as glycine betaine (Perroud and Le Rudulier, 1985; Lucht and Bremer, 1994) that E. coli does not efficiently synthesize de novo (von Weymarn et al, 2001). The promoter region of yijO, a putative transcriptional regulator with unknown function also bore a SNP mutation, which suggests that this putative regulator may be implicated in stress response.
Table 1. Mutations identified in the evolved E. coli strains.
| Strain | Type | Gene | Positiona | Mutation or change | Comments | ||
|---|---|---|---|---|---|---|---|
| Reference | Gene | ||||||
| G500 | SNP | ybcS | 577 278 | 443 | G→T | DLP12 prophage lysozyme | |
| SNP | ynfL | 1 666 765 | 852 | A→C | Neutral | Neutral Ala→Ala substitution | |
| SNP | pykF | 1 754 158 | 437 | T→A | Pyruvate kinase I | ||
| SNP | rpoB | 4 180 843 | 1576 | C→T | β Subunit of RNA polymerase | ||
| O500 | Amp | 606 179–614 717 | 8.5 kbp | 12 × Amplification | Includes fepA, entD, fes, hokE, ybdZ | ||
| SNP | proV | 2 802 894 | 58 | C→T | Premature stop | proVWX osmoprotectant transporter | |
| Ins | yijO | 4 146 344 | −4 | 151 bp | Promoter, IS5 ins. | Putative transcriptional regulator | |
| SNP | rpoB | 4 180 972 | 1705 | A→C | β Subunit of RNA polymerase | ||
| B500 | SNP | acrA | 483 876 | 968 | A→T | Component of multidrug efflux pump | |
| SNP | fepA, entD | 611 871 | −154 | G→T | Promoter, fepA-entD operon | Enterobactin-iron transport | |
| SNP | marC | 1 616 292 | 641 | A→C | Inner membrane protein | ||
| SNP | rpoB | 4 181 399 | 2132 | A→G | β Subunit of RNA polymerase | ||
| H500 | SNP | rsxD | 1 707 591 | 426 | G→T | Member of the rsxABCDGE cluster, encoding the soxR reducing system | |
| SNP | ccmD | 2 293 421 | 188 | G→T | Integral membrane protein | ||
| SNP | katG | 4 131 777 | −81 | C→T | Promoter | Bifunctional hydroperoxidase I | |
| SNP | rpoB | 4 181 129 | 1862 | C→A | β Subunit of RNA polymerase | ||
| P500 | Ins | evgS | 2 485 683 | 3288 | >1334, bp | IS3 insertion | Membrane sensor-histidine kinase |
| Amp | ≈3 617 000–3 764 000 | ≈147 kbp | 2 × Amplification | Includes Acid Fitness Island (13 gens); contained between homologous rhsB and rhsA genes | |||
| Del | rph | 3 813 964 | 609 | 82 bp | Ribonuclease PH | ||
| Ins | yjjM | 4 593 800 | 75 | >1424, bp | IS186 insertion | Predicted transcriptional regulator | |
Abbreviations: Amp, amplification; Del, deletion; Ins, insertion; SNP, single nucleotide polymorphism.
aPositions are shown according to the MG1655 reference genome map and relative to the translational start of the genes.
Interestingly, the butanol-adapted strain B500 has undertaken an alternative path to alter fepA expression by introducing a mutation in its promoter region (Table I), which affects both the fepA and entD genes. We also found a mutation on the acrA gene that is involved in the acr multidrug efflux system for n- and iso-butanol resistance (Atsumi et al, 2010). A SNP mutation was identified in the marC gene, which was recently identified to also play a role in butanol tolerance (McDermott et al, 2008; Atsumi et al, 2010).
In the H2O2-adapted strain (H500) we find mutations in rsxD, ccmD, katG and rpoB. The rsxD gene is a member of the rsxABCDGE cluster, encoding the soxR reducing system. The deletion of any member of this gene cluster results in constitutive activation of soxS transcription (Koo et al, 2003), which was found to participate in superoxide removal (Demple, 1996), a finding that suggests an overlap of the hydrogen peroxide and superoxide stress response of E. coli. The ccmD gene is a membrane protein and involved in heme transport and cytochrome c maturation by its interaction with ccmC and ccmE (Ahuja and Thony-Meyer, 2005). Since H2O2 can destroy heme prosthetic groups and therefore inactivate cytochrome c (Villegas et al, 2000), ccmD changes can influence cellular cytochrome c production and thereby compensate increased cytochrome c turnover in oxidative stress conditions. We also identified a mutation in the promoter region of katG which is an oxyR-dependent gene that was found to be differentially expressed under H2O2-induced oxidative stress (Zheng et al, 2001). As such, mutations in its promoter region can influence intracellular catalase levels and further detoxification of hydrogen peroxide.
Finally, in the acidic-adapted strain P500, one of the insertions affected the evgS membrane sensor kinase, which together with the evgA response regulator has already been described to function in acid and drug resistance in E. coli (Itou et al, 2009). The strain also contains a duplication of a 147-kb fragment, which includes the acid fitness island region involved in the acid response (Supplementary Figure S17B), and is enclosed between homologous rhsB and rhsA genes of the same transcriptional orientation. Additionally, a deletion in rph, a ribonuclease involved in tRNA processing (Kelly et al, 1992) and an insertion in yjjM, essential for the growth on L-galactonate (Reed et al, 2006) was found, but the importance of these mutations is unclear. Surprisingly, rpoB mutations were not found in the P500 strain, indicating that adaptation to low pH and to the medium may be under antagonistic pleiotropy, something that is further supported by the fact that the P500 strains are unfit when compared with most of the other strains on M9 glucose media and in the absence of any stressor (Figure 2).
Differential gene expression under cross-stress protection
To further elucidate the genes and pathways that are implicated in cross-stress protection, we performed transcriptional profiling (RNA-Seq, verification by qRT–PCR, see Supplementary Materials) of the five strains under the osmotic stress, an environment where cross-stress protection was clearly observed. Populations evolved under butanol and osmotic stress showed almost the same fitness in this environment, while the fitness of populations evolved under acidic and oxidative stress was increased under osmotic stress when compared to the control populations. We analyzed the data by measuring statistically significant differential expression (DE) of genes between each stress-evolved strain and the G500 strain (Figure 4). In the H500, O500, P500 and B500 strains we identified 98, 79, 150, and 8 DE genes, respectively (P-value <0.05 after correction for multiple hypothesis testing). Among all DE genes in at least one of the stress-evolved strains (267 genes), several GO terms are statistically over-represented and include functions related to the ribosome, membrane, protein binding, flagella, peroxidase, RNA biosynthesis, and glycolysis (Figure 5A). We observed a significantly larger number of genes simultaneously DE in multiple strains versus what is expected by a pure chance: 49 versus 4 in 2 strains simultaneously, 8 versus 0.06 in 3 strains, and 1 versus 0.001 in 4 strains (see also Supplementary Results). However, despite the significant number of differentially expressed genes in at least one strain, only 9 DE genes (related to protein, membrane, and RNA binding; Figures 4 and 5B) where shared in at least three out of the four strains. It is a remarkably small overlap (by the absolute gene count) that is consistent with recent observations on acquired stress response in yeast (Berry et al, 2011). The majority of DE genes (53% of all DE genes and 83% of genes that do not belong in an amplified DNA region) are under-expressed relative to the control in at least one of the experimental strains, which is apparently an attempt to shut down non-essential pathways in stressful environments. There is a strong overlap between DE genes in H500 and O500 strains (52 genes, P-value <0.05), all of which are downregulated and they encode for parts of the transcription/translation machinery or are involved in carbon metabolism (Figure 5E). In the P500 strain, DE genes include protein binding, flagella and membrane-related genes (Figure 5G and H). The strongly upregulated flagella cluster was unexpected, and it suggests that flagella genes are implicated in acid response, a link that has been hypothesized before (Maurer et al, 2005). Interestingly, B500 expression profile is very similar to that of the G500 control strain with only eight genes being differentially expressed, which is a stark contrast to their fitness dissimilarity in the various stress environments.
Figure 4.
Differentially expressed genes across all strains under osmotic stress. (A) Genes differentially expressed in the evolved strains relative to the G500 strains tested by RNA-Seq under osmotic stress conditions. Venn diagrams (upper left) depict genes differentially expressed in one of four stress-evolved strains: B500, H500, O500, and P500 relative to the G500 strain. Region highlighted with red contains 10 genes, which are simultaneously DE in at least three strains. Expression profiles are shown (middle) for a subset of genes differentially expressed in at least one of the stress-evolved strains. Venn diagrams depict the overlap between genes differentially expressed in H500 and O500 strains (upper right) and the overlap between differentially expressed and 2 × amplified genes in the P500 strain (lower right). (B) Summary of the expression levels (relative to G500) of the top DE genes and any corresponding mutations. Values represent log-fold change; genes with a DE P-value <0.05 appear in color (green/red denoting up/downregulation, respectively). Full list of DE genes with P-values is provided in Supplementary Table S-XI. Source data for this figure is available on the online supplementary information page.
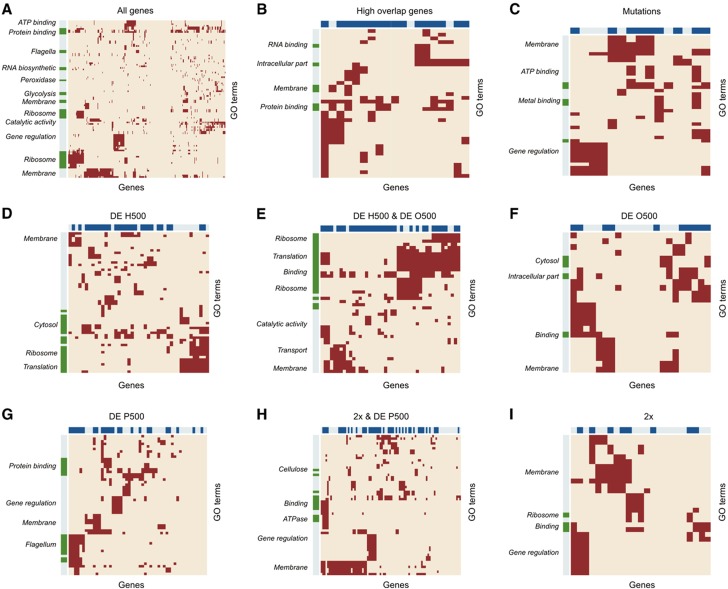
Figure 5.
Functional analysis of differentially expressed genes. (A) All genes that are differentially expressed relative to G500, amplified genes, and genes with mutations in at least one of the strains; (B) genes which are differentially expressed in at least two strains simultaneously; (C) genes with mutations; (D) genes differentially expressed only in the H500 strain; (E) genes differentially expressed in the H500 and O500 strains simultaneously; (F) genes differentially expressed only in the O500 strain; (G) not amplified genes, which are differentially expressed in the P500 strain; (H) amplified genes in P500, which are differentially expressed and (I) genes amplified in the P500 strain, but not differentially expressed. Dark red points indicate the correlation between genes and GO terms in each group. Dark green labels on the vertical sidebars show significantly overrepresented GO terms in each group, and dark blue labels on the horizontal sidebars show genes that are in the currently known gene regulatory network of E. coli (Keseler et al, 2011).
Several of the differential transcript expression levels relative to the G500 strain can be explained by the resequencing results. The SNP mutation at the promoter of katG in H500 results in its 3.6-fold over-expression in that strain (P-value=2 × 10−8), as does the SNP mutation in the fepA promoter in B500 (2.3-fold upregulation, P-value=2.5 × 10−15). An average 2.2-fold upregulation (P-value <10−3), of Acid Fitness Island (AFI) genes in the P500 strain is consistent with the average 1.6-fold DNA AFI amplification that we identified. Similarly, we observe a 10-fold upregulation of the fepA gene (P-value <10−130) in the osmotic stress-evolved clone as expected from the 12-fold DNA amplification of several genes from the enterobactin cassette that includes fepA. The proVWX operon was found to have a premature stop codon mutation in the proV gene of the O500 strain, and the operon was found to be significantly downregulated (2.7–4.4-fold, P-value <10−30), although we have no explanation on what the role of the mutation is and why the operon is downregulated in the O500 strain. Notably, our analysis did not reveal any major regulators other than rpoB, which has been implicated with increased evolvability in mutant cells (Barrick et al, 2010).
Effect of individual mutations on cross stress protection
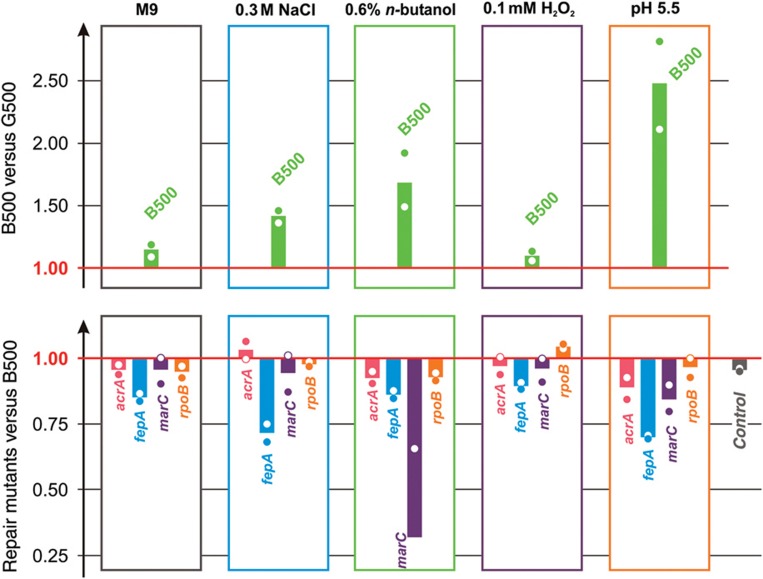
To associate the mutations identified with the phenotype observed, we reversed individual mutations in the evolved B500 sequenced clone (see Materials and methods) and again performed direct competition assays (Supplementary Figure S22–S25). Since a single B500 clone was used for repair of the mutations, we first assessed its phenotypic profile through competition assays and we confirmed its similarity to the population averages that were used in the previous analysis (Figure 6; Supplementary Figure S27). One striking result was that mutations to the ferric transporter fepA were found to be adaptive in all environments and confer its upregulation (Figure 4; Supplementary Table S-XII). This is partially explained by the fact that fepA is responsible for medium adaptation, as fepA reverse mutants had a significant disadvantage against the G500 population (w=0.850±0.005) in M9 glucose competition, while the B500 clone mutants without the repair outcompeted the G500 strains in the same environment (w=1.130±0.019, Supplementary Figure S26). However, in osmotic and acidic stress, the effect of fepA extends above whatever is expected from medium adaptation only, as the reverse mutants were significantly less fit than the original B500 clones (w=0.72±0.03 and w=0.70±0.0006, respectively). The marC mutation was validated to be the dominant driver for the increased fitness of B500 cells under n-butanol stress, with acrA and rpoB mutations contributing significantly lower. Interestingly, both the marC and acrA mutations were advantageous in the case of cross-stress protection of B500 in the acidic environment (w=0.85±0.03 and w=0.89±0.03, respectively). Finally, the rpoB mutation was found to have a negative impact to the B500 fitness under oxidative stress and is the main reason of the poor performance of these strains under that environment. Similarly, the lack of an rpoB mutation in the P500 strains can explain the increase fitness of the latter when compared with both O500 and B500 strains.
Figure 6.
Relative fitness of B500-evolved clones relative to the reference G500 population (top) and fitness of B500 repair mutants relative to the original B500-evolved clone (bottom) under all environmental conditions. Relative fitness of strains was assessed through competition assays in all five environments (see Materials and methods). Fitness refers to Darwinian fitness (Supplementary Text) as measured through competition. Competition of E. coli MG1655 strain with added chloramphenicol resistance gene relative to E. coli MG1655 (chloramphenicol sensitive) was added as a control (gray bar). Bars represent averages of two independent competitions (shown with dots); counts for each competition were averaged over three plates in each experiment. Competition assays were obtained over 48 h of growth with one transfer (1:50 dilution) at 24 h Supplementary Table S-XII summarizes the competition assay results. Source data for this figure is available on the online supplementary information page.
Discussion
Through integration of sequencing and transcriptional profiles, functional gene analysis, and measured Darwinian fitness, we are able to get a glimpse of the evolutionary trade-offs and dependencies that underlie microbial stress resistance. Cross-stress protection emerged at various degrees in all evolved strains, suggesting a ubiquitous phenomenon that may stem from high pleiotropic dependencies in the underlying genes and pathways. The rapidly emerging and reproducible adaptation patterns argue that bacterial populations occupy a genotypic space that allows high phenotypic plasticity within short timeframes. Adaptation to any specific stress and the growth media was found to be generally independent, as only a few trade-offs were observed between the control and stress-evolved strains. Surprisingly, our results argue that long-term adaptation to an environment does not necessarily lead to the fittest phenotype for that particular environment, as it is the case of cross-stress protection in acidic stress. This result suggests that the environment in which a cell evolves plays a significant role in the fixation rate of advantageous mutations, possibly by altering the selection dynamics.
We found several different types of mutations, such as SNPs in the coding and non-coding regions that can potentially affect different biological processes. Although rare, such point mutations are rapidly fixed in a population when they confer a significant fitness advantage. Insertion of transposable elements and the amplification of several genetic loci were also observed in various clones. Transposable elements and their random insertion represent a way for a given population to rapidly explore the genetic network space and develop new phenotypes and can be used to decouple evolutionary conserved responses even on a laboratory time scale (Tagkopoulos et al, 2008). In the current analysis, gene amplifications result in increased gene dosage and have an impact to the organism's fitness in a stressful environment but our data also indicate minor fitness trade-offs upon removal of the stressor and transfer to a less selective environment. Our observations are in accordance with recent theoretical analyses on the role of gene duplication in adaptation and evolution of novel functions (Kondrashov and Kondrashov, 2006; Bergthorsson et al, 2007). Furthermore, our data on low pH evolved populations indicate that the emergence of optimal fitness can be slow under certain circumstances and that improved phenotypes are hardly accessible from a given point in network space. As such, evolutionary adaptation to low pH may represent a suitable model system to further analyze the importance of historical contingency and population size in molecular evolution and neutral network theory (Wagner, 2008).
Two of the strains, O500 and B500, were found to have similar profiles and they exhibited the most profound cases of cross-stress behavior. A common denominator in both these strains is the upregulation of genes that are related to iron acquisition, either through mutation in the respective regulatory region (B500) or through DNA amplification (O500), which can partially explain the observed phenotypic similarity in the osmotic and acidic environments. Indeed, the fepA mutation was found to be the dominant factor behind the high B500 fitness in both these environments, as reversal of this mutation significantly decreased the fitness of the B500 mutants. This link that we establish in this work is in support of indirect prior evidence that iron-related pathways are generally over-expressed under osmotic stress (Snyder et al, 2004; Hagan and Mobley, 2007; Muela et al, 2008; Nielubowicz et al, 2008) and the high correlation of iron-regulation and acid response pathways in Helicobacter pylori (Bijlsma et al, 2002); Salmonella typhimurium (Hall and Foster, 1996), Shigella flexneri (Oglesby et al, 2005), Corynebacterium glutamicum (Jakob et al, 2007) and E. coli (Zhu et al, 2002). The role of iron transport in n-butanol and more generally in stress tolerance is not clear although it has been shown that iron is critical to the control of genes implicated in lipopolysaccharide biosynthesis, which in turn is the most prominent mechanism of toxic tolerance (Hagiwara et al, 2004; Reyes et al, 2012). Our results show that mutations in iron-related genes are a common genetic factor that drives bacterial tolerance across multiple stresses.
Both osmotic-evolved and butanol-evolved cells performed poorly under oxidative stress, which is in contrast to their superior performance in all other stresses. One hypothesis may be that the upregulation of iron-related genes in both strains may lead to an excess of iron inside the cell that can be toxic under oxidative stress (Touati et al, 1995; Andrews et al, 2003) as it facilitates the creation of reactive hydroxyl radicals that damage nucleic acids. While our mutation reversal experiments support this hypothesis, the effect of the iron transporter fepA was found to be secondary to the effect of the rpoB mutation, which was maladaptive in the case of oxidative stress. In addition, since the two strains have very different mechanisms to alter iron-related cells (i.e., mutation of the fepA promoter in B500 versus 12 × amplification in O500), the conservation of the regulatory control in the O500 can attribute to the increased fitness of the O500 to B500 strains when compared in oxidative stress. This is also supported from the transcriptional analysis: fepA was found to be significantly downregulated in G500 strains under oxidative stress (qRT–PCR, 3.8 × downregulation), upregulated in O500 cells (8.2 × upregulation), and strongly upregulated in B500 strains (16 × upregulation). Another interesting finding of the transcriptional analysis is the strong upregulation of the flagella cluster in the acidic environment. It would be interesting to further investigate the importance of this finding in the organism’s physiology within its ecological context, for example as a mechanism to maintain pH homeostasis during its transition through the mammalian GI tract.
Given its abundance, it is important to further investigate what are the underlying causes of evolutionary cross-stress protection and trade-offs for different stress combinations. If specific stresses have similar effects at a cellular level, then a high degree of cross-stress protection can be expected since responses to these stresses have to protect the cell versus common perturbations. Alternatively, the environmental context under which cells have evolved may be responsible for cross-stress protection or trade-off, even in the absence of common underlying mechanism. Indeed, E. coli and baker’s yeast were recently shown to exhibit cases of cross-stress protection when stresses were applied in the order that exist in their natural environment, while this coupling was lost quite rapidly when adapted in environments with the inverse dynamics (Tagkopoulos et al, 2008; Mitchell et al, 2009). Concomitantly, the absence of cross-stress protection or strong trade-offs may imply that the respective stresses did not co-exist in the organism’s natural habitat, nor was the latter exposed to rapid fluctuations across these dimensions. The expansion of laboratory evolution experiments beyond what we perceive to be their natural habitat parameters is relevant to other areas, including the evolution and treatment of food-borne and nosocomial pathogens. Due to their fast rate of evolutionary adaptation, bacteria pose a major problem in those areas and a better understanding of their adaptive potential in natural and non-natural stress conditions can lead to techniques that aim to control their proliferation. Furthermore, besides its significance in the development of sterilization techniques, analyses similar to what presented here may also greatly impact the development of microbial strains in industrial biotechnology. The biotechnological production of biofuels and other organic substances (including non-natural metabolites) depends on microbial cells as biocatalysts. In most cases such systems depend on bacterial growth and microbial cells can suffer stress during cultivation, including variations in pH, aeration and temperature (e.g., during cultivation in large bioreactors due to improper mixing of the culture broth). Additionally, cells are often engineered to produce chemical compounds that occur only in very small quantities or not at all in their natural habitats. As such, the interplay and compatibility of ubiquitous stress parameters and artificial stresses is of great interest for the optimization of bacterial growth in complex industrial environments. As our knowledge of bacterial physiology and ecology continues to expand, so will our understanding of the underlying causes of cross-stress potential and behavior. Until then, evolutionary experiments such as the one presented here will provide a unique perspective on the emergence of complex microbial behavior.
Materials and methods
Bacterial strains and growth media and conditions
E. coli MG1655 and MG1655 ΔlacZ strains were used in all experiments of this study. The inclusion of ΔlacZ mutants allowed us to perform competition assays using X-Gal and IPTG staining. The neutrality of the ΔlacZ mutation was confirmed in all environments prior to adaptation (Supplementary Table S-I; Supplementary Figure S4). Minimal M9 salt medium with 0.4% (w/v) glucose as carbon source was used for evolutionary adaptation, growth tests, and competition assays. All cultivations were performed in 10 ml volume in 125 ml shake flasks at 37°C on an orbital shaker at 150 r.p.m.
Evolutionary adaptation by serial dilution experiments
Adaptive evolution experiments were performed by daily serial dilutions after 24 h of growth in fourfold replication (2 × E. coli MG1655 and 2 × E. coli MG1655 ΔlacZ) for each growth condition. For each of the daily transfers, the OD600 was monitored and cultures were diluted (1:500) in fresh growth medium yielding ∼7–9 generations per day. Serial dilutions were performed for a total of 500 generations.
Stress conditions
E. coli strains were evolved in M9 salts medium with glucose as the sole carbon source and the following stressors: osmotic stress (0.3 M NaCl, O500 strains), acidic stress (pH 5.5, P500 strains), oxidative stress (100 μM H2O2, H500 strains), n-butanol stress (0.6% n-butanol, B500 strains), and control (no-stress, G500 strains). The OD600 of each culture was measured each day before the daily transfers to ensure that the estimated nine generations per day were reached (Supplementary Material).
Growth curves
Growth curves of the ancestral and evolved cell lines were performed in the growth media described above. For growth curves, the starter cultures of all strains were grown and therefore adapted (∼7–9 generations) to the medium to be tested, for 18 h. Cultures were started at OD600=0.1 or 0.05, depending on the growth condition. Samples were taken in regular intervals and OD600 was measured on a biophotometer (Bio-Rad). Two independent replicate growth tests were performed for each strain and population.
Competition assays
For direct competition assays, the two competing populations (E. coli MG1655 and E. coli MG1655 ΔlacZ) were grown under adaptive conditions overnight and inoculated at an approximate 1:1 ratio at a starting OD600=0.004. After 24 h of growth, cultures were diluted 1:500 in fresh medium and competed for additionally 24 h. Samples were taken at 0, 8, 24, and 48 h. Two independent replicates were performed for each individual competition. Cell counts were determined on LB agar plates containing 0.25 mM IPTG (Isopropyl β-D-1-thiogalactopyranoside) and 40 μg/ml X-gal (bromo-chloro-indolyl-galactopyranoside). Plates were incubated at 37°C overnight. Darwinian fitness (W) was calculated as described in the Supplementary Methods section.
Maximum stressor concentration
Starter cultures of evolved and ancestral populations were grown in M9 medium at 37°C overnight and a main culture (M9 and LB medium with different stressor concentrations) was inoculated at OD600=0.1. Growth potential was monitored by measuring optical density after 6 and 24 h. Maximum stressor concentration is referred to as the concentration of NaCl and butanol in the growth medium, which exceeded the concentrations that were used for the initial evolutionary adaptation experiment to test whether the maximum level of abiotic stressor was increased as compared with the ancestral strains.
Genome resequencing of evolved clones
Selected clones were grown on LB medium overnight and genomic DNA (gDNA) was isolated using a Wizard Genomic DNA Purification Kit (Promega). gDNA was quantified photometrically and 7 μg were used for fragmentation in a bioruptor (Diagenode) sonication apparatus. DNA was blunt ended using T4 DNA polymerase and Klenow fragment. A tailing was performed using Klenow fragment (exo). After A tailing DNA linkers were ligated using fast ligase. PCR enrichment and DNA size selection was performed on 2% agarose gels. DNA purification steps were performed using a NucleoSpin Extract II purification kit (Machery and Nagel). DNA libraries were sequenced by a paired end run with 85 bp read length on an Illumina Genome Analyzer GAII. The shotgun sequence data was assembled, using the IDBA assembler (Peng et al, 2010), after preliminary filtration and quality-trimming of the Illumina reads using SGA (Dinh and Rajasekaran, 2011). Scaffolding was performed with SSPACE script (Boetzer et al, 2011). Parameters for the assembly and scaffolding were selected based on the estimations provided by an automated pipeline (Darling et al, unpublished data). Scaffolds were reordered using Mauve Contig Mover (Rissman et al, 2009) and aligned to the reference E. coli strain K12 substrain MG1655 genome (Hayashi et al, 2006) using progressive Mauve genome aligner (Darling et al, 2010). SNPs and variances between sequenced strains and the reference E. coli genome were obtained for each strain by analyzing both: (i) shotgun reads mapped to the reference genome using BWA (Li and Durbin, 2010) and SAM tools (Li et al, 2009) and (ii) the de novo assembly followed by alignment to the reference genome. Variances that are found in all six strains are attributed to the ancestral strain; unique variances are the result of the independent evolution of the strains.
Gene expression analysis
For transcriptional profiling, the resequenced clones were grown in M9-gluc medium with 0.3 M NaCl (osmotic stress medium) to mid-log phase (OD600 approx. 0.8). In all, 1 ml aliquots were harvested and mixed with 0.5 ml Phenol/ethanol (5% (v/v) Phenol in EtOHabs). Cells were collected by centrifugation and stored at −80°C until use. RNA was extracted using an RNeasy kit (Qiagen) and after first- and second-strand cDNA synthesis, linker ligation, and size selection subjected to shotgun sequencing using single read 40 bp read length runs on an Illumina Genome Analyzer GAII. llumina reads for RNA-Seq libraries were mapped to the reference E. coli strain K12 substrain MG1655 genome (GenBank accession no. U00096.2) by BWA toolkit (Li and Durbin, 2010). Differentially expressed genes were identified by processing raw counts of mapped reads with edgeR library. Gene ontology analysis was performed by goseq R package. Differentially expressed (DE) genes in all stress-evolved strains were found relative to the expression levels in the reference G500 strain using the edgeR R package. Genes with BH (Benjamini and Hochberg) adjusted P-values below 0.05 threshold were selected as DE genes. Genes from the amplified regions in O500 (12-fold) and P500 (twofold) strains are significantly over-expressed relative to the reference: average log Fold Change for concentrations is +3.6 and +0.8 (both with P-values <10−20), respectively. Full list of genes DE in at least one of the strains and sorted by the expression patterns is shown in Supplementary Table S-XI. The RNA-seq data set can be obtained from the GEO repository, record no. GSE39926.
Mutation repairs in B500-evolved strain
To perform single mutation repairs, DNA fragments from wild-type E. coli MG1655 were used to replace each of the mutations in the B500-evolved strain by P1 transduction (Miller, 1992) using a chloramphenicol resistance gene as a selectable marker. PCR-amplified chloramphenicol resistance cassettes were introduced into wild-type E. coli chromosome in sites corresponding to genomic regions close to the mutation sites present in the B500 strain (described in Table I) by linear DNA transformation (Poteete and Fenton, 1984, 2000; Murphy, 1998; El Karoui et al, 1999; Datsenko and Wanner, 2000). The newly transformed strains carrying one chloramphenicol cassette linked to one of the mutations were used as donors to replace the B500 strain mutated sequences. New B500 mutants repaired a single mutation site, while still carrying three of the original four mutations. The repaired fragments were verified by Sanger sequencing.
Fitness effect of individual mutations
Repaired B500 mutants were competed against the original B500-evolved strain on M9 salts medium with glucose under several conditions. The two competing strains were grown overnight in 2 ml M9 salts medium with glucose, washed and inoculated at an approximate 1:1 ratio for direct competition under adaptive conditions. After 24 h, they were diluted 1:50 into fresh medium and competed for an additional 24 h. Two independent replicates were performed for each individual competition. Culture samples at 0, 24, and 48 h were diluted and plated in LB medium and LB medium with 20 μg/ml chloramphenicol, and incubated at 37°C overnight. The number of colonies present on the LB medium represents the total number of all cell types in the population. The number of colonies on the LB chloramphenicol medium represents the total number of chloramphenicol resistant cells in the population (repaired mutant). The difference between colony numbers from the two plate types will show the total number of chloramphenicol susceptible cells in the population (original B500 strain). From this experiment, we determined the relative fitness of each repair mutant under different growth conditions relative to the original B500 mutant. E. coli MG1655 strain with an added chloramphenicol resistance gene was competed to wild-type E. coli MG1655 (chloramphenicol sensitive) as a control.
Verification of mutations and transcriptional abundance
All mutations were independently verified by sequencing. To test gene expression fold changes, evolved E. coli MG1655 cells were grown in M9 salt glucose medium. The overnight cultures were diluted 1:10 in fresh M9 salt glucose medium with different stress (no stress, 0.3 M NaCl, 0.6% n-butanol, or 100 μM H2O2) and grown at 37°C with shaking (150 r.p.m.) to the log phase. Total RNA was prepared according to the manufacturer’s instructions (RNeasy mini kit, Qiagen). cDNA was synthesized from 1 μg of total RNA by using the iScript cDNA synthesis kit (Bio-Rad). Cells were grown in Luria Broth (LB) media overnight, and genomic DNA was isolated via the Wizard Genomic DNA Purification Kit (Promega). Samples were then analyzed by Chromo4 instrument (Bio-Rad). Primers were designed using the Primer Quest (IDT DNA). Primers were made for the target genes and σ70 gene, which was used as reference. qPCR was performed in 50 μl mix containing 1 × SsoAdvanced SYBR Green Supermix (Bio-Rad), 40 ng genomic DNA or cDNA, and 5 pmol primers. Experiments were done in triplicate and qPCR data were analyzed by Opticon Monitor 3 (Bio-Rad).
Supplementary Material
Supplementary Tables SI–XII, Supplementary Figures S1–27
De novo assembly breaks analysis file
Log-fold change and p-values for transcriptional profiling (RNA-Seq) data
Acknowledgments
We thank Aaron Darling, Jonathan Eisen, Marc Facciotti, Artyom Kopp, John Roth, and Saeed Tavazoie for the manuscript comments and helpful discussions. This work was supported by the UC Davis Opportunity Fund and DoD award W911NF1210231-0.
Author contributions: MD performed the strain evolution, strain characterization, and library preparation for genome sequencing and RNA-Seq. VM performed the data analysis, the genome assembly and RNA-Seq. SQS repaired the mutations and performed competition assays for validating the fitness effect of the mutations. JP performed qRT–PCR validation experiments. MD, VM, SQS and IT evaluated data and prepared the manuscript. IT conceived of the study and supervised all aspects of the project.
Footnotes
The authors declare no conflict of interest.
References
- Ahuja U, Thony-Meyer L (2005) CcmD is involved in complex formation between CcmC and the heme chaperone CcmE during cytochrome c maturation. J Biol Chem 280: 236–243 [DOI] [PubMed] [Google Scholar]
- Alcántara-Díaz D, Breña-Valle M, Serment-Guerrero J (2004) Divergent adaptation of Escherichia coli to cyclic ultraviolet light exposures. Mutagenesis 19: 349–354 [DOI] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F (2003) Bacterial iron homeostasis. FEMS Microbiol Rev 27: 215–237 [DOI] [PubMed] [Google Scholar]
- Araya CL, Payen C, Dunham MJ, Fields S (2010) Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomics 11: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451: 86–89 [DOI] [PubMed] [Google Scholar]
- Atsumi S, Wu TY, Machado IM, Huang WC, Chen PY, Pellegrini M, Liao JC (2010) Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol Syst Biol 6: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Kauth MR, Strelioff CC, Lenski RE (2010) Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol Biol Evol 27: 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M, Gahan C, Hill C (2002) Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl Environ Microbiol 68: 6005–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett AF, Lenski RE (2007) An experimental test of evolutionary trade-offs during temperature adaptation. Proc Natl Acad Sci USA 104: Suppl 18649–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Andersson DI, Roth JR (2007) Ohno’s dilemma: evolution of new genes under continuous selection. Proc Nat Acad Sci Usa 104: 17004–17009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DB, Gasch AP (2008) Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell 19: 4580–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DB, Guan Q, Hose J, Haroon S, Gebbia M, Heisler LE, Nislow C, Giaever G, Gasch AP (2011) Multiple means to the same end: the genetic basis of acquired stress resistance in yeast. PLoS Genet 7: e1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma JJ, Waidner B, Vliet AH, Hughes NJ, Hag S, Bereswill S, Kelly DJ, Vandenbroucke-Grauls CM, Kist M, Kusters JG (2002) The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect Immun 70: 606–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Borland CZ, Lenski RE (2008) Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA 105: 7899–7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W (2011) Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27: 578. [DOI] [PubMed] [Google Scholar]
- Cánovas D, Fletcher S, Hayashi M, Csonka L (2001) Role of trehalose in growth at high temperature of Salmonella enterica serovar Typhimurium. J Bacteriol 183: 3365–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad TM, Frazier M, Joyce AR, Cho BK, Knight EM, Lewis NE, Landick R, Palsson BØ (2010) RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci 107: 20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad TM, Lewis NE, Palsson BO (2011) Microbial laboratory evolution in the era of genome-scale science. Mol Syst Biol 7: 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T, Lenski R (2010) Experimental evolution with E. coli in diverse resource environments. I. Fluctuating environments promote divergence of replicate populations. BMC Evol Biol 10: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS one 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Nat Acad Sci Usa 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel E, Alon U (2005) Optimality and evolutionary tuning of the expression level of a protein. Nature 436: 588–592 [DOI] [PubMed] [Google Scholar]
- Demple B (1996) Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene 179: 53–57 [DOI] [PubMed] [Google Scholar]
- Dinh H, Rajasekaran S (2011) A memory-efficient data structure representing exact-match overlap graphs with application for next-generation DNA assembly. Bioinformatics 27: 1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- El Karoui M, Amundsen SK, Dabert P, Gruss A (1999) Gene replacement with linear DNA in electroporated wild-type Escherichia coli. Nucleic Acids Res 27: 1296–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci T (2008) The spread of a beneficial mutation in experimental bacterial populations: the influence of the environment and genotype on the fixation of rpoS mutations. Heredity 100: 446–452 [DOI] [PubMed] [Google Scholar]
- Fletcher S, Csonka L (1998) Characterization of the induction of thermotolerance by osmotic stress in Salmonella typhimurium. Food Microbiol 15: 307–317 [Google Scholar]
- Freddolino PL, Amini S, Tavazoie S (2012) Newly identified genetic variations in common Escherichia coli MG1655 stock cultures. J Bacteriol 194: 303–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren D, Harrison M, Russell S (1998) Acid tolerance and acid shock response of Escherichia coli O157:H7 and non-O157:H7 isolates provide cross protection to sodium lactate and sodium chloride. J Food Prot 61: 158–161 [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Bennett BD, Amini S, Reaves ML, Hottes AK, Rabinowitz JD, Tavazoie S (2010) Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol Syst Biol 6: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera T, Csonka L, Paliy O (2008) Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J Bacteriol 190: 3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan EC, Mobley HL (2007) Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infect Immun 75: 3941–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D, Yamashino T, Mizuno T (2004) A genome-wide view of the Escherichia coli BasS-BasR two-component system implicated in iron-responses. Biosci Biotechnol Biochem 68: 1758–1767 [DOI] [PubMed] [Google Scholar]
- Hall HK, Foster JW (1996) The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J Bacteriol 178: 5683–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T (2006) Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2: 2006.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes B, Cullum A, Bennett A (2007) Evolutionary adaptation to environmental pH in experimental lineages of Escherichia coli. Evolution 61: 1725–1734 [DOI] [PubMed] [Google Scholar]
- Itou J, Eguchi Y, Utsumi R (2009) Molecular mechanism of transcriptional cascade initiated by the EvgS/EvgA system in Escherichia coli K-12. Biosci Biotechnol Biochem 73: 870. [DOI] [PubMed] [Google Scholar]
- Jakob K, Satorhelyi P, Lange C, Wendisch VF, Silakowski B, Scherer S, Neuhaus K (2007) Gene expression analysis of Corynebacterium glutamicum subjected to long-term lactic acid adaptation. J Bacteriol 189: 5582–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins D, Auger E, Matin A (1991) Role of RpoH, a heat shock regulator protein, in Escherichia coli carbon starvation protein synthesis and survival. J Bacteriol 173: 1992–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins D, Chaisson S, Matin A (1990) Starvation-induced cross protection against osmotic challenge in Escherichia coli. J Bacteriol 172: 2779–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins D, Schultz J, Matin A (1988) Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J Bacteriol 170: 3910–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly KO, Reuven NB, Li Z, Deutscher MP (1992) RNase PH is essential for tRNA processing and viability in RNase-deficient Escherichia coli cells. J Biol Chem 267: 16015–16018 [PubMed] [Google Scholar]
- Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muniz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, Kaipa P, Spaulding A, Pacheco J, Latendresse M, Fulcher C, Sarker M, Shearer AG, Mackie A, Paulsen I, Gunsalus RP et al. (2011) EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res 39: D583–D590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Marcuschamer D, Santos CN, Yu H, Stephanopoulos G (2009) Mutagenesis of the bacterial RNA polymerase alpha subunit for improvement of complex phenotypes. Appl Environ Microbiol 75: 2705–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Kondrashov AS (2006) Role of selection in fixation of gene duplications. J Theor Biol 239: 141–151 [DOI] [PubMed] [Google Scholar]
- Koo MS, Lee JH, Rah SY, Yeo WS, Lee JW, Lee KL, Koh YS, Kang SO, Roe JH (2003) A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J 22: 2614–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyer G, Johnson E (1993) Acid adaptation induces cross-protection against environmental stresses in Salmonella typhimurium. Appl Environ Microbiol 59: 1842–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Yousef A (1997) Adaptation to sublethal environmental stresses protects Listeria monocytogenes against lethal preservation factors. Appl Environ Microbiol 63: 1252–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucht JM, Bremer E (1994) Adaptation of Escherichia coli to high osmolarity environments: osmoregulation of the high-affinity glycine betaine transport system proU. FEMS Microbiol Rev 14: 3–20 [DOI] [PubMed] [Google Scholar]
- Machielsen R, van Alen-Boerrigter I, Koole L, Bongers R, Kleerebezem M, Van Hylckama Vlieg J (2010) Indigenous and environmental modulation of frequencies of mutation in Lactobacillus plantarum. Appl Environ Microbiol 76: 1587–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattimore V, Battista JR (1996) Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J Bacteriol 178: 633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL (2005) pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol 187: 304–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott PF, McMurry LM, Podglajen I, Dzink-Fox JL, Schneiders T, Draper MP, Levy SB (2008) The marC gene of Escherichia coli is not involved in multiple antibiotic resistance. Antimicrob Agents Chemother 52: 382–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH (1992) A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Miller, J H A short course in bacterial genetics: A laboratory manual and handbook for Escherichia coli and related bacteria xviii+Pagination varies; xix+456p Cold Spring Harbor Laboratory Press: Plainview, New York, USA. Illus Paper ISBN 0-87969-349-5
- Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, Dahan O, Pilpel Y (2009) Adaptive prediction of environmental changes by microorganisms. Nature 460: 220–224 [DOI] [PubMed] [Google Scholar]
- Muela A, Seco C, Camafeita E, Arana I, Orruno M, López JA, Barcina I (2008) Changes in Escherichia coli outer membrane subproteome under environmental conditions inducing the viable but nonculturable state. FEMS Microbiol Ecol 64: 28–36 [DOI] [PubMed] [Google Scholar]
- Murphy KC (1998) Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol 180: 2063–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielubowicz GR, Smith SN, Mobley HL (2008) Outer membrane antigens of the uropathogen Proteus mirabilis recognized by the humoral response during experimental murine urinary tract infection. Infect Immun 76: 4222–4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby AG, Murphy ER, Iyer VR, Payne SM (2005) Fur regulates acid resistance in Shigella flexneri via RyhB and ydeP. Mol Microbiol 58: 1354–1367 [DOI] [PubMed] [Google Scholar]
- Patten CL, Kirchhof MG, Schertzberg MR, Morton RA, Schellhorn HE (2004) Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol Genet Genomics 272: 580–591 [DOI] [PubMed] [Google Scholar]
- Peng Y, Leung H, Yiu S, Chin F (2010) IDBA–A practical iterative de Bruijn graph de novo assembler. LNBI 6044: 426–440 [Google Scholar]
- Perroud B, Le Rudulier D (1985) Glycine betaine transport in Escherichia coli: osmotic modulation. J Bacteriol 161: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete AR, Fenton AC (1984) Lambda red-dependent growth and recombination of phage-22. Virology 134: 161–167 [DOI] [PubMed] [Google Scholar]
- Poteete AR, Fenton AC (2000) Genetic requirements of phage lambda red-mediated gene replacement in Escherichia coli K-12. J Bacteriol 182: 2336–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaghello L, Lee C, Safdie FM, Wei M, Madia F, Bianchi G, Longo VD (2008) Starvation-dependent differential stress resistance protects normal but not cancer cells against high-dose chemotherapy. Proc Natl Acad Sci 105: 8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JL, Patel TR, Chen KH, Joyce AR, Applebee MK, Herring CD, Bui OT, Knight EM, Fong SS, Palsson BO (2006) Systems approach to refining genome annotation. Proc Natl Acad Sci USA 103: 17480–17484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes LH, Almario MP, Winkler J, Orozco MM, Kao KC (2012) Visualizing evolution in real time to determine the molecular mechanisms of n-butanol tolerance in Escherichia coli. Metab Eng 14: 579–590 [DOI] [PubMed] [Google Scholar]
- Riehle MM, Bennett AF, Lenski RE, Long AD (2003) Evolutionary changes in heat-inducible gene expression in lines of Escherichia coli adapted to high temperature. Physiol Genomics 14: 47–58 [DOI] [PubMed] [Google Scholar]
- Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT (2009) Reordering contigs of draft genomes using the Mauve Aligner. Bioinformatics 25: 2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford B, Dahl R, Price R, Szmidt H, Benke P, Mukhopadhyay A, Keasling J (2010) Functional genomic study of exogenous n-butanol stress in Escherichia coli. Appl Environ Microbiol 76: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J, Beuchat L (1998) Influence of acid tolerance responses on survival, growth, and thermal cross-protection of Escherichia coli O157:H7 in acidified media and fruit juices. Int J Food Microbiol 45: 185–193 [DOI] [PubMed] [Google Scholar]
- Sansom MSP (1999) Membrane proteins: a tale of barrels and corks. Curr Biol 9: R254–R257 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci 97: 11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sghaier H, Narumi I, Satoh K, Ohba H, Mitomo H (2007) Problems with the current deinococcal hypothesis: an alternative theory. Theory Biosci 126: 43–45 [DOI] [PubMed] [Google Scholar]
- Silver R, Mateles R (1969) Control of mixed-substrate utilization in continuous cultures of Escherichia coli. J Bacteriol 97: 535–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleight S, Orlic C, Schneider D, Lenski R (2008) Genetic basis of evolutionary adaptation by Escherichia coli to stressful cycles of freezing, thawing and growth. Genetics 180: 431–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HL (2004) Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect Immun 72: 6373–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srividhya KV, Krishnaswamy S (2007) Subclassification and targeted characterization of prophage-encoded two-component cell lysis cassette. J Biosci 32: 979–990 [DOI] [PubMed] [Google Scholar]
- Storz G, Hengge-Aronis R (2000) Bacterial stress responses Washington, USA: American Society for Microbiology, [Google Scholar]
- Tagkopoulos I, Liu YC, Tavazoie S (2008) Predictive behavior within microbial genetic networks. Science 320: 1313–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesone S, Hughes A, Hurst A (1981) Salt extends the upper temperature limit for growth of food-poisoning bacteria. Can J Microbiol 27: 970–972 [DOI] [PubMed] [Google Scholar]
- Touati D, Jacques M, Tardat B, Bouchard L, Despied S (1995) Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol 177: 2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas JA, Mauk AG, Vazquez-Duhalt R (2000) A cytochrome c variant resistant to heme degradation by hydrogen peroxide. Chem Biol 7: 237–244 [DOI] [PubMed] [Google Scholar]
- von Weymarn N, Nyyssola A, Reinikainen T, Leisola M, Ojamo H (2001) Improved osmotolerance of recombinant Escherichia coli by de novo glycine betaine biosynthesis. Appl Microbiol Biotechnol 55: 214–218 [DOI] [PubMed] [Google Scholar]
- Wachi M, Takada A, Nagai K (1999) Overproduction of the outer-membrane proteins FepA and FhuE responsible for iron transport in Escherichia coli hfq::cat mutant. Biochem Biophys Res Commun 264: 525–529 [DOI] [PubMed] [Google Scholar]
- Wagner A (2008) Robustness and evolvability: a paradox resolved. Proc Roy Soc B-Biol Sci 275: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Spira B, Zhou Z, Feng L, Maharjan RP, Li X, Li F, McKenzie C, Reeves PR, Ferenci T (2010) Divergence involving global regulatory gene mutations in an Escherichia coli population evolving under phosphate limitation. Genome Biol Evol 2: 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Polen T, Heuveling J, Wendisch V, Hengge R (2005) Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187: 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Ziegler C, Um S, Pérez N, Berns A, Malhowski A, Young S (2008) Low temperature (23 degrees C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology 154: 148–166 [DOI] [PubMed] [Google Scholar]
- Zhao H (2009) Ischemic postconditioning as a novel avenue to protect against brain injury after stroke. J Cereb Blood Flow Metab 29: 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Wang X, Templeton L, Smulski D, LaRossa R, Storz G (2001) DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol 183: 4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Ngeleka M, Potter AA, Allan BJ (2002) Effect of fur mutation on acid-tolerance response and in vivo virulence of avian septicemic Escherichia coli. Can J Microbiol 48: 458–462 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables SI–XII, Supplementary Figures S1–27
De novo assembly breaks analysis file
Log-fold change and p-values for transcriptional profiling (RNA-Seq) data