Abstract
The asymmetric unit of the title compound, C14H11NO5, contains two independent molecules in which the dihedral angles between the benzene rings are 89.27 (16) and 77.14 (12)°. In the crystal, molecules are linked by O—H⋯O hydrogen bonds, generating C(8) chains propagating in [010] for one molecule and [001] C(8) chains for the other. The chains are connected by C—H⋯O hydrogen bonds and π–π interactions [shortest centroid–centroid distance = 3.5908 (12)°], generating a three-dimensional network.
Related literature
For general background to aromatic nitro groups, see: Ghosh et al. (2012 ▶); Sugiyama et al. (2002 ▶).
Experimental
Crystal data
C14H11NO5
M r = 273.24
Monoclinic,
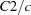
a = 42.313 (6) Å
b = 8.0047 (11) Å
c = 16.1078 (18) Å
β = 105.819 (4)°
V = 5249.2 (12) Å3
Z = 16
Mo Kα radiation
μ = 0.11 mm−1
T = 298 K
0.24 × 0.20 × 0.16 mm
Data collection
Bruker SMART CCD diffractometer
Absorption correction: ψ scan (SADABS; Sheldrick, 2007 ▶) T min = 0.975, T max = 0.983
28460 measured reflections
4602 independent reflections
3349 reflections with I > 2σ(I)
R int = 0.054
Refinement
R[F 2 > 2σ(F 2)] = 0.047
wR(F 2) = 0.143
S = 1.02
4602 reflections
361 parameters
H-atom parameters constrained
Δρmax = 0.26 e Å−3
Δρmin = −0.20 e Å−3
Data collection: SMART (Bruker, 2001 ▶); cell refinement: SAINT (Bruker, 2001 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 2012 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812048271/hb6999sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812048271/hb6999Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812048271/hb6999Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1A—H1A⋯O2A i | 0.82 | 1.94 | 2.753 (2) | 172 |
| O1B—H1B⋯O2B ii | 0.82 | 1.94 | 2.727 (3) | 160 |
| C7B—H7B1⋯O4A iii | 0.96 | 2.50 | 3.418 (3) | 159 |
| C12A—H12A⋯O2A i | 0.93 | 2.54 | 3.245 (2) | 132 |
| C19B—H19B⋯O5A iv | 0.93 | 2.58 | 3.476 (4) | 163 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii) 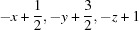 ; (iv)
; (iv)  .
.
Acknowledgments
The authors thank Professor T. N. Guru Row, Solid State and Structural Chemistry Unit, Indian Institute of Science, Bangalore, and G. B. Sadananda, Department of Studies and Research in Physics, U.C.S. Tumkur University, Tumkur for their help and suggestions.
supplementary crystallographic information
Comment
Electron withdrawing nitro group is an important structural element having good synthetic value in organic synthesis to achieve a wide variety of natural and biological active molecules (Ghosh et al., 2012). Formation of two-component molecular crystals from nitro benzoic acid and aromatic or heterocyclic bases leads to discovery of new functional solid materials for nonlinear optics (Sugiyama et al., 2002).
The asymmetric unit of 4-Nitrophenyl 4-hydroxy-3-methylbenzoate crystallographically two independent molecules are shown in Fig.1. Each independent molecule (a and b) is approximately perpendicular to each other; the dihedral angle is 89.17 (14)°. The dihedral angles between the benzene rings in the two molecules [(C8a–C13a and C15a–C20a) and C8b–C13b and C15b–C20b)] are 89.27 (16)° and 77.14 (12)° respectively. The crystal structure features O—H···O and C—H···O interactions (Table 1) and π—π interactions.
Experimental
A mixture of 4-nitrophenol (0.100 g, 0.072 mol) and 4-hydroxy-3-methylbenzoic acid (0.109 g, 0.072 mol) and dicyclohexyldicarbodimide (0.150 g, 0.11 mol) in 1 ml of dry dimethylsufoxide (DMSO) was irradiated under microwave (600 MHz) for 5 x 60sec. Reaction was monitored by TLC, after completion of reaction; the reaction mixture was poured into ice cold, dilute hydrochloric acid which precipitated as ester. The crude product was filtered through Buckner funnel with vacuum, washed with water, crude precipitate was stirred for 1 hr with a saturated sodium bicarbonate solution to remove excess acid. The ester was again collected by filtration, washed repeatedly with water, air dried, and then repeatedly crystallized to get colourless plates from an ethanol-chloroform solvent mixture.
Refinement
All H atoms were positioned geometrically, with O—H = 0.82, C—H = 0.93 Å for aromatic H, and C—H = 0.96 Å for methyl H,and refined using a riding model with Uiso(H) = 1.5Ueq(C) for methyl H and Uiso(H) = 1.2Ueq(C) for all other H.
Figures
Fig. 1.
The molecular structure of the title compound. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
The packing of molecules.
Crystal data
| C14H11NO5 | F(000) = 2272 |
| Mr = 273.24 | Dx = 1.383 Mg m−3 |
| Monoclinic, C2/c | Melting point: 453 K |
| Hall symbol: -C 2yc | Mo Kα radiation, λ = 0.71073 Å |
| a = 42.313 (6) Å | Cell parameters from 4602 reflections |
| b = 8.0047 (11) Å | θ = 2.0–25.1° |
| c = 16.1078 (18) Å | µ = 0.11 mm−1 |
| β = 105.819 (4)° | T = 298 K |
| V = 5249.2 (12) Å3 | Plate, colourless |
| Z = 16 | 0.24 × 0.20 × 0.16 mm |
Data collection
| Bruker SMART CCD diffractometer | 4602 independent reflections |
| Radiation source: fine-focus sealed tube | 3349 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.054 |
| ω and φ scans | θmax = 25.1°, θmin = 2.0° |
| Absorption correction: ψ scan (SADABS; Sheldrick, 2007) | h = −49→50 |
| Tmin = 0.975, Tmax = 0.983 | k = −9→9 |
| 28460 measured reflections | l = −19→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.047 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.143 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0757P)2 + 2.2456P] where P = (Fo2 + 2Fc2)/3 |
| 4602 reflections | (Δ/σ)max < 0.001 |
| 361 parameters | Δρmax = 0.26 e Å−3 |
| 0 restraints | Δρmin = −0.20 e Å−3 |
Special details
| Experimental. IR (cm-1); 2923, 2851, 1739, 1603, 1513, 1246, 1199;. 1H-NMR (400 MHz, CDCl3):8.27 (d, 2H, J = 8.56 Hz, Ar—H), 7.79 (m, 2H, Ar—H), 7.43 (m, 2H, Ar—H), 6.78 (m, 1H, Ar—H), 5.01 (s, 1H, Ar—OH), 2.34 (t, 3H, J = 6.5 Hz, Ar—CH3); Elemental analysis: C14H11NO5 requires C, 61.54; H, 4.06; N, 5.13; found C, 61.95;H, 4.34; N, 4.79. |
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1A | −0.03888 (3) | −0.32059 (16) | 0.40683 (10) | 0.0550 (4) | |
| H1A | −0.0263 | −0.3968 | 0.4036 | 0.082* | |
| O2A | 0.00229 (4) | 0.43257 (18) | 0.37864 (11) | 0.0652 (4) | |
| O3A | 0.04796 (3) | 0.29667 (16) | 0.37629 (10) | 0.0542 (4) | |
| O4A | 0.12917 (6) | 0.9576 (3) | 0.45292 (13) | 0.1007 (7) | |
| O5A | 0.11233 (7) | 0.9690 (3) | 0.31718 (14) | 0.1168 (9) | |
| N6A | 0.11378 (5) | 0.9012 (2) | 0.38481 (14) | 0.0611 (5) | |
| C7A | −0.07695 (5) | −0.0418 (3) | 0.41356 (16) | 0.0605 (6) | |
| H7A1 | −0.0824 | −0.1574 | 0.4175 | 0.091* | |
| H7A2 | −0.0930 | 0.0096 | 0.3666 | 0.091* | |
| H7A3 | −0.0767 | 0.0139 | 0.4665 | 0.091* | |
| C8A | −0.04364 (5) | −0.0287 (2) | 0.39804 (12) | 0.0432 (5) | |
| C9A | −0.02951 (5) | 0.1231 (2) | 0.38957 (12) | 0.0446 (5) | |
| H9A | −0.0412 | 0.2204 | 0.3921 | 0.053* | |
| C10A | 0.00168 (5) | 0.1357 (2) | 0.37741 (12) | 0.0418 (4) | |
| C11A | 0.01863 (5) | −0.0096 (2) | 0.37014 (13) | 0.0462 (5) | |
| H11A | 0.0392 | −0.0033 | 0.3598 | 0.055* | |
| C12A | 0.00513 (5) | −0.1623 (2) | 0.37818 (13) | 0.0489 (5) | |
| H12A | 0.0165 | −0.2595 | 0.3729 | 0.059* | |
| C13A | −0.02537 (5) | −0.1723 (2) | 0.39407 (12) | 0.0423 (4) | |
| C14A | 0.01605 (5) | 0.3012 (2) | 0.37702 (12) | 0.0440 (5) | |
| C15A | 0.06391 (5) | 0.4505 (2) | 0.37895 (13) | 0.0462 (5) | |
| C16A | 0.08228 (5) | 0.5103 (3) | 0.45681 (14) | 0.0542 (5) | |
| H16A | 0.0835 | 0.4515 | 0.5074 | 0.065* | |
| C17A | 0.09898 (5) | 0.6592 (3) | 0.45916 (13) | 0.0526 (5) | |
| H17A | 0.1117 | 0.7023 | 0.5112 | 0.063* | |
| C18A | 0.09643 (5) | 0.7423 (3) | 0.38305 (13) | 0.0465 (5) | |
| C19A | 0.07829 (5) | 0.6819 (3) | 0.30503 (13) | 0.0530 (5) | |
| H19A | 0.0771 | 0.7404 | 0.2544 | 0.064* | |
| C20A | 0.06183 (5) | 0.5328 (3) | 0.30305 (14) | 0.0544 (5) | |
| H20A | 0.0495 | 0.4886 | 0.2509 | 0.065* | |
| O1B | 0.29237 (4) | 0.4579 (3) | 0.43358 (10) | 0.0803 (6) | |
| H1B | 0.2789 | 0.4362 | 0.4602 | 0.120* | |
| O2B | 0.24794 (5) | 0.5330 (3) | 0.02839 (11) | 0.1045 (8) | |
| O3B | 0.20070 (4) | 0.4732 (2) | 0.05604 (9) | 0.0719 (5) | |
| O4B | 0.13548 (7) | 0.4123 (4) | −0.33906 (14) | 0.1226 (9) | |
| O5B | 0.11288 (6) | 0.6376 (3) | −0.31360 (14) | 0.1102 (8) | |
| N6B | 0.13174 (6) | 0.5217 (3) | −0.29087 (14) | 0.0750 (6) | |
| C7B | 0.33294 (6) | 0.5292 (5) | 0.33073 (18) | 0.0921 (10) | |
| H7B1 | 0.3386 | 0.5163 | 0.3923 | 0.138* | |
| H7B2 | 0.3383 | 0.6403 | 0.3168 | 0.138* | |
| H7B3 | 0.3450 | 0.4497 | 0.3068 | 0.138* | |
| C8B | 0.29669 (5) | 0.4997 (3) | 0.29354 (13) | 0.0557 (6) | |
| C9B | 0.28152 (5) | 0.5074 (3) | 0.20657 (14) | 0.0582 (6) | |
| H9B | 0.2942 | 0.5300 | 0.1689 | 0.070* | |
| C10B | 0.24797 (5) | 0.4827 (3) | 0.17292 (13) | 0.0515 (5) | |
| C11B | 0.22927 (6) | 0.4457 (3) | 0.22887 (14) | 0.0628 (6) | |
| H11B | 0.2068 | 0.4271 | 0.2075 | 0.075* | |
| C12B | 0.24386 (6) | 0.4367 (4) | 0.31560 (14) | 0.0663 (7) | |
| H12B | 0.2312 | 0.4119 | 0.3531 | 0.080* | |
| C13B | 0.27708 (5) | 0.4640 (3) | 0.34785 (13) | 0.0553 (5) | |
| C14B | 0.23353 (6) | 0.4987 (3) | 0.08041 (14) | 0.0615 (6) | |
| C15B | 0.18461 (5) | 0.4871 (3) | −0.03191 (14) | 0.0592 (6) | |
| C16B | 0.18584 (6) | 0.3559 (3) | −0.08597 (16) | 0.0668 (6) | |
| H16B | 0.1981 | 0.2609 | −0.0650 | 0.080* | |
| C17B | 0.16863 (6) | 0.3677 (3) | −0.17177 (15) | 0.0654 (6) | |
| H17B | 0.1693 | 0.2816 | −0.2100 | 0.078* | |
| C18B | 0.15043 (6) | 0.5094 (3) | −0.19964 (14) | 0.0591 (6) | |
| C19B | 0.14870 (6) | 0.6388 (3) | −0.14563 (16) | 0.0646 (6) | |
| H19B | 0.1360 | 0.7325 | −0.1661 | 0.077* | |
| C20B | 0.16627 (6) | 0.6272 (3) | −0.06012 (15) | 0.0647 (6) | |
| H20B | 0.1657 | 0.7136 | −0.0220 | 0.078* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1A | 0.0550 (9) | 0.0383 (7) | 0.0738 (10) | −0.0022 (6) | 0.0213 (8) | 0.0009 (6) |
| O2A | 0.0512 (9) | 0.0370 (8) | 0.1076 (13) | 0.0056 (7) | 0.0219 (9) | 0.0054 (8) |
| O3A | 0.0462 (8) | 0.0380 (7) | 0.0792 (10) | −0.0012 (6) | 0.0183 (7) | 0.0010 (7) |
| O4A | 0.1216 (18) | 0.0931 (14) | 0.0767 (13) | −0.0565 (13) | 0.0087 (12) | −0.0182 (11) |
| O5A | 0.167 (2) | 0.0962 (16) | 0.0801 (14) | −0.0701 (16) | 0.0215 (15) | 0.0111 (11) |
| N6A | 0.0621 (12) | 0.0575 (11) | 0.0652 (13) | −0.0157 (9) | 0.0198 (10) | −0.0026 (10) |
| C7A | 0.0465 (12) | 0.0540 (13) | 0.0842 (16) | −0.0002 (10) | 0.0234 (12) | −0.0030 (11) |
| C8A | 0.0384 (11) | 0.0444 (11) | 0.0441 (11) | 0.0027 (8) | 0.0068 (9) | −0.0019 (8) |
| C9A | 0.0422 (11) | 0.0360 (10) | 0.0531 (12) | 0.0072 (8) | 0.0089 (9) | −0.0016 (8) |
| C10A | 0.0418 (11) | 0.0379 (10) | 0.0429 (11) | 0.0038 (8) | 0.0068 (9) | 0.0014 (8) |
| C11A | 0.0419 (11) | 0.0412 (11) | 0.0563 (12) | 0.0032 (8) | 0.0149 (10) | −0.0017 (9) |
| C12A | 0.0483 (12) | 0.0353 (10) | 0.0647 (13) | 0.0063 (8) | 0.0183 (10) | −0.0025 (9) |
| C13A | 0.0448 (11) | 0.0379 (10) | 0.0403 (10) | −0.0009 (8) | 0.0050 (9) | −0.0016 (8) |
| C14A | 0.0408 (11) | 0.0393 (10) | 0.0486 (12) | 0.0047 (9) | 0.0069 (9) | 0.0028 (8) |
| C15A | 0.0390 (11) | 0.0406 (10) | 0.0591 (13) | −0.0001 (8) | 0.0134 (10) | 0.0009 (9) |
| C16A | 0.0580 (13) | 0.0565 (13) | 0.0459 (12) | −0.0025 (10) | 0.0105 (11) | 0.0070 (10) |
| C17A | 0.0548 (13) | 0.0571 (13) | 0.0420 (11) | −0.0066 (10) | 0.0064 (10) | −0.0048 (9) |
| C18A | 0.0410 (11) | 0.0476 (11) | 0.0508 (12) | −0.0037 (9) | 0.0120 (9) | −0.0037 (9) |
| C19A | 0.0543 (13) | 0.0583 (13) | 0.0443 (12) | −0.0088 (10) | 0.0100 (10) | 0.0053 (9) |
| C20A | 0.0507 (13) | 0.0594 (13) | 0.0470 (12) | −0.0101 (10) | 0.0029 (10) | −0.0025 (10) |
| O1B | 0.0576 (10) | 0.1384 (17) | 0.0412 (9) | 0.0057 (10) | 0.0071 (8) | 0.0077 (9) |
| O2B | 0.0605 (11) | 0.207 (3) | 0.0457 (10) | −0.0223 (13) | 0.0144 (9) | 0.0125 (12) |
| O3B | 0.0469 (9) | 0.1225 (15) | 0.0421 (9) | −0.0040 (9) | 0.0049 (7) | 0.0036 (9) |
| O4B | 0.135 (2) | 0.151 (2) | 0.0596 (13) | 0.0139 (17) | −0.0116 (13) | −0.0215 (14) |
| O5B | 0.1202 (18) | 0.1087 (17) | 0.0772 (14) | 0.0137 (15) | −0.0145 (13) | 0.0291 (12) |
| N6B | 0.0712 (15) | 0.0898 (17) | 0.0529 (13) | −0.0161 (13) | −0.0015 (11) | 0.0063 (12) |
| C7B | 0.0491 (15) | 0.161 (3) | 0.0627 (16) | −0.0108 (17) | 0.0101 (12) | 0.0000 (17) |
| C8B | 0.0443 (12) | 0.0739 (15) | 0.0475 (12) | 0.0047 (10) | 0.0105 (10) | −0.0010 (10) |
| C9B | 0.0485 (13) | 0.0830 (16) | 0.0463 (12) | 0.0003 (11) | 0.0183 (11) | 0.0036 (11) |
| C10B | 0.0441 (12) | 0.0677 (14) | 0.0423 (11) | 0.0020 (10) | 0.0114 (9) | −0.0009 (10) |
| C11B | 0.0436 (12) | 0.0952 (18) | 0.0487 (13) | −0.0038 (12) | 0.0112 (10) | 0.0032 (12) |
| C12B | 0.0507 (14) | 0.1058 (19) | 0.0449 (13) | −0.0032 (13) | 0.0173 (11) | 0.0041 (12) |
| C13B | 0.0486 (13) | 0.0741 (15) | 0.0409 (12) | 0.0076 (11) | 0.0083 (10) | 0.0022 (10) |
| C14B | 0.0481 (13) | 0.0926 (18) | 0.0444 (12) | −0.0005 (12) | 0.0137 (11) | −0.0012 (11) |
| C15B | 0.0446 (12) | 0.0875 (17) | 0.0426 (12) | −0.0035 (11) | 0.0069 (10) | 0.0025 (11) |
| C16B | 0.0604 (15) | 0.0727 (16) | 0.0593 (15) | 0.0090 (12) | 0.0022 (12) | 0.0038 (12) |
| C17B | 0.0601 (14) | 0.0746 (16) | 0.0561 (14) | −0.0014 (12) | 0.0067 (12) | −0.0092 (12) |
| C18B | 0.0507 (13) | 0.0717 (15) | 0.0484 (13) | −0.0107 (11) | 0.0023 (11) | 0.0071 (11) |
| C19B | 0.0586 (14) | 0.0637 (14) | 0.0648 (16) | 0.0007 (11) | 0.0056 (12) | 0.0063 (12) |
| C20B | 0.0606 (15) | 0.0724 (16) | 0.0572 (15) | −0.0012 (12) | 0.0097 (12) | −0.0074 (12) |
Geometric parameters (Å, º)
| O1A—C13A | 1.357 (2) | O1B—C13B | 1.357 (3) |
| O1A—H1A | 0.8200 | O1B—H1B | 0.8200 |
| O2A—C14A | 1.205 (2) | O2B—C14B | 1.195 (3) |
| O3A—C14A | 1.354 (2) | O3B—C14B | 1.352 (3) |
| O3A—C15A | 1.399 (2) | O3B—C15B | 1.399 (3) |
| O4A—N6A | 1.201 (3) | O4B—N6B | 1.209 (3) |
| O5A—N6A | 1.204 (3) | O5B—N6B | 1.214 (3) |
| N6A—C18A | 1.465 (3) | N6B—C18B | 1.470 (3) |
| C7A—C8A | 1.501 (3) | C7B—C8B | 1.505 (3) |
| C7A—H7A1 | 0.9600 | C7B—H7B1 | 0.9600 |
| C7A—H7A2 | 0.9600 | C7B—H7B2 | 0.9600 |
| C7A—H7A3 | 0.9600 | C7B—H7B3 | 0.9600 |
| C8A—C9A | 1.377 (3) | C8B—C9B | 1.374 (3) |
| C8A—C13A | 1.396 (3) | C8B—C13B | 1.390 (3) |
| C9A—C10A | 1.390 (3) | C9B—C10B | 1.389 (3) |
| C9A—H9A | 0.9300 | C9B—H9B | 0.9300 |
| C10A—C11A | 1.388 (3) | C10B—C11B | 1.384 (3) |
| C10A—C14A | 1.459 (3) | C10B—C14B | 1.454 (3) |
| C11A—C12A | 1.371 (3) | C11B—C12B | 1.367 (3) |
| C11A—H11A | 0.9300 | C11B—H11B | 0.9300 |
| C12A—C13A | 1.385 (3) | C12B—C13B | 1.377 (3) |
| C12A—H12A | 0.9300 | C12B—H12B | 0.9300 |
| C15A—C16A | 1.369 (3) | C15B—C20B | 1.369 (3) |
| C15A—C20A | 1.370 (3) | C15B—C16B | 1.374 (3) |
| C16A—C17A | 1.381 (3) | C16B—C17B | 1.378 (3) |
| C16A—H16A | 0.9300 | C16B—H16B | 0.9300 |
| C17A—C18A | 1.373 (3) | C17B—C18B | 1.376 (3) |
| C17A—H17A | 0.9300 | C17B—H17B | 0.9300 |
| C18A—C19A | 1.370 (3) | C18B—C19B | 1.367 (3) |
| C19A—C20A | 1.378 (3) | C19B—C20B | 1.379 (3) |
| C19A—H19A | 0.9300 | C19B—H19B | 0.9300 |
| C20A—H20A | 0.9300 | C20B—H20B | 0.9300 |
| C13A—O1A—H1A | 109.5 | C13B—O1B—H1B | 109.5 |
| C14A—O3A—C15A | 116.73 (15) | C14B—O3B—C15B | 117.38 (17) |
| O4A—N6A—O5A | 122.6 (2) | O4B—N6B—O5B | 123.4 (2) |
| O4A—N6A—C18A | 119.2 (2) | O4B—N6B—C18B | 117.7 (3) |
| O5A—N6A—C18A | 118.2 (2) | O5B—N6B—C18B | 118.9 (2) |
| C8A—C7A—H7A1 | 109.5 | C8B—C7B—H7B1 | 109.5 |
| C8A—C7A—H7A2 | 109.5 | C8B—C7B—H7B2 | 109.5 |
| H7A1—C7A—H7A2 | 109.5 | H7B1—C7B—H7B2 | 109.5 |
| C8A—C7A—H7A3 | 109.5 | C8B—C7B—H7B3 | 109.5 |
| H7A1—C7A—H7A3 | 109.5 | H7B1—C7B—H7B3 | 109.5 |
| H7A2—C7A—H7A3 | 109.5 | H7B2—C7B—H7B3 | 109.5 |
| C9A—C8A—C13A | 117.47 (17) | C9B—C8B—C13B | 117.3 (2) |
| C9A—C8A—C7A | 122.07 (17) | C9B—C8B—C7B | 122.7 (2) |
| C13A—C8A—C7A | 120.43 (17) | C13B—C8B—C7B | 120.0 (2) |
| C8A—C9A—C10A | 122.14 (17) | C8B—C9B—C10B | 122.36 (19) |
| C8A—C9A—H9A | 118.9 | C8B—C9B—H9B | 118.8 |
| C10A—C9A—H9A | 118.9 | C10B—C9B—H9B | 118.8 |
| C11A—C10A—C9A | 118.95 (17) | C11B—C10B—C9B | 118.8 (2) |
| C11A—C10A—C14A | 122.36 (17) | C11B—C10B—C14B | 122.1 (2) |
| C9A—C10A—C14A | 118.62 (16) | C9B—C10B—C14B | 119.12 (19) |
| C12A—C11A—C10A | 120.06 (18) | C12B—C11B—C10B | 119.9 (2) |
| C12A—C11A—H11A | 120.0 | C12B—C11B—H11B | 120.1 |
| C10A—C11A—H11A | 120.0 | C10B—C11B—H11B | 120.1 |
| C11A—C12A—C13A | 120.17 (17) | C11B—C12B—C13B | 120.5 (2) |
| C11A—C12A—H12A | 119.9 | C11B—C12B—H12B | 119.8 |
| C13A—C12A—H12A | 119.9 | C13B—C12B—H12B | 119.8 |
| O1A—C13A—C12A | 122.05 (17) | O1B—C13B—C12B | 122.19 (19) |
| O1A—C13A—C8A | 116.87 (17) | O1B—C13B—C8B | 116.57 (19) |
| C12A—C13A—C8A | 121.08 (17) | C12B—C13B—C8B | 121.2 (2) |
| O2A—C14A—O3A | 120.82 (17) | O2B—C14B—O3B | 120.8 (2) |
| O2A—C14A—C10A | 126.02 (18) | O2B—C14B—C10B | 125.9 (2) |
| O3A—C14A—C10A | 113.15 (16) | O3B—C14B—C10B | 113.31 (18) |
| C16A—C15A—C20A | 122.09 (19) | C20B—C15B—C16B | 122.1 (2) |
| C16A—C15A—O3A | 118.98 (18) | C20B—C15B—O3B | 118.5 (2) |
| C20A—C15A—O3A | 118.86 (19) | C16B—C15B—O3B | 119.3 (2) |
| C15A—C16A—C17A | 119.05 (19) | C15B—C16B—C17B | 118.8 (2) |
| C15A—C16A—H16A | 120.5 | C15B—C16B—H16B | 120.6 |
| C17A—C16A—H16A | 120.5 | C17B—C16B—H16B | 120.6 |
| C18A—C17A—C16A | 118.60 (19) | C18B—C17B—C16B | 118.6 (2) |
| C18A—C17A—H17A | 120.7 | C18B—C17B—H17B | 120.7 |
| C16A—C17A—H17A | 120.7 | C16B—C17B—H17B | 120.7 |
| C19A—C18A—C17A | 122.41 (19) | C19B—C18B—C17B | 122.6 (2) |
| C19A—C18A—N6A | 118.49 (18) | C19B—C18B—N6B | 118.6 (2) |
| C17A—C18A—N6A | 119.10 (19) | C17B—C18B—N6B | 118.8 (2) |
| C18A—C19A—C20A | 118.71 (19) | C18B—C19B—C20B | 118.5 (2) |
| C18A—C19A—H19A | 120.6 | C18B—C19B—H19B | 120.8 |
| C20A—C19A—H19A | 120.6 | C20B—C19B—H19B | 120.8 |
| C15A—C20A—C19A | 119.1 (2) | C15B—C20B—C19B | 119.3 (2) |
| C15A—C20A—H20A | 120.4 | C15B—C20B—H20B | 120.4 |
| C19A—C20A—H20A | 120.4 | C19B—C20B—H20B | 120.4 |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1A—H1A···O2Ai | 0.82 | 1.94 | 2.753 (2) | 172 |
| O1B—H1B···O2Bii | 0.82 | 1.94 | 2.727 (3) | 160 |
| C7B—H7B1···O4Aiii | 0.96 | 2.50 | 3.418 (3) | 159 |
| C12A—H12A···O2Ai | 0.93 | 2.54 | 3.245 (2) | 132 |
| C19B—H19B···O5Aiv | 0.93 | 2.58 | 3.476 (4) | 163 |
Symmetry codes: (i) x, y−1, z; (ii) x, −y+1, z+1/2; (iii) −x+1/2, −y+3/2, −z+1; (iv) x, −y+2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB6999).
References
- Bruker (2001). SMART and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Ghosh, P. P., Pal, G., Paul, S. & Das, A. R. (2012). Green Chem. 14, 2691–2698.
- Sheldrick, G. M. (2007). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sugiyama, T., Meng, J., Wen, Z., Li, J. & Matsuura, T. (2002). Mol. Cryst. Liq. Cryst. 389, 17–23.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812048271/hb6999sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812048271/hb6999Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812048271/hb6999Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




