Abstract
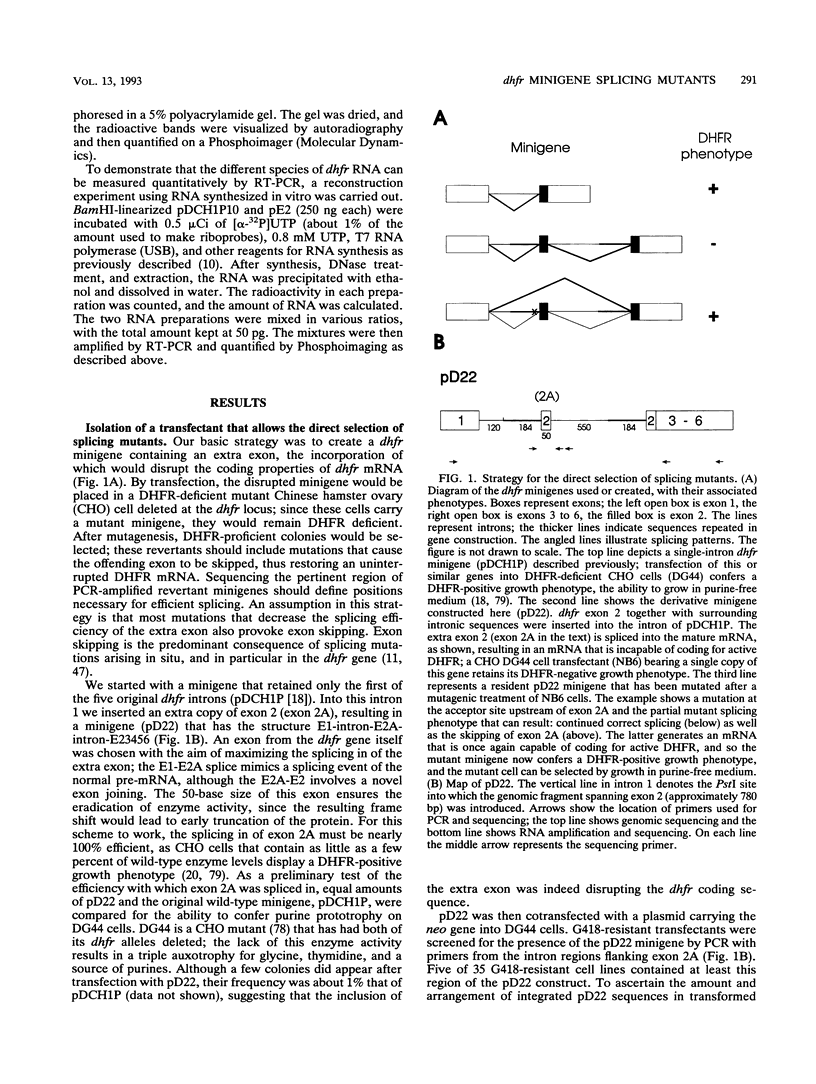
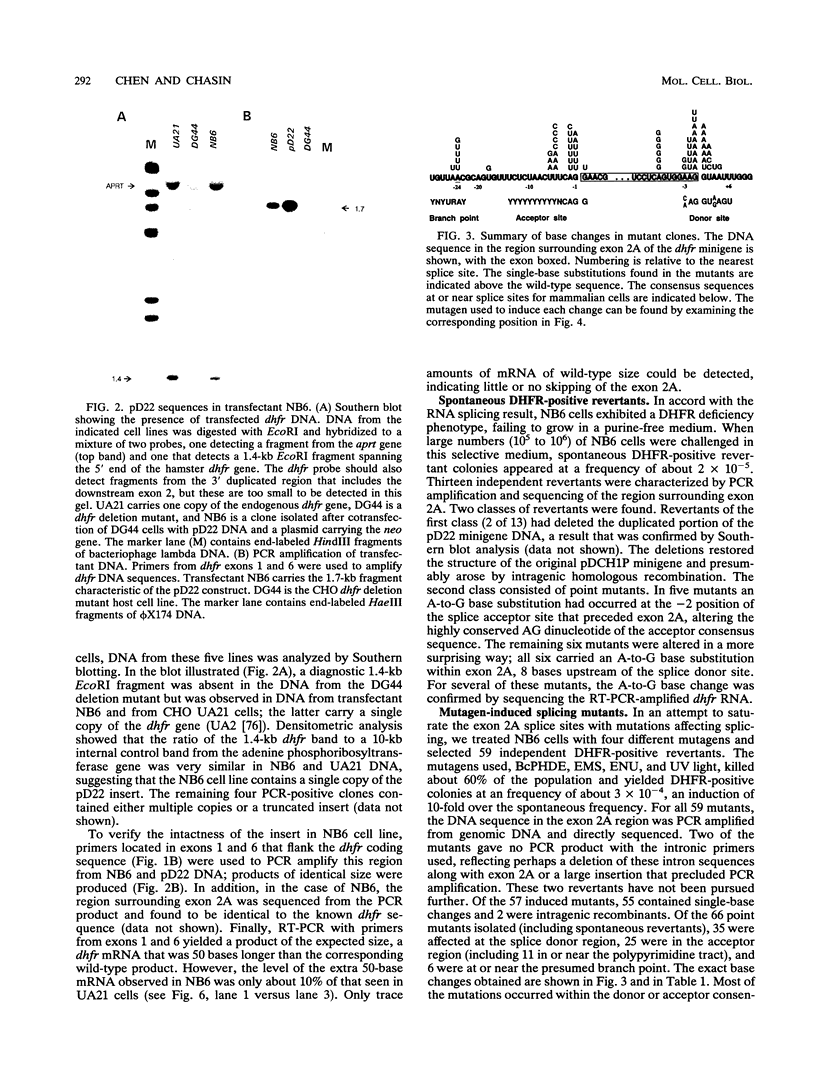
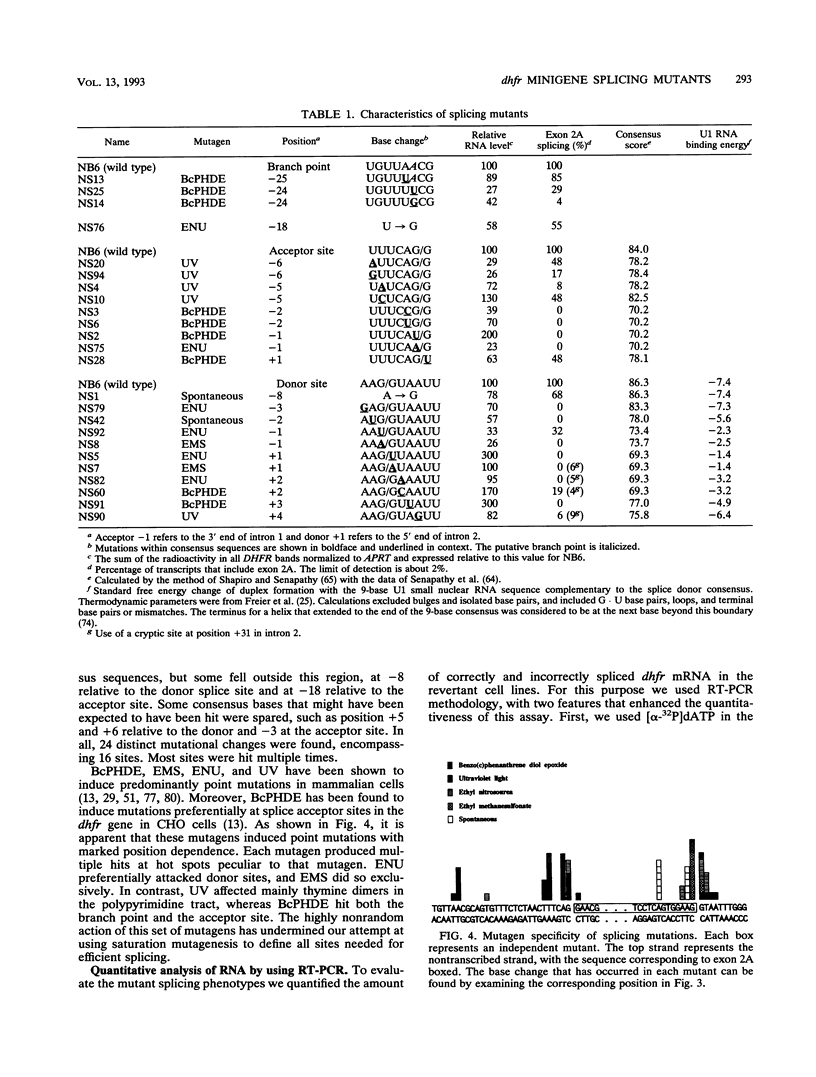
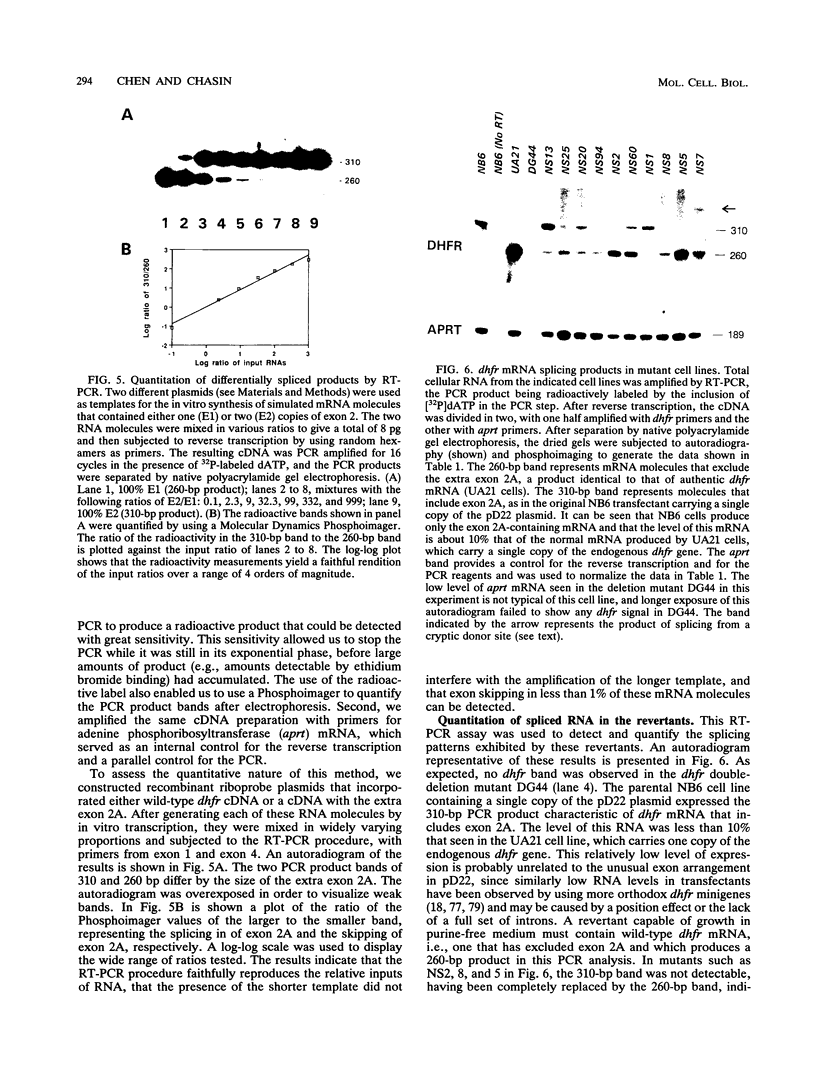
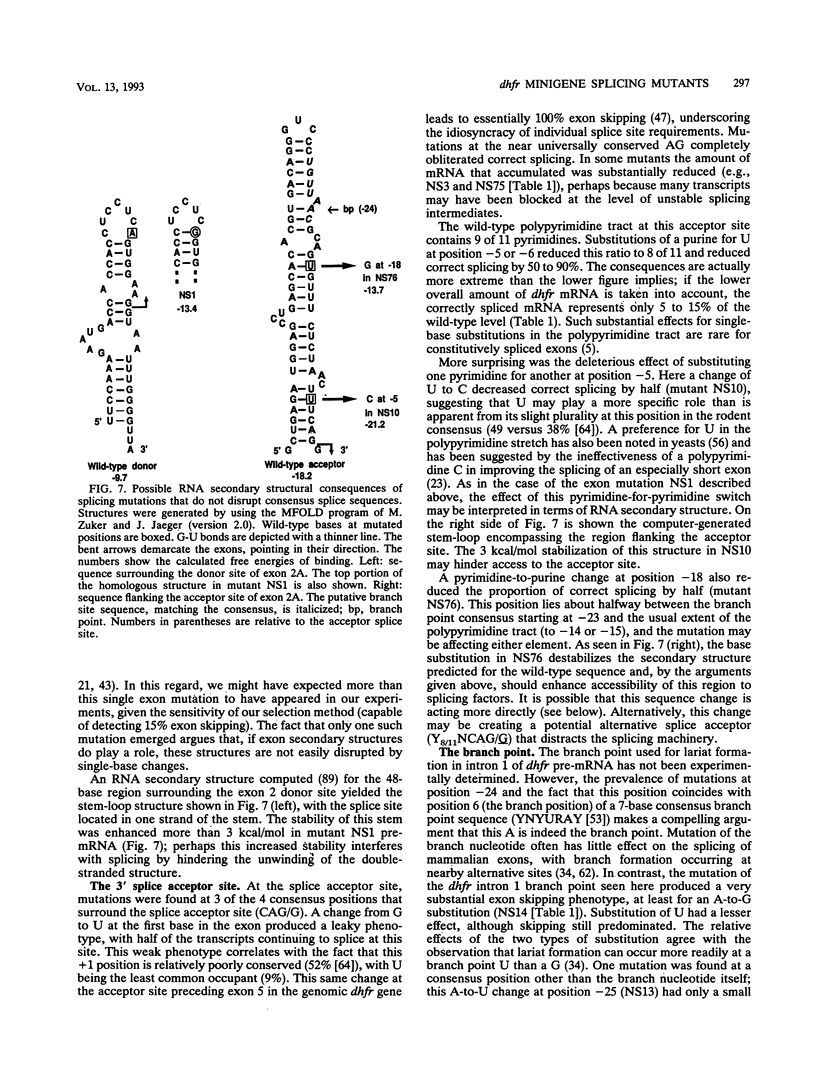
A Chinese hamster cell line containing an extra exon 2 (50 bp) inserted into a single intron of a dihydrofolate reductase (dhfr) minigene was constructed. The extra exon 2 was efficiently spliced into the RNA, resulting in an mRNA that is incapable of coding for the DHFR enzyme. Mutations that decreased splicing of this extra exon 2 caused it to be skipped and so produced normal dhfr mRNA. In contrast to the parental cell line, the splicing mutants display a DHFR-positive growth phenotype. Splicing mutants were isolated from this cell line after treatment with four different mutagens (racemic benzo[c]phenanthrene diol epoxide, ethyl methanesulfonate, ethyl nitrosourea, and UV irradiation). By polymerase chain reaction amplification and direct DNA sequencing, we determined the base changes in 66 mutants. Each of the mutagens generated highly specific base changes. All mutations were single-base substitutions and comprised 24 different changes distributed over 16 positions. Most of the mutations were within the consensus sequences at the exon 2 splice donor, acceptor, and branch sites. The RNA splicing patterns in the mutants were analyzed by quantitative reverse transcription-polymerase chain reaction. The recruitment of cryptic sites was rarely seen; simple exon skipping was the predominant mutant phenotype. The wide variety of mutations that produced exon skipping suggests that this phenotype is the typical consequence of splice site damage and supports the exon definition model of splice site selection. A few mutations were located outside the consensus sequences, in the exon or between the branch point and the polypyrimidine tract, identifying additional positions that play a role in splice site definition. That most of these 66 mutations fell within consensus sequences in this near-saturation mutagenesis suggests that splicing signals beyond the consensus may consist of robust RNA structures.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adema G. J., van Hulst K. L., Baas P. D. Uridine branch acceptor is a cis-acting element involved in regulation of the alternative processing of calcitonin/CGRP-l pre-mRNA. Nucleic Acids Res. 1990 Sep 25;18(18):5365–5373. doi: 10.1093/nar/18.18.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Weissmann C. 5' cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5' splice region, not by the conserved 5' GU. Cell. 1987 Jul 17;50(2):237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Archibald A. L., Thompson N. A., Kvist S. A single nucleotide difference at the 3' end of an intron causes differential splicing of two histocompatibility genes. EMBO J. 1986 May;5(5):957–965. doi: 10.1002/j.1460-2075.1986.tb04309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashman C. R., Davidson R. L. DNA base sequence changes induced by ethyl methanesulfonate in a chromosomally integrated shuttle vector gene in mouse cells. Somat Cell Mol Genet. 1987 Sep;13(5):563–568. doi: 10.1007/BF01534497. [DOI] [PubMed] [Google Scholar]
- Bigger C. A., Strandberg J., Yagi H., Jerina D. M., Dipple A. Mutagenic specificity of a potent carcinogen, benzo[c]phenanthrene (4R,3S)-dihydrodiol (2S,1R)-epoxide, which reacts with adenine and guanine in DNA. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2291–2295. doi: 10.1073/pnas.86.7.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers A. M., Mucha J., Grunberger D. DNA strand-specific mutations induced by (+/-)-3 alpha,4 beta-dihydroxy- 1 alpha,2 alpha-epoxy-1,2,3,4-tetrahydrobenzo[c]phenanthrene in the dihydrofolate reductase gene. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5749–5753. doi: 10.1073/pnas.88.13.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Ellis N., Chasin L. A. Structure of the dihydrofolate reductase gene in Chinese hamster ovary cells. Nucleic Acids Res. 1983 Apr 11;11(7):1997–2012. doi: 10.1093/nar/11.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Grunberger D., Chasin L. A. Mapping and characterization of mutations induced by benzo[a]pyrene diol epoxide at dihydrofolate reductase locus in CHO cells. Somat Cell Mol Genet. 1988 Mar;14(2):169–183. doi: 10.1007/BF01534402. [DOI] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Mucha J., Grunberger D., Chasin L. A. Point mutation analysis in a mammalian gene: rapid preparation of total RNA, PCR amplification of cDNA, and Taq sequencing by a novel method. Biotechniques. 1989 May;7(5):494-6, 498-9. [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Mucha J., Harvey R. G., Chasin L. A., Grunberger D. Splicing mutations in the CHO DHFR gene preferentially induced by (+/-)-3 alpha,4 beta-dihydroxy-1 alpha,2 alpha-epoxy-1,2,3,4- tetrahydrobenzo[c]phenanthrene. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5464–5468. doi: 10.1073/pnas.87.14.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens R. P., Fenton W. A., Rosenberg L. R. Identification of RNA splicing errors resulting in human ornithine transcarbamylase deficiency. Am J Hum Genet. 1991 Jun;48(6):1105–1114. [PMC free article] [PubMed] [Google Scholar]
- Chen R. H., Maher V. M., McCormick J. J. Effect of excision repair by diploid human fibroblasts on the kinds and locations of mutations induced by (+/-)-7 beta,8 alpha-dihydroxy-9 alpha,10 alpha-epoxy-7,8,9,10- tetrahydrobenzo[a]pyrene in the coding region of the HPRT gene. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8680–8684. doi: 10.1073/pnas.87.21.8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Ciudad C. J., Urlaub G., Chasin L. A. Deletion analysis of the Chinese hamster dihydrofolate reductase gene promoter. J Biol Chem. 1988 Nov 5;263(31):16274–16282. [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991 Jun 28;252(5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., McEwan R. N., Pearson M. L. Expression and amplification of engineered mouse dihydrofolate reductase minigenes. Mol Cell Biol. 1983 Feb;3(2):257–266. doi: 10.1128/mcb.3.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler J. O., Rossi J. J. Unexpected point mutations activate cryptic 3' splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 1991 Jul;5(7):1252–1263. doi: 10.1101/gad.5.7.1252. [DOI] [PubMed] [Google Scholar]
- Dipple A., Pigott M. A., Agarwal S. K., Yagi H., Sayer J. M., Jerina D. M. Optically active benzo[c]phenanthrene diol epoxides bind extensively to adenine in DNA. Nature. 1987 Jun 11;327(6122):535–536. doi: 10.1038/327535a0. [DOI] [PubMed] [Google Scholar]
- Dominski Z., Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991 Dec;11(12):6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. D., Katz R. A., Skalka A. M., Maniatis T. The role of branchpoint and 3'-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 1991 Feb;5(2):211–220. doi: 10.1101/gad.5.2.211. [DOI] [PubMed] [Google Scholar]
- Fu X. D., Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990 Feb 1;343(6257):437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Fu X. Y., Manley J. L. Factors influencing alternative splice site utilization in vivo. Mol Cell Biol. 1987 Feb;7(2):738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuscoe J. C., Fenwick R. G., Jr, Ledbetter D. H., Caskey C. T. Deletion and amplification of the HGPRT locus in Chinese hamster cells. Mol Cell Biol. 1983 Jun;3(6):1086–1096. doi: 10.1128/mcb.3.6.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hampson R. K., La Follette L., Rottman F. M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989 Apr;9(4):1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka Y., Palella T. D., O'Toole T. E., Tarlé S. A., Kelley W. N. Human adenine phosphoribosyltransferase. Identification of allelic mutations at the nucleotide level as a cause of complete deficiency of the enzyme. J Clin Invest. 1987 Nov;80(5):1409–1415. doi: 10.1172/JCI113219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig H., Aebi M., Weissmann C. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature. 1986 Dec 11;324(6097):589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- Huang S., Spector D. L. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 1991 Dec;5(12A):2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- Jackson I. J. A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 1991 Jul 25;19(14):3795–3798. doi: 10.1093/nar/19.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes D. H., Steitz J. A. Accurate 5' splice-site selection in mouse kappa immunoglobulin light chain premessenger RNAs is not cell-type-specific. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7928–7932. doi: 10.1073/pnas.84.22.7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes D. H., Steitz J. A. Correct in vivo splicing of the mouse immunoglobulin kappa light-chain pre-mRNA is dependent on 5' splice-site position even in the absence of transcription. Genes Dev. 1988 Nov;2(11):1448–1459. doi: 10.1101/gad.2.11.1448. [DOI] [PubMed] [Google Scholar]
- Kuivaniemi H., Kontusaari S., Tromp G., Zhao M. J., Sabol C., Prockop D. J. Identical G+1 to A mutations in three different introns of the type III procollagen gene (COL3A1) produce different patterns of RNA splicing in three variants of Ehlers-Danlos syndrome. IV. An explanation for exon skipping some mutations and not others. J Biol Chem. 1990 Jul 15;265(20):12067–12074. [PubMed] [Google Scholar]
- Lawrence J. B., Singer R. H., Marselle L. M. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989 May 5;57(3):493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Lebkowski J. S., Miller J. H., Calos M. P. Determination of DNA sequence changes induced by ethyl methanesulfonate in human cells, using a shuttle vector system. Mol Cell Biol. 1986 May;6(5):1838–1842. doi: 10.1128/mcb.6.5.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Piseri A., Fiszman M. Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991 Jun 28;252(5014):1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Lossi A. M., Bergé-Lefranc J. L. The mRNA transcripts from a mutant beta-globin gene derived from splicing at preferential cryptic sites. FEBS Lett. 1989 Oct 9;256(1-2):163–166. doi: 10.1016/0014-5793(89)81740-5. [DOI] [PubMed] [Google Scholar]
- Lowery D. E., Van Ness B. G. In vitro splicing of kappa immunoglobulin precursor mRNA. Mol Cell Biol. 1987 Apr;7(4):1346–1351. doi: 10.1128/mcb.7.4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy I., Pellicer A., Jackson J. F., Sim G. K., Silverstein S., Axel R. Isolation of transforming DNA: cloning the hamster aprt gene. Cell. 1980 Dec;22(3):817–823. doi: 10.1016/0092-8674(80)90558-9. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Carothers A. M., Han J. H., Harding J. D., Kas E., Venolia L., Chasin L. A. Multiple transcription start sites, DNase I-hypersensitive sites, and an opposite-strand exon in the 5' region of the CHO dhfr gene. Mol Cell Biol. 1986 Feb;6(2):425–440. doi: 10.1128/mcb.6.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Urlaub G., Chasin L. Spontaneous splicing mutations at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1986 Jun;6(6):1926–1935. doi: 10.1128/mcb.6.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan R. C., Berg P. Expression of a bacterial gene in mammalian cells. Science. 1980 Sep 19;209(4463):1422–1427. doi: 10.1126/science.6251549. [DOI] [PubMed] [Google Scholar]
- Nalbantoglu J., Goncalves O., Meuth M. Structure of mutant alleles at the aprt locus of Chinese hamster ovary cells. J Mol Biol. 1983 Jul 5;167(3):575–594. doi: 10.1016/s0022-2836(83)80099-0. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mammalian U2 snRNP has a sequence-specific RNA-binding activity. Genes Dev. 1989 Oct;3(10):1562–1571. doi: 10.1101/gad.3.10.1562. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev. 1988 Mar;2(3):319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Prives C., Manley J. L. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. Genes Dev. 1988 Nov;2(11):1460–1475. doi: 10.1101/gad.2.11.1460. [DOI] [PubMed] [Google Scholar]
- Orkin S. H., Kazazian H. H., Jr The mutation and polymorphism of the human beta-globin gene and its surrounding DNA. Annu Rev Genet. 1984;18:131–171. doi: 10.1146/annurev.ge.18.120184.001023. [DOI] [PubMed] [Google Scholar]
- Patterson B., Guthrie C. A U-rich tract enhances usage of an alternative 3' splice site in yeast. Cell. 1991 Jan 11;64(1):181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- Penotti F. E. Human pre-mRNA splicing signals. J Theor Biol. 1991 Jun 7;150(3):385–420. doi: 10.1016/s0022-5193(05)80436-9. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989 Dec;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Greene J. M., Green M. R. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985 Jul;41(3):833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Seetharam S., Dicker I. B. A rapid and complete 4-step protocol for obtaining nucleotide sequence from E. coli genomic DNA from overnight cultures. Biotechniques. 1991 Jul;11(1):32–34. [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano P. G., Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988 Oct;2(10):1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steingrimsdottir H., Rowley G., Dorado G., Cole J., Lehmann A. R. Mutations which alter splicing in the human hypoxanthine-guanine phosphoribosyltransferase gene. Nucleic Acids Res. 1992 Mar 25;20(6):1201–1208. doi: 10.1093/nar/20.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli M., Saito H. Regulation of tissue-specific alternative splicing: exon-specific cis-elements govern the splicing of leukocyte common antigen pre-mRNA. EMBO J. 1989 Mar;8(3):787–796. doi: 10.1002/j.1460-2075.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M., Berget S. M. Effect of 5' splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990 Dec;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull-Ross A. D., Else A. J., Eperon I. C. The dependence of splicing efficiency on the length of 3' exon. Nucleic Acids Res. 1988 Jan 25;16(2):395–411. doi: 10.1093/nar/16.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Urlaub G., Chasin L. A. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4216–4220. doi: 10.1073/pnas.77.7.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Käs E., Carothers A. M., Chasin L. A. Deletion of the diploid dihydrofolate reductase locus from cultured mammalian cells. Cell. 1983 Jun;33(2):405–412. doi: 10.1016/0092-8674(83)90422-1. [DOI] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Ciudad C. J., Chasin L. A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989 Jul;9(7):2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Kas E., Chasin L. A., Funanage V. L., Myoda T. T., Hamlin J. Effect of gamma rays at the dihydrofolate reductase locus: deletions and inversions. Somat Cell Mol Genet. 1986 Nov;12(6):555–566. doi: 10.1007/BF01671941. [DOI] [PubMed] [Google Scholar]
- Venolia L., Urlaub G., Chasin L. A. Polyadenylation of Chinese hamster dihydrofolate reductase genomic genes and minigenes after gene transfer. Somat Cell Mol Genet. 1987 Sep;13(5):491–504. doi: 10.1007/BF01534491. [DOI] [PubMed] [Google Scholar]
- Vrieling H., Niericker M. J., Simons J. W., van Zeeland A. A. Molecular analysis of mutations induced by N-ethyl-N-nitrosourea at the HPRT locus in mouse lymphoma cells. Mutat Res. 1988 Mar;198(1):99–106. doi: 10.1016/0027-5107(88)90045-0. [DOI] [PubMed] [Google Scholar]
- Vrieling H., Van Rooijen M. L., Groen N. A., Zdzienicka M. Z., Simons J. W., Lohman P. H., van Zeeland A. A. DNA strand specificity for UV-induced mutations in mammalian cells. Mol Cell Biol. 1989 Mar;9(3):1277–1283. doi: 10.1128/mcb.9.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S., Aebi M. In vitro splicing of mRNA precursors: 5' cleavage site can be predicted from the interaction between the 5' splice region and the 5' terminus of U1 snRNA. Nucleic Acids Res. 1988 Jan 25;16(2):471–486. doi: 10.1093/nar/16.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R., Urlaub G., Chasin L. DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zhang L. H., Vrieling H., van Zeeland A. A., Jenssen D. Spectrum of spontaneously occurring mutations in the hprt gene of V79 Chinese hamster cells. J Mol Biol. 1992 Feb 5;223(3):627–635. doi: 10.1016/0022-2836(92)90979-t. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]