Abstract
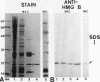
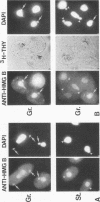
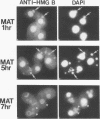
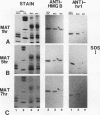
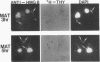
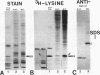
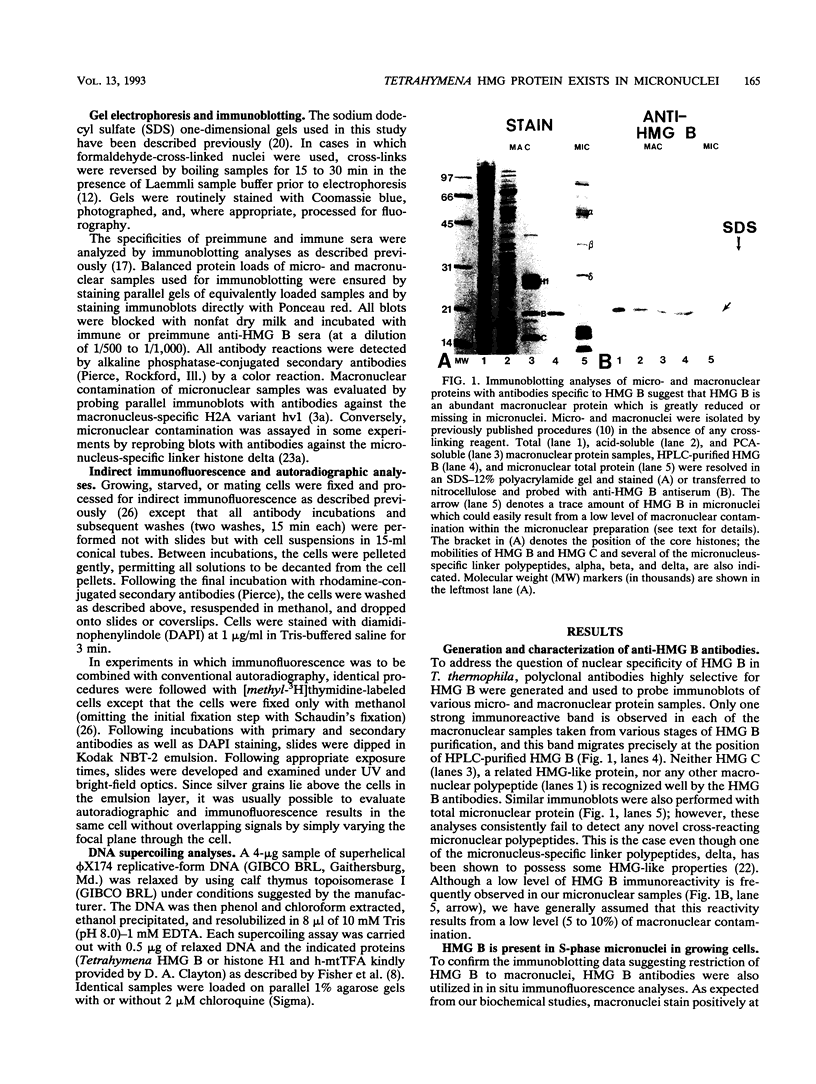
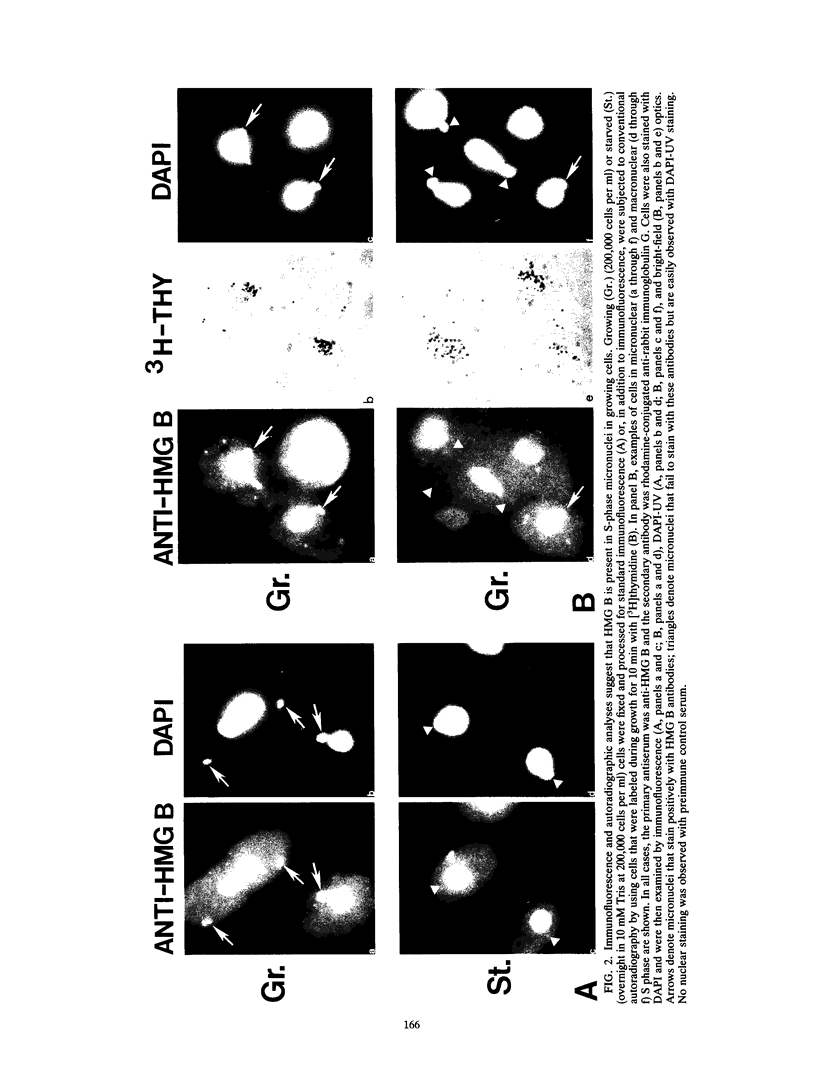
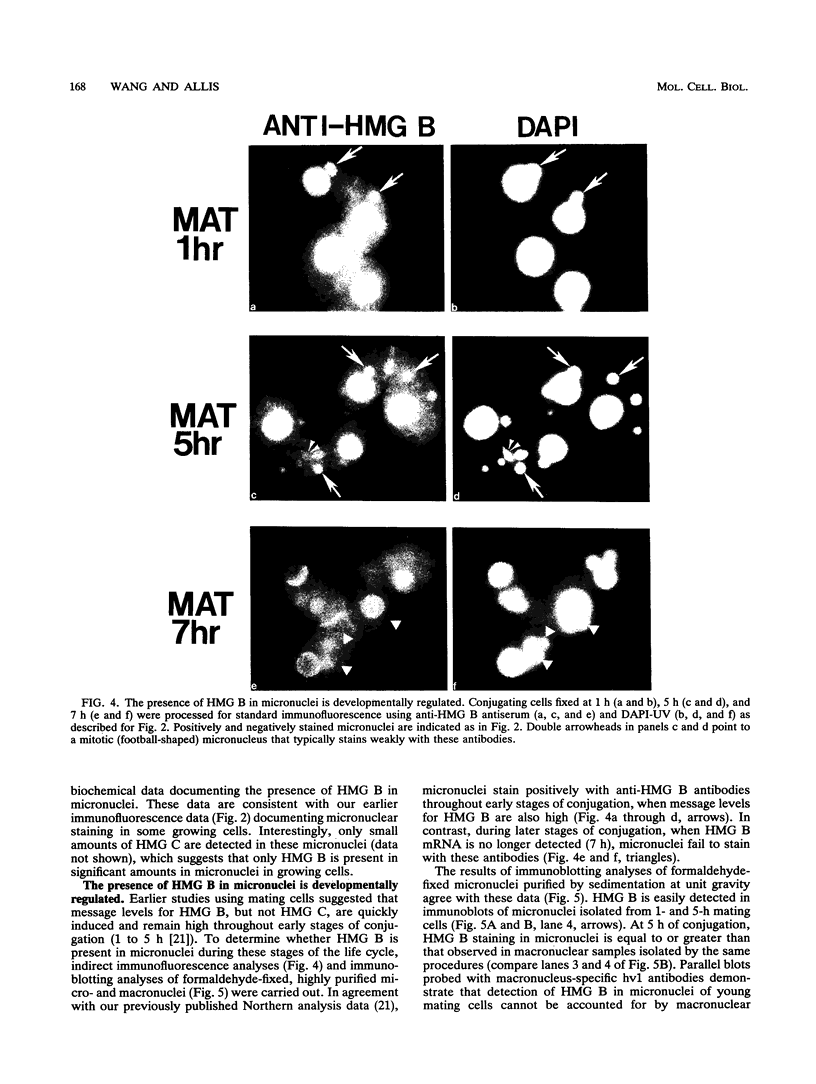
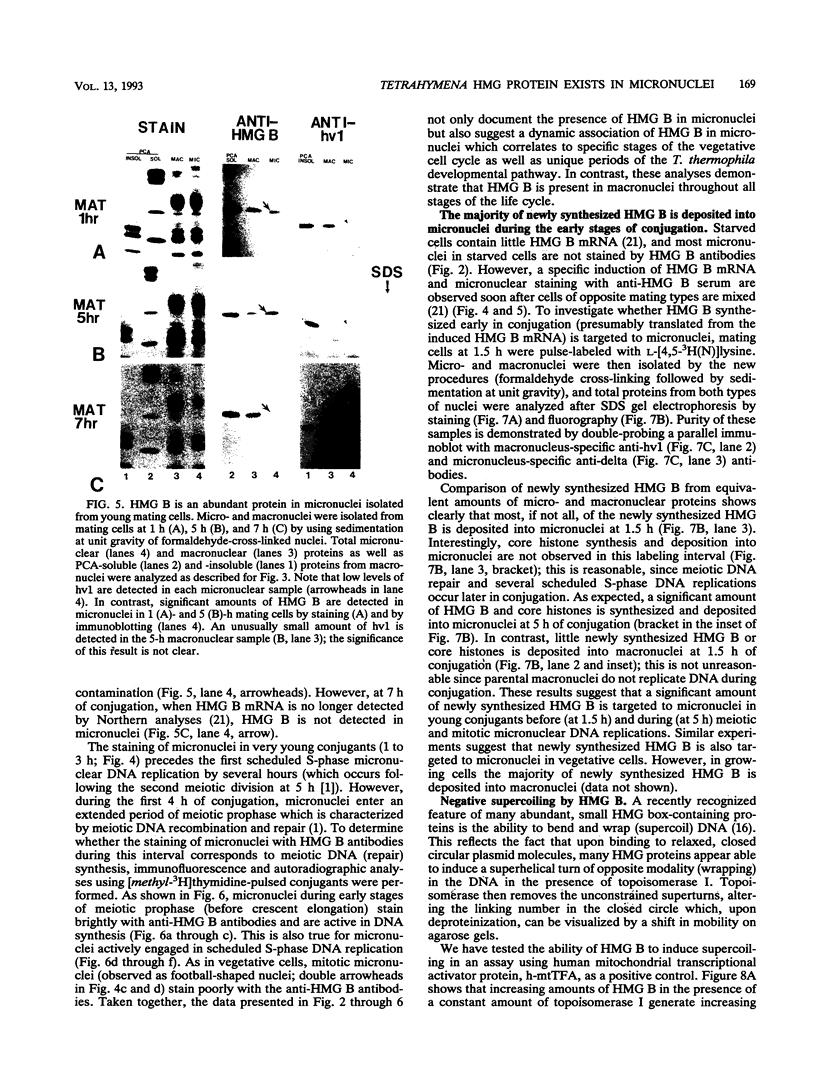
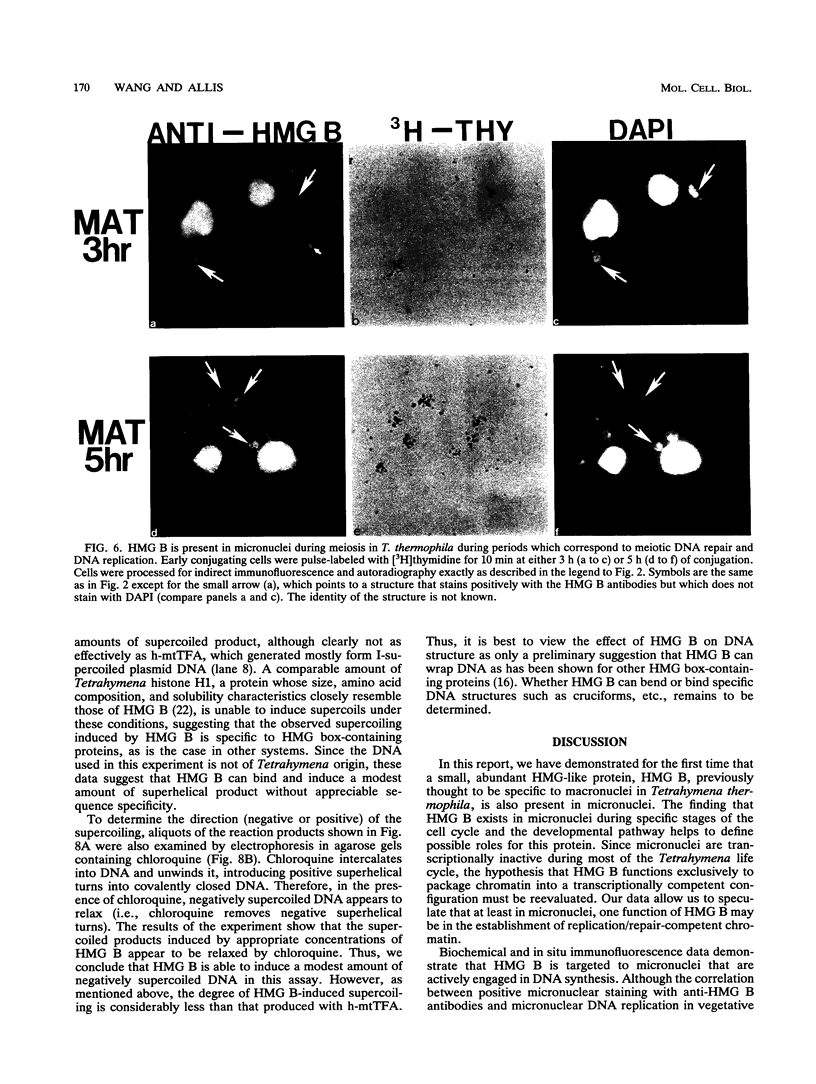
In this report, we have demonstrated for the first time that an abundant high-mobility-group (HMG)-like protein, HMG B, previously thought to be specific to macronuclei in Tetrahymena thermophila, is also present in micronuclei. Biochemical data document the fact that HMG B is extremely labile in micronuclei. Unless extreme precautions are taken during the isolation of nuclei (addition of 1% formaldehyde to the nucleus isolation buffer), HMG B is not detected in micronuclei. Using polyclonal antibodies highly selective for HMG B, immunoblotting and immunofluorescence analyses show that the presence of HMG B in micronuclei is dynamic, correlating well with known periods of micronuclear DNA replication. This is the case not only during the vegetative cell cycle but also during early stages of the sexual cycle, conjugation, when the presence of HMG B in micronuclei is also closely correlated with meiotic DNA recombination and repair. Since micronuclei are transcriptionally inactive during vegetative growth, our data lend support to the idea that HMG B does not function exclusively in the establishment of transcriptionally competent chromatin. However, micronuclei are transcriptionally active during early stages of conjugation. Evidence that HMG B is strongly synthesized and deposited into micronuclei during this stage is presented. Therefore, it is tempting to suggest that HMG B may play an important role in remodeling micronuclear chromatin into an "active," more open configuration. We favor a model wherein HMG B, like other abundant, low-specificity HMG box-containing proteins, functions to wrap DNA, presumably modulating higher-order chromatin structure for a broad range of biological processes, including transcription and replication.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allis C. D., Colavito-Shepanski M., Gorovsky M. A. Scheduled and unscheduled DNA synthesis during development in conjugating Tetrahymena. Dev Biol. 1987 Dec;124(2):469–480. doi: 10.1016/0012-1606(87)90500-8. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Dennison D. K. Identification and purification of young macronuclear anlagen from conjugating cells of Tetrahymena thermophila. Dev Biol. 1982 Oct;93(2):519–533. doi: 10.1016/0012-1606(82)90139-7. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Wiggins J. C. Histone rearrangements accompany nuclear differentiation and dedifferentiation in Tetrahymena. Dev Biol. 1984 Feb;101(2):282–294. doi: 10.1016/0012-1606(84)90142-8. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Ziegler Y. S., Gorovsky M. A., Olmsted J. B. A conserved histone variant enriched in nucleoli of mammalian cells. Cell. 1982 Nov;31(1):131–136. doi: 10.1016/0092-8674(82)90412-3. [DOI] [PubMed] [Google Scholar]
- Bruns P. J., Brussard T. B. Pair formation in tetrahymena pyriformis, an inducible developmental system. J Exp Zool. 1974 Jun;188(3):337–344. doi: 10.1002/jez.1401880309. [DOI] [PubMed] [Google Scholar]
- Bustin M., Lehn D. A., Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990 Jul 30;1049(3):231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. DNA binding properties of an HMG1-related protein from yeast mitochondria. J Biol Chem. 1992 Feb 15;267(5):3368–3374. [PubMed] [Google Scholar]
- Fisher R. P., Lisowsky T., Parisi M. A., Clayton D. A. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992 Feb 15;267(5):3358–3367. [PubMed] [Google Scholar]
- Giese K., Cox J., Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992 Apr 3;69(1):185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Hamana K., Iwai K. High mobility group nonhistone chromosomal proteins also exist in Tetrahymena. J Biochem. 1979 Sep;86(3):789–794. doi: 10.1093/oxfordjournals.jbchem.a132586. [DOI] [PubMed] [Google Scholar]
- Jackson V. Deposition of newly synthesized histones: new histones H2A and H2B do not deposit in the same nucleosome with new histones H3 and H4. Biochemistry. 1987 Apr 21;26(8):2315–2325. doi: 10.1021/bi00382a037. [DOI] [PubMed] [Google Scholar]
- Kolodrubetz D., Burgum A. Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J Biol Chem. 1990 Feb 25;265(6):3234–3239. [PubMed] [Google Scholar]
- Levy-Wilson B., Denker M. S., Ito E. Isolation, characterization, and postsynthetic modifications of tetrahymena high mobility group proteins. Biochemistry. 1983 Mar 29;22(7):1715–1721. doi: 10.1021/bi00276a030. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. DNA--protein interactions. HMG has DNA wrapped up. Nature. 1992 May 28;357(6376):282–283. doi: 10.1038/357282a0. [DOI] [PubMed] [Google Scholar]
- Lin R., Leone J. W., Cook R. G., Allis C. D. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J Cell Biol. 1989 May;108(5):1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W., Allis C. D., Bruns P. J. RNA and protein synthesis during meiotic prophase in Tetrahymena thermophila. J Protozool. 1985 Nov;32(4):644–649. doi: 10.1111/j.1550-7408.1985.tb03094.x. [DOI] [PubMed] [Google Scholar]
- Ner S. S. HMGs everywhere. Curr Biol. 1992 Apr;2(4):208–210. doi: 10.1016/0960-9822(92)90541-h. [DOI] [PubMed] [Google Scholar]
- Schulman I. G., Cook R. G., Richman R., Allis C. D. Tetrahymena contain two distinct and unusual high mobility group (HMG)-like proteins. J Cell Biol. 1987 Jun;104(6):1485–1494. doi: 10.1083/jcb.104.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman I. G., Wang T. T., Stargell L. A., Gorovsky M. A., Allis C. D. Cell-cell interactions trigger the rapid induction of a specific high mobility group-like protein during early stages of conjugation in Tetrahymena. Dev Biol. 1991 Feb;143(2):248–257. doi: 10.1016/0012-1606(91)90075-e. [DOI] [PubMed] [Google Scholar]
- Schulman I. G., Wang T., Wu M., Bowen J., Cook R. G., Gorovsky M. A., Allis C. D. Macronuclei and micronuclei in Tetrahymena thermophila contain high-mobility-group-like chromosomal proteins containing a highly conserved eleven-amino-acid putative DNA-binding sequence. Mol Cell Biol. 1991 Jan;11(1):166–174. doi: 10.1128/mcb.11.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai T., Hiwatashi K. Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in Tetrahymena pyriformis. J Protozool. 1974 Oct;21(4):542–548. doi: 10.1111/j.1550-7408.1974.tb03695.x. [DOI] [PubMed] [Google Scholar]
- Wagner C. R., Hamana K., Elgin S. C. A high-mobility-group protein and its cDNAs from Drosophila melanogaster. Mol Cell Biol. 1992 May;12(5):1915–1923. doi: 10.1128/mcb.12.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkert D., Allis C. D. Timing of the appearance of macronuclear-specific histone variant hv1 and gene expression in developing new macronuclei of Tetrahymena thermophila. J Cell Biol. 1984 Jun;98(6):2107–2117. doi: 10.1083/jcb.98.6.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. M., Allis C. D., Goldfarb D. S., Srivastva A., Weir J. W., Gorovsky M. A. Nucleus-specific and temporally restricted localization of proteins in Tetrahymena macronuclei and micronuclei. J Cell Biol. 1989 Nov;109(5):1983–1992. doi: 10.1083/jcb.109.5.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. M., Gorovsky M. A. Localization and expression of mRNA for a macronuclear-specific histone H2A variant (hv1) during the cell cycle and conjugation of Tetrahymena thermophila. Mol Cell Biol. 1988 Nov;8(11):4780–4786. doi: 10.1128/mcb.8.11.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Allis C. D., Gorovsky M. A. Cell-cycle regulation as a mechanism for targeting proteins to specific DNA sequences in Tetrahymena thermophila. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2205–2209. doi: 10.1073/pnas.85.7.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]