Abstract
In the title compound, C20H20O4 {systematic name: 4-[(3R)-8,8-dimethyl-3,4-dihydro-2H-pyrano[2,3-f]chromen-3-yl]benzene-1,3-diol}, the hydropyran ring linked to the pendant benzene ring adopts an envelope conformation, with the methyne C atom forming the flap. In the crystal, the –OH group at the 3-position of the benzene ring forms an O—H⋯O hydrogen bond to a chromene O-atom acceptor, whereas the –OH group at the 1-position forms an O—H⋯π interaction with a neighboring benzene ring. The O—H⋯O hydrogen bonds form [001] chains and the O—H⋯π bonds cross-link the chains into (101) sheets. The absolute structure was assumed to be the same as that deduced from previous studies for the natural product.
Related literature
For background to the pharmacological activity of the title compound, see: Fukai et al. (2000 ▶); Messier & Grenier; (2011 ▶); Thiyagarajan et al. (2011 ▶); Ahn et al. (2012 ▶); Choi (2005 ▶). For the assignment of the absolute structure, see: Kim et al. (2009 ▶).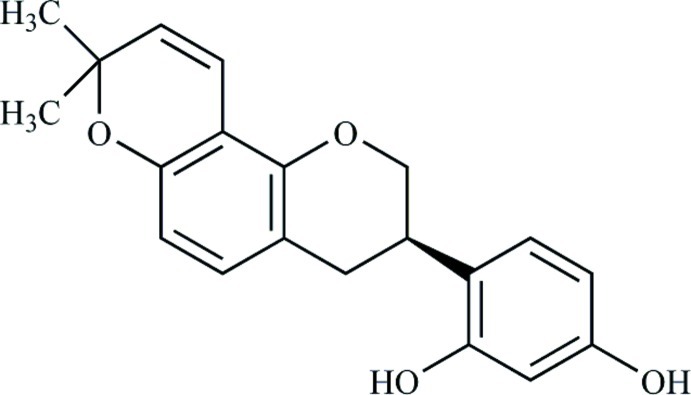
Experimental
Crystal data
C20H20O4
M r = 324.36
Orthorhombic,
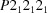
a = 6.4301 (4) Å
b = 12.0307 (7) Å
c = 21.0690 (13) Å
V = 1629.87 (17) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 293 K
0.22 × 0.14 × 0.07 mm
Data collection
Bruker APEX CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2003 ▶) T min = 0.984, T max = 0.994
15459 measured reflections
2866 independent reflections
2551 reflections with I > 2σ(I)
R int = 0.045
Refinement
R[F 2 > 2σ(F 2)] = 0.046
wR(F 2) = 0.095
S = 1.16
2866 reflections
225 parameters
2 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.15 e Å−3
Δρmin = −0.17 e Å−3
Data collection: SMART (Bruker, 1998 ▶); cell refinement: SAINT (Bruker, 2003 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: SHELXTL (Sheldrick, 2008 ▶) and publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812048647/hb6989sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812048647/hb6989Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812048647/hb6989Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg4 is the centroid of the C13–C18 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O4—H4A⋯O1i | 0.82 (2) | 2.02 (2) | 2.841 (3) | 177 (3) |
| O3—H3A⋯Cg4ii | 0.80 (2) | 2.51 (2) | 3.213 (2) | 148 (3) |
Symmetry codes: (i) 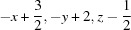 ; (ii)
; (ii) 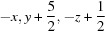 .
.
Acknowledgments
Financial support by the Nanotechnology Center (NANOTEC), NSTDA, Ministry of Science and Technology, Thailand, through Center of Excellence Network program is gratefully acknowledged.
supplementary crystallographic information
Comment
4-[(3R)-8,8-Dimethyl-3,4-dihydro-2H-pyrano[2,3-f]chromen-3-yl]benzene-1,3-diol (Glabridin) is a pyranoisoflavan isolated from licorice. It has various pharmacological activities such as cytotoxic activity (Fukai et al., 2000), antimicrobial activity (Messier & Grenier, 2011), anti-inflammation (Thiyagarajan et al., 2011), anti-obesity effect (Ahn et al., 2012) and prevention for osteoporosis and inflammatory bone diseases (Choi, 2005). For the assignment of its absolute structure, see: Kim et al. (2009).
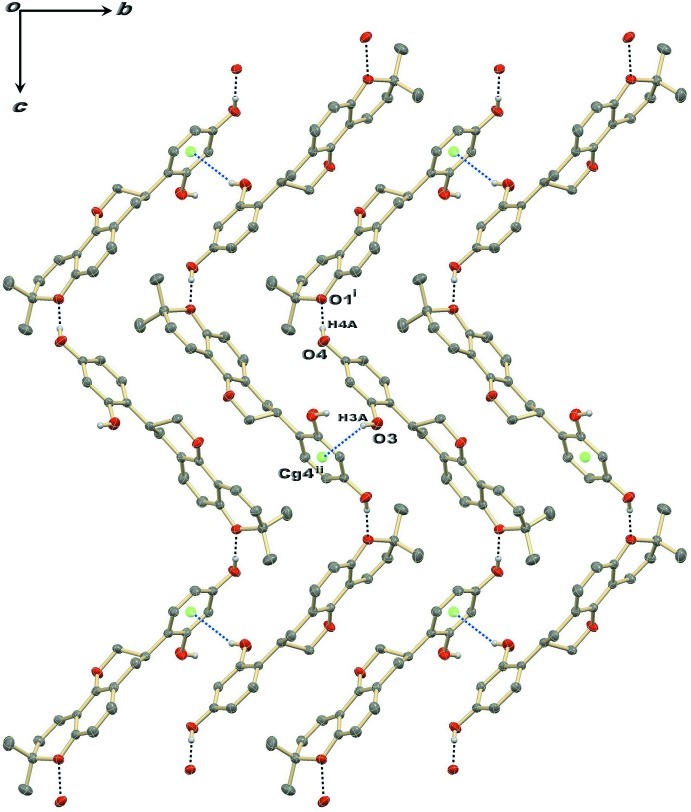
The molecular structure of the title compound is shown in Fig. 1. The packing features O—H···O intermolecular hydrogen bonding between hydroxyl group at 3 position of benzene ring with the donor-acceptor distance of 2.841 (3) Å. (O4—H4A···O1i; i: -x + 3/2, -y + 2, z - 1/2) forming a zigzag chains running parallel to the [001] direction. Besides, the O—H···π interactions with O···centroid distances of 3.213 (2) Å are observed between hydroxyl group at the 1 position of benzene ring and the nearby benzene ring of adjacent molecule, O3—H3A···Cg4ii (Cg4 is the centroid of C13—C14—C15—C16—C17—C18 and the symmetry code ii is -x, y + 5/2, -z + 1/2), linking among the zigzag chains generating two-dimensional layer parallel to (101) plane. The crystal packing of interaction is depicted in Fig. 2.
Experimental
The title compound was obtained from Nanjing Zelang Medical Technology Co. Ltd. Colourless blocks were obtained by dissolving the compound in methanol followed by a slow evaporation of the solvent.
Refinement
Amonalous dispersion was found to be negligible and the absolute structure is indeterminate. Friedel pairs were merged before the final refinement. H atoms on carbon atoms were positioned geometrically and constrained to ride on their parent atoms with Uiso(H) = 1.2Ueq(C-sp2) with C—H = 0.93 Å and 1.5Ueq(C-sp3) with the distances ranging from 0.96 to 0.98 Å, respectively. The H atoms on the oxygen atoms were located in a difference Fourier map and restrained with Uiso(H) = 1.2Ueq(OH). (O—H = 0.80 (2) and 0.82 (2) Å).
Figures
Fig. 1.
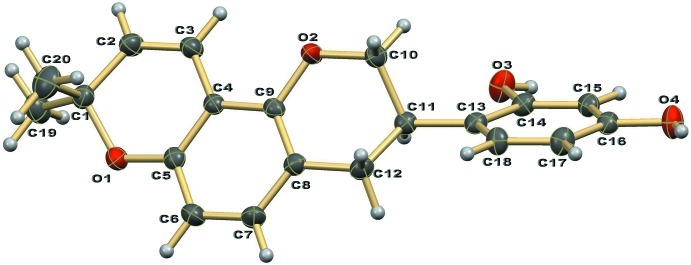
The molecular structure of the title compound.
Fig. 2.
The packing of the intermolecular interactions of the title compound is plotted down a axis.
Crystal data
| C20H20O4 | F(000) = 688 |
| Mr = 324.36 | Dx = 1.322 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 2104 reflections |
| a = 6.4301 (4) Å | θ = 3.3–21.0° |
| b = 12.0307 (7) Å | µ = 0.09 mm−1 |
| c = 21.0690 (13) Å | T = 293 K |
| V = 1629.87 (17) Å3 | Block, colourless |
| Z = 4 | 0.22 × 0.14 × 0.07 mm |
Data collection
| Bruker APEX CCD diffractometer | 2866 independent reflections |
| Radiation source: fine-focus sealed tube | 2551 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.045 |
| Frames, each covering 0.3 ° in ω scans | θmax = 25.0°, θmin = 1.9° |
| Absorption correction: multi-scan (SADABS; Bruker, 2003) | h = −7→7 |
| Tmin = 0.984, Tmax = 0.994 | k = −14→14 |
| 15459 measured reflections | l = −25→25 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.046 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.095 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.16 | w = 1/[σ2(Fo2) + (0.0365P)2 + 0.2296P] where P = (Fo2 + 2Fc2)/3 |
| 2866 reflections | (Δ/σ)max < 0.001 |
| 225 parameters | Δρmax = 0.15 e Å−3 |
| 2 restraints | Δρmin = −0.17 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against all reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on all data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.7767 (3) | 0.66156 (13) | 0.23880 (7) | 0.0443 (4) | |
| O2 | 1.0695 (3) | 0.80853 (13) | 0.05050 (7) | 0.0477 (5) | |
| C1 | 0.9085 (4) | 0.56254 (19) | 0.23598 (11) | 0.0417 (6) | |
| C2 | 1.1009 (4) | 0.5878 (2) | 0.19907 (11) | 0.0458 (6) | |
| H2 | 1.2221 | 0.5486 | 0.2080 | 0.055* | |
| C3 | 1.1042 (4) | 0.6644 (2) | 0.15409 (11) | 0.0424 (6) | |
| H3 | 1.2243 | 0.6754 | 0.1303 | 0.051* | |
| C4 | 0.9211 (4) | 0.73112 (18) | 0.14167 (10) | 0.0326 (5) | |
| C5 | 0.7586 (4) | 0.72632 (18) | 0.18500 (10) | 0.0370 (6) | |
| C6 | 0.5850 (4) | 0.7914 (2) | 0.17877 (12) | 0.0479 (6) | |
| H6 | 0.4783 | 0.7876 | 0.2085 | 0.057* | |
| C7 | 0.5712 (4) | 0.8631 (2) | 0.12745 (12) | 0.0466 (6) | |
| H7 | 0.4530 | 0.9070 | 0.1230 | 0.056* | |
| C8 | 0.7277 (4) | 0.87146 (18) | 0.08254 (11) | 0.0366 (5) | |
| C9 | 0.9013 (4) | 0.80460 (18) | 0.09051 (10) | 0.0340 (5) | |
| C10 | 1.0643 (4) | 0.88701 (19) | −0.00121 (10) | 0.0431 (6) | |
| H10A | 1.0019 | 0.8517 | −0.0380 | 0.052* | |
| H10B | 1.2055 | 0.9076 | −0.0123 | 0.052* | |
| C11 | 0.9426 (4) | 0.99118 (18) | 0.01485 (10) | 0.0357 (5) | |
| H11 | 1.0017 | 1.0217 | 0.0540 | 0.043* | |
| C12 | 0.7201 (4) | 0.95505 (19) | 0.02970 (12) | 0.0435 (6) | |
| H12A | 0.6376 | 1.0188 | 0.0424 | 0.052* | |
| H12B | 0.6567 | 0.9223 | −0.0076 | 0.052* | |
| C13 | 0.9633 (4) | 1.07917 (18) | −0.03561 (10) | 0.0353 (5) | |
| C14 | 1.1391 (4) | 1.14759 (18) | −0.03656 (10) | 0.0364 (6) | |
| C15 | 1.1636 (4) | 1.23098 (19) | −0.08114 (11) | 0.0378 (5) | |
| H15 | 1.2811 | 1.2760 | −0.0802 | 0.045* | |
| C16 | 1.0135 (4) | 1.24727 (18) | −0.12703 (10) | 0.0372 (6) | |
| C17 | 0.8386 (4) | 1.1812 (2) | −0.12775 (11) | 0.0449 (6) | |
| H17 | 0.7366 | 1.1918 | −0.1585 | 0.054* | |
| C18 | 0.8159 (4) | 1.09882 (19) | −0.08240 (11) | 0.0425 (6) | |
| H18 | 0.6970 | 1.0549 | −0.0833 | 0.051* | |
| C19 | 0.9574 (5) | 0.5352 (3) | 0.30460 (12) | 0.0639 (8) | |
| H19A | 1.0368 | 0.5946 | 0.3230 | 0.096* | |
| H19B | 1.0363 | 0.4675 | 0.3065 | 0.096* | |
| H19C | 0.8300 | 0.5261 | 0.3278 | 0.096* | |
| C20 | 0.7812 (6) | 0.4713 (2) | 0.20419 (15) | 0.0721 (9) | |
| H20A | 0.6555 | 0.4593 | 0.2278 | 0.108* | |
| H20B | 0.8608 | 0.4038 | 0.2031 | 0.108* | |
| H20C | 0.7472 | 0.4934 | 0.1617 | 0.108* | |
| O3 | 1.2858 (3) | 1.12753 (14) | 0.00917 (9) | 0.0536 (5) | |
| H3A | 1.362 (4) | 1.1797 (18) | 0.0133 (14) | 0.064* | |
| O4 | 1.0477 (3) | 1.33015 (16) | −0.16975 (8) | 0.0551 (5) | |
| H4A | 0.957 (4) | 1.334 (2) | −0.1969 (11) | 0.066* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0529 (11) | 0.0443 (9) | 0.0356 (9) | 0.0025 (8) | 0.0083 (8) | 0.0037 (8) |
| O2 | 0.0485 (11) | 0.0493 (10) | 0.0452 (9) | 0.0166 (9) | 0.0169 (9) | 0.0160 (8) |
| C1 | 0.0460 (15) | 0.0360 (13) | 0.0431 (13) | −0.0006 (11) | 0.0002 (13) | 0.0026 (11) |
| C2 | 0.0429 (16) | 0.0503 (15) | 0.0443 (14) | 0.0106 (13) | 0.0013 (12) | 0.0064 (12) |
| C3 | 0.0391 (14) | 0.0493 (14) | 0.0389 (13) | 0.0042 (12) | 0.0071 (12) | 0.0038 (12) |
| C4 | 0.0349 (12) | 0.0318 (11) | 0.0312 (11) | −0.0027 (10) | 0.0009 (11) | −0.0031 (10) |
| C5 | 0.0418 (14) | 0.0341 (12) | 0.0351 (12) | −0.0048 (11) | 0.0001 (11) | −0.0035 (10) |
| C6 | 0.0435 (15) | 0.0524 (15) | 0.0479 (14) | 0.0039 (13) | 0.0142 (13) | 0.0032 (12) |
| C7 | 0.0337 (14) | 0.0463 (14) | 0.0598 (15) | 0.0073 (12) | 0.0054 (13) | 0.0070 (13) |
| C8 | 0.0328 (13) | 0.0336 (12) | 0.0433 (13) | −0.0014 (11) | −0.0025 (12) | −0.0027 (11) |
| C9 | 0.0347 (13) | 0.0339 (11) | 0.0335 (12) | −0.0036 (10) | 0.0048 (11) | −0.0061 (10) |
| C10 | 0.0521 (15) | 0.0446 (13) | 0.0326 (12) | 0.0064 (12) | 0.0058 (12) | 0.0053 (11) |
| C11 | 0.0381 (13) | 0.0372 (12) | 0.0319 (12) | 0.0005 (11) | −0.0050 (11) | −0.0018 (10) |
| C12 | 0.0403 (15) | 0.0419 (13) | 0.0482 (14) | 0.0067 (12) | −0.0014 (12) | 0.0038 (12) |
| C13 | 0.0405 (14) | 0.0350 (12) | 0.0304 (12) | 0.0018 (11) | −0.0035 (11) | −0.0038 (10) |
| C14 | 0.0383 (14) | 0.0353 (12) | 0.0355 (12) | 0.0014 (11) | −0.0093 (11) | −0.0044 (11) |
| C15 | 0.0370 (14) | 0.0352 (12) | 0.0413 (12) | −0.0063 (11) | −0.0006 (11) | −0.0031 (11) |
| C16 | 0.0463 (15) | 0.0352 (12) | 0.0302 (12) | 0.0006 (12) | −0.0021 (11) | −0.0016 (10) |
| C17 | 0.0506 (16) | 0.0469 (14) | 0.0372 (13) | −0.0027 (13) | −0.0152 (12) | 0.0017 (12) |
| C18 | 0.0447 (15) | 0.0426 (13) | 0.0402 (13) | −0.0106 (12) | −0.0083 (13) | 0.0005 (12) |
| C19 | 0.067 (2) | 0.076 (2) | 0.0484 (16) | 0.0029 (18) | 0.0022 (16) | 0.0185 (15) |
| C20 | 0.086 (2) | 0.0478 (16) | 0.082 (2) | −0.0106 (18) | −0.008 (2) | −0.0059 (16) |
| O3 | 0.0471 (11) | 0.0522 (11) | 0.0616 (11) | −0.0086 (9) | −0.0261 (10) | 0.0097 (10) |
| O4 | 0.0654 (13) | 0.0539 (11) | 0.0462 (10) | −0.0134 (11) | −0.0119 (9) | 0.0168 (9) |
Geometric parameters (Å, º)
| O1—C5 | 1.380 (3) | C11—C12 | 1.528 (3) |
| O1—C1 | 1.463 (3) | C11—H11 | 0.9800 |
| O2—C9 | 1.372 (3) | C12—H12A | 0.9700 |
| O2—C10 | 1.442 (2) | C12—H12B | 0.9700 |
| C1—C2 | 1.493 (4) | C13—C18 | 1.388 (3) |
| C1—C19 | 1.516 (3) | C13—C14 | 1.398 (3) |
| C1—C20 | 1.524 (4) | C14—O3 | 1.369 (3) |
| C2—C3 | 1.321 (3) | C14—C15 | 1.383 (3) |
| C2—H2 | 0.9300 | C15—C16 | 1.380 (3) |
| C3—C4 | 1.449 (3) | C15—H15 | 0.9300 |
| C3—H3 | 0.9300 | C16—O4 | 1.361 (3) |
| C4—C5 | 1.389 (3) | C16—C17 | 1.377 (3) |
| C4—C9 | 1.400 (3) | C17—C18 | 1.384 (3) |
| C5—C6 | 1.370 (3) | C17—H17 | 0.9300 |
| C6—C7 | 1.386 (3) | C18—H18 | 0.9300 |
| C6—H6 | 0.9300 | C19—H19A | 0.9600 |
| C7—C8 | 1.385 (3) | C19—H19B | 0.9600 |
| C7—H7 | 0.9300 | C19—H19C | 0.9600 |
| C8—C9 | 1.386 (3) | C20—H20A | 0.9600 |
| C8—C12 | 1.501 (3) | C20—H20B | 0.9600 |
| C10—C11 | 1.516 (3) | C20—H20C | 0.9600 |
| C10—H10A | 0.9700 | O3—H3A | 0.802 (17) |
| C10—H10B | 0.9700 | O4—H4A | 0.820 (17) |
| C11—C13 | 1.506 (3) | ||
| C5—O1—C1 | 118.37 (17) | C13—C11—H11 | 107.3 |
| C9—O2—C10 | 117.92 (18) | C10—C11—H11 | 107.3 |
| O1—C1—C2 | 109.58 (18) | C12—C11—H11 | 107.3 |
| O1—C1—C19 | 105.0 (2) | C8—C12—C11 | 108.17 (19) |
| C2—C1—C19 | 111.7 (2) | C8—C12—H12A | 110.1 |
| O1—C1—C20 | 107.1 (2) | C11—C12—H12A | 110.1 |
| C2—C1—C20 | 111.3 (2) | C8—C12—H12B | 110.1 |
| C19—C1—C20 | 112.0 (2) | C11—C12—H12B | 110.1 |
| C3—C2—C1 | 121.9 (2) | H12A—C12—H12B | 108.4 |
| C3—C2—H2 | 119.0 | C18—C13—C14 | 116.2 (2) |
| C1—C2—H2 | 119.0 | C18—C13—C11 | 124.1 (2) |
| C2—C3—C4 | 120.2 (2) | C14—C13—C11 | 119.7 (2) |
| C2—C3—H3 | 119.9 | O3—C14—C15 | 121.8 (2) |
| C4—C3—H3 | 119.9 | O3—C14—C13 | 116.3 (2) |
| C5—C4—C9 | 117.7 (2) | C15—C14—C13 | 121.9 (2) |
| C5—C4—C3 | 118.02 (19) | C16—C15—C14 | 119.9 (2) |
| C9—C4—C3 | 124.2 (2) | C16—C15—H15 | 120.0 |
| C6—C5—O1 | 118.1 (2) | C14—C15—H15 | 120.0 |
| C6—C5—C4 | 121.8 (2) | O4—C16—C17 | 123.2 (2) |
| O1—C5—C4 | 120.0 (2) | O4—C16—C15 | 117.0 (2) |
| C5—C6—C7 | 118.9 (2) | C17—C16—C15 | 119.8 (2) |
| C5—C6—H6 | 120.6 | C16—C17—C18 | 119.5 (2) |
| C7—C6—H6 | 120.6 | C16—C17—H17 | 120.3 |
| C8—C7—C6 | 122.1 (2) | C18—C17—H17 | 120.3 |
| C8—C7—H7 | 118.9 | C17—C18—C13 | 122.7 (2) |
| C6—C7—H7 | 118.9 | C17—C18—H18 | 118.7 |
| C7—C8—C9 | 117.4 (2) | C13—C18—H18 | 118.7 |
| C7—C8—C12 | 122.1 (2) | C1—C19—H19A | 109.5 |
| C9—C8—C12 | 120.3 (2) | C1—C19—H19B | 109.5 |
| O2—C9—C8 | 122.7 (2) | H19A—C19—H19B | 109.5 |
| O2—C9—C4 | 115.1 (2) | C1—C19—H19C | 109.5 |
| C8—C9—C4 | 122.2 (2) | H19A—C19—H19C | 109.5 |
| O2—C10—C11 | 112.64 (17) | H19B—C19—H19C | 109.5 |
| O2—C10—H10A | 109.1 | C1—C20—H20A | 109.5 |
| C11—C10—H10A | 109.1 | C1—C20—H20B | 109.5 |
| O2—C10—H10B | 109.1 | H20A—C20—H20B | 109.5 |
| C11—C10—H10B | 109.1 | C1—C20—H20C | 109.5 |
| H10A—C10—H10B | 107.8 | H20A—C20—H20C | 109.5 |
| C13—C11—C10 | 112.19 (18) | H20B—C20—H20C | 109.5 |
| C13—C11—C12 | 115.3 (2) | C14—O3—H3A | 111 (2) |
| C10—C11—C12 | 107.10 (19) | C16—O4—H4A | 113 (2) |
| C5—O1—C1—C2 | 40.3 (3) | C3—C4—C9—O2 | 1.3 (3) |
| C5—O1—C1—C19 | 160.4 (2) | C5—C4—C9—C8 | 0.3 (3) |
| C5—O1—C1—C20 | −80.5 (3) | C3—C4—C9—C8 | −176.3 (2) |
| O1—C1—C2—C3 | −28.0 (3) | C9—O2—C10—C11 | 31.4 (3) |
| C19—C1—C2—C3 | −143.8 (3) | O2—C10—C11—C13 | 171.8 (2) |
| C20—C1—C2—C3 | 90.2 (3) | O2—C10—C11—C12 | −60.7 (3) |
| C1—C2—C3—C4 | 3.5 (4) | C7—C8—C12—C11 | 147.1 (2) |
| C2—C3—C4—C5 | 11.3 (3) | C9—C8—C12—C11 | −28.3 (3) |
| C2—C3—C4—C9 | −172.2 (2) | C13—C11—C12—C8 | −177.54 (18) |
| C1—O1—C5—C6 | 156.1 (2) | C10—C11—C12—C8 | 56.8 (2) |
| C1—O1—C5—C4 | −29.1 (3) | C10—C11—C13—C18 | 99.2 (3) |
| C9—C4—C5—C6 | −0.6 (3) | C12—C11—C13—C18 | −23.7 (3) |
| C3—C4—C5—C6 | 176.2 (2) | C10—C11—C13—C14 | −81.4 (3) |
| C9—C4—C5—O1 | −175.14 (19) | C12—C11—C13—C14 | 155.7 (2) |
| C3—C4—C5—O1 | 1.7 (3) | C18—C13—C14—O3 | −179.3 (2) |
| O1—C5—C6—C7 | 175.3 (2) | C11—C13—C14—O3 | 1.2 (3) |
| C4—C5—C6—C7 | 0.6 (4) | C18—C13—C14—C15 | 0.7 (3) |
| C5—C6—C7—C8 | −0.4 (4) | C11—C13—C14—C15 | −178.7 (2) |
| C6—C7—C8—C9 | 0.2 (4) | O3—C14—C15—C16 | 179.0 (2) |
| C6—C7—C8—C12 | −175.3 (2) | C13—C14—C15—C16 | −1.1 (3) |
| C10—O2—C9—C8 | 0.9 (3) | C14—C15—C16—O4 | −179.5 (2) |
| C10—O2—C9—C4 | −176.63 (19) | C14—C15—C16—C17 | 0.8 (3) |
| C7—C8—C9—O2 | −177.5 (2) | O4—C16—C17—C18 | −179.9 (2) |
| C12—C8—C9—O2 | −1.9 (3) | C15—C16—C17—C18 | −0.2 (3) |
| C7—C8—C9—C4 | −0.2 (3) | C16—C17—C18—C13 | −0.2 (4) |
| C12—C8—C9—C4 | 175.5 (2) | C14—C13—C18—C17 | −0.1 (3) |
| C5—C4—C9—O2 | 177.88 (19) | C11—C13—C18—C17 | 179.3 (2) |
Hydrogen-bond geometry (Å, º)
Cg4 is the centroid of the C13–C18 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| O4—H4A···O1i | 0.82 (2) | 2.02 (2) | 2.841 (3) | 177 (3) |
| O3—H3A···Cg4ii | 0.80 (2) | 2.51 (2) | 3.213 (2) | 148 (3) |
Symmetry codes: (i) −x+3/2, −y+2, z−1/2; (ii) −x, y+5/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB6989).
References
- Ahn, J., Lee, H., Jang, J., Kim, S. & Ha, T. (2012). Food Chem Toxicol 51, 439–445. [DOI] [PubMed]
- Bruker (1998). SMART Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2003). SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Choi, E. M. (2005). Biochem. Pharmacol. 70, 363–368. [DOI] [PubMed]
- Fukai, T., Sakagami, H., Toguchi, M., Takayama, F., Iwakura, I., Atsumi, T., Ueha, T., Nakashima, H. & Nomura, T. (2000). Anticancer Res. 20, 2525–2536. [PubMed]
- Kim, M., Kim, S.-N., Kim, Y.-U. & Han, J. (2009). Bull. Kor. Chem. Soc. 30, 415–418.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Messier, C. & Grenier, D. (2011). Mycoses, 54, 801–806. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Thiyagarajan, P., Chandrasekaran, C. V., Deepak, H. B. & Agarwal, A. (2011). Inflammopharmacology, 19, 235–241. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812048647/hb6989sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812048647/hb6989Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812048647/hb6989Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report