Abstract
The molecular mechanisms regulating tissue size represent an unsolved puzzle in developmental biology. One signalling pathway controlling growth of the Drosophila wing is Dpp. Dpp promotes growth by repression of the transcription factor Brk. The transcriptional targets of Brk that control cell growth and proliferation, however, are not yet fully elucidated. We report here a genome-wide ChIP-Seq of endogenous Brk from wing imaginal discs. We identify the growth regulator Myc as a target of Brk and show that repression of Myc and of the miRNA bantam explains a significant fraction of the growth inhibition caused by Brk. This work sheds light on the effector mechanisms by which Dpp signalling controls tissue growth.
Keywords: Drosophila , growth control, Brk, Myc, Dpp
INTRODUCTION
During animal development, tissue growth rates and final sizes are precisely regulated. The molecular mechanisms controlling this process, however, are not well understood. The Drosophila wing has become an important model system for studying tissue growth control [1,2,3]. Three signalling pathways are thought to control wing growth: insulin, hippo/yorkie and Dpp. The insulin pathway modulates overall animal size in response to nutrient conditions [3,4,5,6]. The hippo/yorkie signalling pathway senses several inputs including information about cell–cell contacts, tissue polarity and cytoskeletal tension, and it controls tissue growth by inhibiting apoptosis and promoting the cell cycle [7, 8]. The third signalling pathway regulating tissue growth is the Dpp pathway, homologous to mammalian transforming growth factor-β and bone morphogenetic protein signalling. The mechanism by which Dpp controls tissue growth is not well understood. The Dpp morphogen is synthesized in a medial region of the wing disc abutting the boundary between anterior and posterior compartments, and secreted to signal to all cells in the disc in a distance-dependent graded manner [9]. Cells far from the Dpp source sense low levels of Dpp and activate ‘low-threshold’ response genes such as omb (bi), whereas cells closer to the Dpp source sense high levels of signalling and additionally activate ‘high-threshold’ response genes such as Spalt (salm) [9]. One Dpp target gene of particular interest is the transcription factor Brinker (Brk). Brk is repressed by Dpp signalling, and hence forms an inverse-gradient to the Dpp gradient [10,11,12,13,14]. Brk inhibits expression of Dpp targets ensuring they are turned off laterally [10, 11, 15]. Indeed, a large part of transcriptional regulation by Dpp occurs using this Brk-mediated double-negative mechanism. Brk often represses Dpp targets by competing for binding to sites overlapping with those of the transcriptional activator Mad [16, 17].
The mechanism by which Dpp controls tissue growth has been an issue of recent interest [18,19,20]. Wing disc size is sensitive to Dpp signalling dosage, with high signalling leading to large tissue size [21]. Almost all of the growth regulation by Dpp can be attributed to Brk, as loss of Brk leads to significant tissue overgrowth [10] and the small size of Dpp loss-of-function wings can be completely rescued by the removal of Brk [19, 22]. Thus, the question of how Dpp controls tissue growth can be rephrased in terms of Brk, and how Brk controls tissue growth. As Brk is a transcription factor, it is presumably regulating expression of genes involved in cell growth and proliferation. These target genes, however, are not well characterized. A recent study identified the miRNA bantam as a Brk target [23]; however, bantam overexpression only partially rescues the undergrowth caused by Brk expression, indicating there are additional uncharacterized targets to be identified.
To understand the mechanisms by which Dpp controls tissue growth through Brk, we performed chromatin immunoprecipitation (ChIP) of endogenous Brk from the wing imaginal discs, followed by high-throughput sequencing. We identify the growth regulator Myc as a target of Brk and show that Myc and bantam combined explain a significant fraction of the growth inhibition caused by Brk. Our data support a simple model whereby Dpp signalling inhibits Brk, thereby inducing expression of Myc and bantam leading to tissue growth.
RESULTS
Brk ChIP-Seq from wing imaginal discs
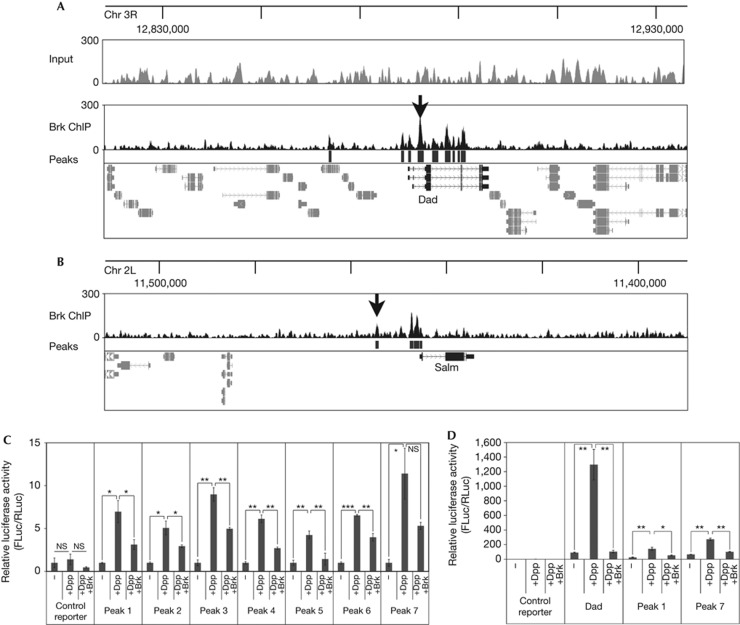
To identify Brk target genes, we raised and immunopurified an antibody that can immunoprecipitate endogenous Brk protein as well as detect it in wing disc stainings (supplementary Fig S1A online). This antibody is very sensitive, as it can detect low levels of Brk protein in medial regions of the wing disc. This signal is gone in BrkXA discs (supplementary Fig S1B,1B′ online) and blunted in discs where Brk expression is reduced in the dorsal compartment by RNA-mediated interference (supplementary Fig S1C online). Using this antibody, we performed ChIP on endogenous Brk from isolated wing discs and subjected the immunoprecipitated material to high-throughput sequencing as previously described [24]. We used PeakSeq (MINFDR=0.05 and PVALTHRESH<10−10) [25] to identify genomic loci that are statistically significantly enriched compared with normalized input control (published in the study by Perez-Lluch et al [24], supplementary Fig S2A online), obtaining 2,547 peaks corresponding to 1,671 genes (supplementary Tables S1, S2 online). Among these regions, previously published Brk binding sites near target genes such as Dad and Spalt (Salm) could easily be recognized as regions rising above the average ‘background’ (black arrows in Fig 1A,B) [17, 26]. In contrast, an enrichment of these regions with above average read density could not be observed when sheared chromatin was sequenced, indicating this is not an artifact of shearing or sequencing (‘Input’ trace, Fig 1A).
Figure 1.
ChIP-Seq of endogenous Brk from larval wing discs. (A,B) ChIP-Seq of endogenous Brk from larval wing discs identifies known Brk binding sites (black arrows) near Dad (A) and Salm (B). Brk binding peaks (black boxes below ChIP-seq trace, ‘Peaks’) identified by PeakSeq relative to normalized input with cutoff P<10−10. ChIP profiles displayed using UCSC genome browser [37]. (C) Luciferase reporter assays on randomly selected Brk ChIP-Seq peaks showing activation by Dpp signalling and repression by Brk. Peaks 1–7 are all the Brk ChIP-Seq peaks on chromosome X between positions 5,590,283 (Peak 1) and 5,795,911 (Peak 7), detailed in supplementary Materials online. Regions were cloned into a firefly luciferase reporter with a basal Hsp70 promoter (‘control reporter’) and assayed relative to a renilla luciferase normalization control. S2 cells were co-transfected to express activated Thickveins+Mad+Medea to activate Dpp signalling (‘+Dpp’) and Brk (‘+Brk’). Values for each reporter are normalized to 1 for the ‘−’ condition n=3. (D) Luciferase assay with Brk ChIP regions #1 and #7 as in C, and a positive control reporter containing a genomic region from Dad. Error bars: standard deviation, *t-test ⩽0.05, **t-test ⩽0.01, ***t-test ⩽0.001. Brk, Brinker; Chr, chromosome; ChIP-Seq, chromatin immunoprecipitation sequence; NS, nonsignificant.
To test the quality of our ChIP-Seq data, we randomly selected seven peaks to test by luciferase assay. We chose all peaks on the X chromosome between positions 5,590,283 and 5,795,911 (Fig 1C, exact sequences in supplementary Materials online). These regions were cloned into a firefly luciferase reporter and transfected together with a renilla normalization control into S2 cells. Transcriptional targets of Dpp are activated by the Mad/Medea complex and repressed by Brk. This is often a consequence of overlapping binding sites for Brk and the Mad/Medea complex, causing Brk to displace Mad/Medea [16, 17]. We first tested if our randomly selected Brk peaks can induce transcription in response to Dpp signalling. As S2 cells do not have endogenous Dpp signalling, we co-transfected plasmids expressing Mad, Medea and activated Thickveins, as previously described [16]. All seven reporters were induced by Dpp signalling (Fig 1C, ‘+Dpp’ versus ‘−’). Coexpression of Brk repressed reporter activity in all cases (Fig 1C, ‘+Dpp +Brk’ versus ‘+Dpp’ and Fig 1D), indicating these regions can respond to Brk.
In addition to the previously reported Brk sites, additional novel binding sites in known target genes such as Dad, Salm and bi (omb) were also found (Fig 1A,B, peaks identified by Peakseq, indicated as black boxes below the ChIP-seq trace). This suggests target genes tend to have several Brk binding sites, which might act cooperatively to regulate gene expression. Interestingly, Brk binding sites are often found near promoters, as seen for Dad and Salm (Fig 1A,B) and genome-wide (supplementary Fig S2A online), and in introns (Fig 1A). To obtain a global view of Brk-regulated genes, we performed a Gene Ontology enrichment analysis using DAVID [27] and found these genes to be enriched for transcriptional regulators and genes involved in patterning (supplementary Fig S2B online).
The Myc genomic region contains Brk binding sites
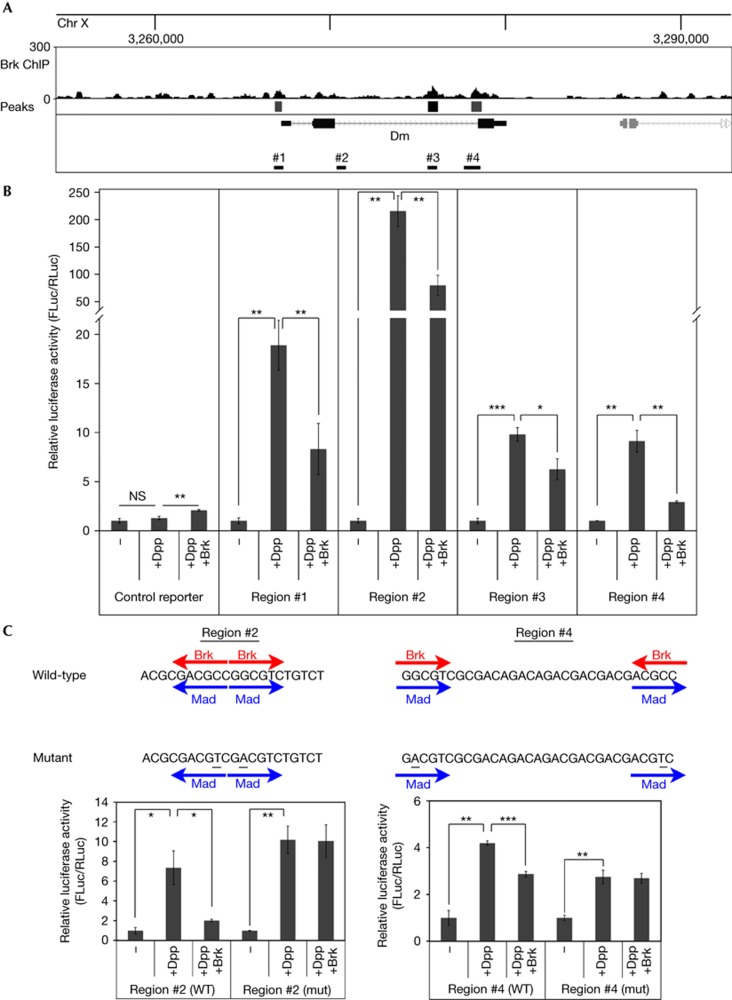
To identify targets of Brk involved in cell growth and proliferation, we studied the list of putative Brk targets and noticed that one peak was in the second intron of the well-described growth regulator Myc (‘diminutive (dm)’ in Drosophila, black box below ChIP-seq trace in Fig 2A). Furthermore, relaxing the statistical threshold of the Peakseq algorithm to its default P<0.01 identified two more regions near Myc (grey boxes, regions #1 and #4 in Fig 2A). Finally, a bioinformatic search using Genome Enhancer [28] identified a region (#2 in Fig 2A) that contains a cluster of Brk binding motifs (GGCGYY, four sites within 350 nt). Although this region was not enriched in our wing disc ChIP-Seq, we decided to also test it as it might respond to Dpp and Brk signalling in other contexts. The sequence of all four regions is detailed in supplementary Materials online. We first tested if these regions have the potential to be induced by Dpp signalling and to be repressed by Brk. We cloned these regions into a luciferase reporter and found that all four are significantly induced by Dpp signalling and repressed by Brk (Fig 2B). Although the baseline expression of the region #4 reporter (reflecting binding of other transcription factors present in S2 cells) is higher than that of the other three reporters, its fold induction in response to Dpp activation and Brk repression is similar to that of the other three. To further analyse activation of these reporters, we selected a short stretch of region 2 and a short stretch of region 4, both of which contain two clustered Brk binding sites (Fig 2C). (The full-length regions 2 and 4 have five and four Brk binding sites, respectively). As previously described for the Dad genomic locus [17], these Brk binding sites overlap with Mad binding sites (Fig 2C). As the consensus binding motif of Brk (GGCGYY) differs slightly from that of Mad (GRCGNC), this allowed us to generate point mutations predicted to abolish Brk binding while retaining Mad binding (underlined nucleotides Fig 2C). Indeed, whereas the wild-type sequences were activated by Dpp signalling and repressed by Brk, the mutated versions retained their activation in response to Dpp signalling but could no longer be repressed by Brk (Fig 2C).
Figure 2.
The Myc genomic region contains elements responsive to Mad/Medea and Brk. (A) Brk binding in the Myc (dm) genomic region by ChIP-Seq. Brk binding peaks identified by the PeakSeq algorithm (‘Peaks’), indicated as boxes below the ChIP-seq trace. One peak (black box, also indicted as region #3 below the gene model) passes the threshold P<10−10. Two more peaks (grey boxes, regions #1 and #4) pass the less stringent default PeakSeq setting of P<0.01. On inspection of the genomic sequence, an extra region (#2) was found to have a clustering of four Brk binding motifs within a 350 nt region. (B) All four regions of the Myc genomic region can be activated by Mad/Medea and repressed by Brk. Relative luminescence of a firefly luciferase reporter containing an Hsp70 basal promoter (‘control reporter’) or the same reporter into which four different Myc genomic regions indicated in A were introduced, relative to a renilla control reporter. Where indicated, S2 cells were co-transfected to express activated Thickveins+Mad+Medea (‘+Dpp’) and Brk (‘+Brk’), n=3. (C) Mutation (mut) of the Brk binding sites renders the Myc genomic regions insensitive to Brk. The indicated genomic sequences were trimerized and cloned into a luciferase reporter as in B. Brk binding motifs (GGCGYY) or Mad binding motifs (GRCGNC) are indicated by red and blue arrows, respectively. Point mutagenesis (underlined) removed the Brk binding motif without disturbing the Mad binding motif, n=3. Values for each reporter are normalized to 1 for the ‘−’ condition (B,C). Error bars: standard deviation. *t-test ⩽0.05, **t-test ⩽0.01, ***t-test ⩽0.001. Brk, Brinker; Chr, chromosome; ChIP-Seq, chromatin immunoprecipitation sequence; NS, nonsignificant; WT, wild-type.
Brk represses Myc expression in wing disc lateral regions
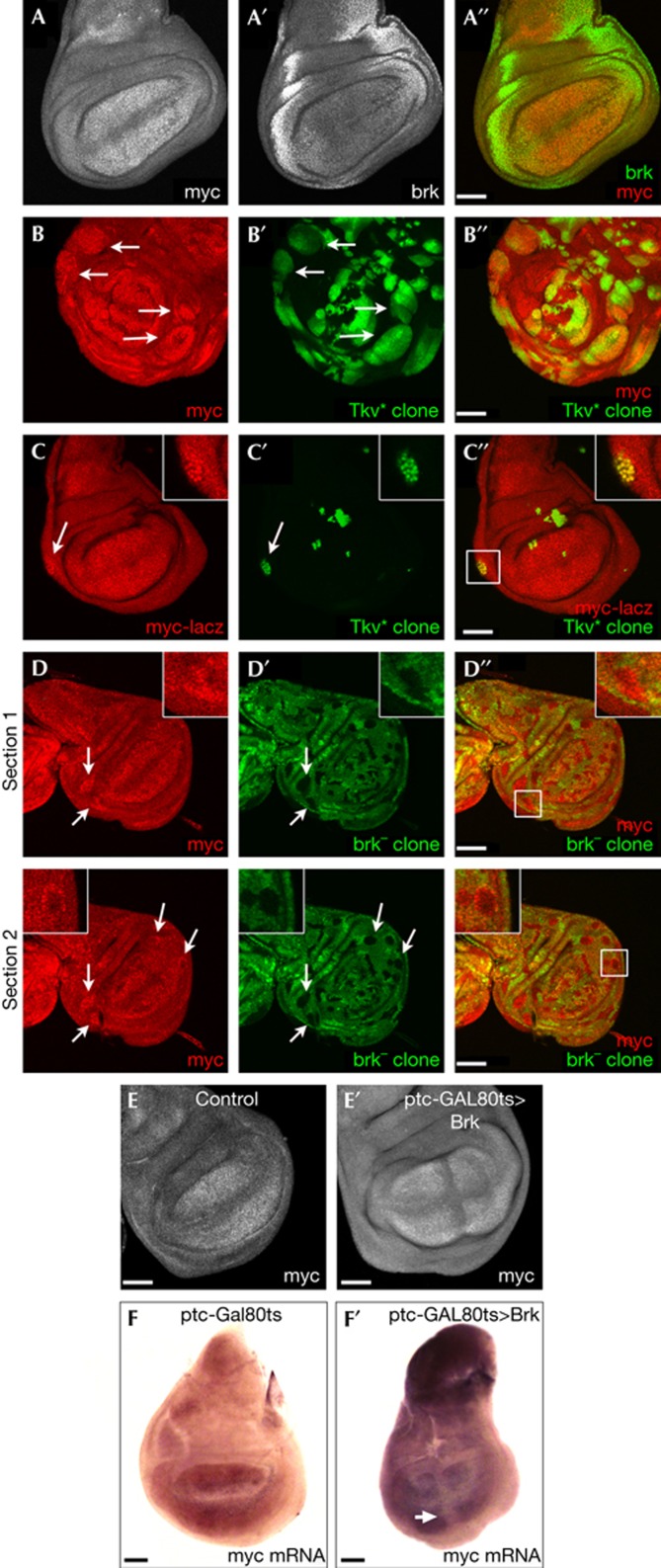
To study if Brk regulates Myc expression in the wing imaginal disc, we first looked at the expression patterns of these two genes. As previously reported [29,30,31], Myc protein is most strongly detected in the wing pouch (Fig 3A), whereas Brk is most strongly detected in the lateral regions of the disc (Fig 3A′). This complementary expression pattern raised the possibility that Brk might be constraining the Myc domain laterally. To test this hypothesis, we first asked if removing Brk laterally results in ectopic Myc. We generated clones expressing activated Thickveins (TkvQ235D) in which Brk expression is inhibited (supplementary Fig S2C online). These clones, marked with green fluorescent protein, showed robust elevation of Myc protein laterally in the wing disc (arrows in Fig 3B–B′′), but not medially in the disc where Brk is not expressed (Fig 3B–B′′). To verify that this occurs at the transcriptional level, we made use of a lacZ insertion near the Myc transcription start site, P{lacW}dmG0359 (‘Myc–lacZ’), which was previously reported to reflect Myc transcription [32]. Indeed, lateral TkvQ235D gain-of-function clones caused increased Myc–lacZ expression (arrow in Fig 3C–C′′). We also tested if Brk represses Myc expression using Brk loss-of-function clones. Loss of Brk resulted in elevated Myc protein levels when the clones were located laterally (white arrows in Fig 3D–D′′), but not medially where Brk is not expressed. Finally, we also performed the converse experiment, testing if expression of Brk medially in the disc can repress Myc expression. Indeed, transient induction of Brk expression medially (using patched-GAL4, Gal80ts) caused a clear reduction in Myc protein levels (Fig 3E–E′) and Myc mRNA (Fig 3F–F′; supplementary Fig S3B,C online), but not in the levels of a panel of nine other negative-control mRNAs (supplementary Fig S3A–C online).
Figure 3.
Brk represses Myc expression in lateral regions of the wing disc. (A–A′′) Myc and Brk are expressed in complementary regions of the wing disc. Wild-type wing disc stained with anti-Myc (A,A′′) and anti-Brk (A′,A′′). (B–B′′) Clones expressing activated Thickveins (TkvQ235D) display elevated levels of Myc protein. Clones marked with green fluorescent protein (GFP) in green and with α-Myc in red. (C–C′′) Clones expressing activated Thickveins (TkvQ235D), marked by GFP (green), display elevated levels of Myc transcription, detected by a lacZ enhancer trap in the Myc locus (P{lacW}dmG0359 in red). Displayed clone is boxed in C′′. (D–D′′) Brk mutant clones (marked by the loss of GFP, green) have elevated levels of Myc protein (α-Myc, red) laterally (arrows) but not medially in the disc where Brk is not expressed. Two sections of the same disc are shown. Displayed clone is boxed in D′′. (E–E′′) Medial expression of Brk represses Myc protein accumulation. Upstream activating sequence (UAS)–Brk was transiently induced in a medial stripe using patched-GAL4, GAL80ts by shifting larvae grown at 19–29°C for 12 h causes reduced Myc protein levels (E′), not observed in control discs (E). (F–F′) Transient, medial expression of Brk, as in E, represses Myc mRNA accumulation, detected by in situ hybridization. Scale bar, 50 μm. Disc genotypes: (A–A′′) w1118 (B–B′′) hsFlp, Act>STOP>GAL4, UAS–GFP, UAS–TkvQ235D (C–C′′) Myc–lacZ(P{lacW}dmG0359), hsFlp, Act>STOP>GAL4, UAS–GFP, UAS–TkvQ235D (D–D′′) hsFlp, FRT brkXA/FRT GFP (E,F) ptc–GAL4, GAL80ts (E′,F′) ptc–GAL4, GAL80ts, UAS–brk. Brk, Brinker; ptc, patched.
Bantam and Myc rescue Brk-induced growth inhibition
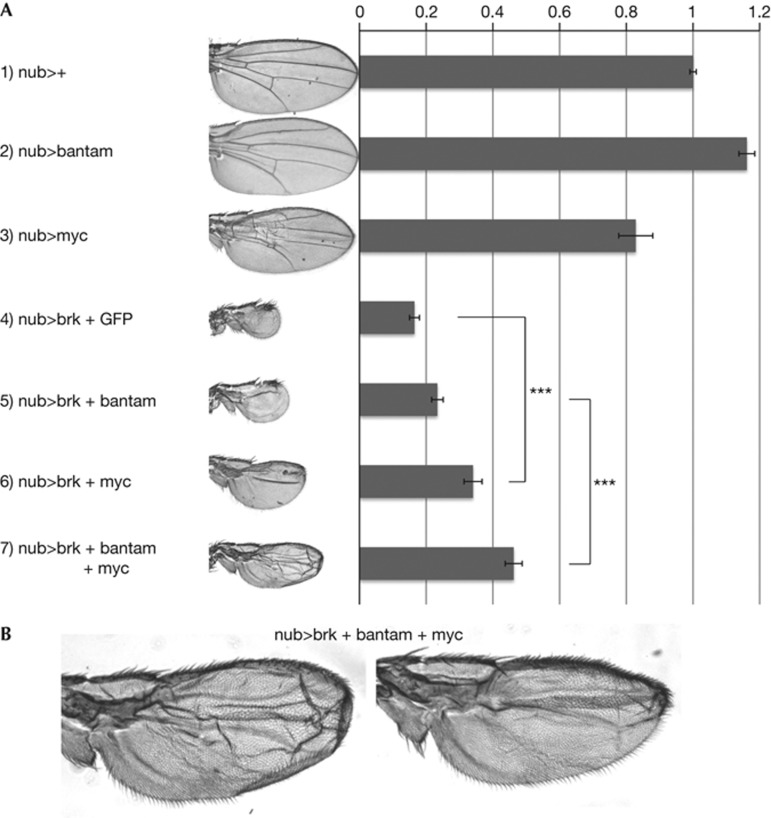
We next tested the functional significance of the inhibition of Myc expression by Brk for wing growth. Ubiquitous expression of Brk in the wing pouch with nubbin–GAL4 leads to adult wings that are significantly reduced in size and have lost Dpp-dependent patterning elements such as veins (row 3, Fig 4A). This is presumably due to Brk inhibiting expression of various target genes, some of which control patterning and some of which control tissue growth. In these wing discs, reconstituting expression of Brk target genes using the upstream activating sequence system should rescue either their growth or patterning. Indeed, expression of bantam, previously reported as a Brk target [23] mildly rescues the growth inhibition caused by Brk expression (row 4, Fig 4A). Likewise, expression of Myc also rescued the undergrowth caused by Brk, indeed to a greater extent than bantam (row 5, Fig 4A). Importantly, Myc expression with nubbin–GAL4 in a wild-type background did not increase the wing size (row 2, Fig 4A; supplementary Fig S4 online), indicating that the rescue of nub>brk wing size by Myc expression is not simply an additive effect, and that Myc levels become limiting when Brk is expressed. Combined expression of bantam and Myc rescued the wing size of Brk-expressing wings to an even larger extent than each alone (row 6, Fig 4A). The size of these wings is almost half the normal wing size, suggesting that a significant fraction of the tissue undergrowth caused by Brk can be attributed to these two target genes. Nonetheless, the lack of complete rescue implies that there might be more targets of Brk which also control tissue growth. In contrast, expression of PI3K (Dp110) did not rescue the undergrowth caused by Brk expression (supplementary Fig S4 online). Expression of Myc and/or bantam in nub>brk wings did not rescue the vein patterning defects (Fig 4B), indicating that the patterning and growth effects of Brk can be attributed to separate subsets of target genes.
Figure 4.
Expression of bantam and Myc rescues Brk-induced proliferation block. (A) Wings expressing Brk ubiquitously during development with nubbin–GAL4 are strongly reduced in size, and lose patterning elements such as veins. Reconstitution of bantam or Myc expression either alone or in combination rescues the undergrowth caused by Brk, but not the patterning defects. Error bars: standard deviation. ***t-test ⩽0.001, n=10 (one wing from 10 flies each). (B) Higher magnification images of two wings resulting from discs overexpressing Brk, bantam and Myc with nubbin–GAL4, showing a rescue of tissue growth but not veins.
DISCUSSION
Dpp signalling regulates tissue growth in the Drosophila wing disc by Brk. We use here a genome-wide approach to pinpoint Brk targets. Surprisingly, we find that Brk has >2,000 binding sites genome-wide. This was unexpected as only a handful of Dpp target genes have been identified so far. This also raises the question what all of these targets are doing? Dpp signalling regulates several aspects of wing development, such as patterning along the anterior/posterior axis, cellular growth rate, cell adhesion and cell competition. Therefore, it is likely that different subsets of target genes are required for these different functions. The ChIP data presented here might serve as a useful resource for studying how Brk regulates these different processes. Interestingly, although most of the Brk binding sites identified so far were fairly distant from transcription start sites (e.g., for Salm, Fig 1B), it appears that Brk also binds to the promoter region of many target genes, such as Dad, Salm and Myc (Figs 1A,B, 2A; supplementary Fig S2A online). This is likely of mechanistic relevance. We also noticed that genomic loci of target genes tend to have many Brk binding sites, in contrast to another transcription factor we studied in the past, FOXO [33]. This suggests that Brk needs several cooperative binding sites to properly repress gene expression, perhaps because Brk is a ‘weak’ repressor. Alternatively, genes with several activating enhancers might need Brk binding sites in each of these enhancers to obtain complete inhibition.
We identify here Myc as a downstream target, by which Dpp regulates tissue growth. Previous work suggested such a link might exist [34]. We show endogenous Brk limits Myc expression in wing disc cells (Fig 3). Furthermore, we find that expressing only two Brk targets, bantam and Myc, is enough to restore a significant fraction of the growth of Brk-overexpressing discs. This is unexpected given the >1,000 target genes of Brk. Nonetheless, expression of bantam and Myc does not completely rescue the wing size back to wild-type size, suggesting other Brk targets might exist that regulate growth. Of note, within Brk loss-of-function clones Myc protein levels are increased but not completely uniform throughout the clone (Fig 3D–D′′), suggesting more regulatory mechanisms might be regulating Myc expression.
Previous studies have suggested that differences in levels of Dpp and Brk signalling lead to cell competition [35]. As differential levels of Myc are a potent inducer of cell competition [36], this could partly explain how Brk affects cell competition. Finally, different models for how Dpp might be regulating tissue growth have been proposed in the recent literature. Our data support a simple, linear model of regulation of tissue growth by Dpp, whereby Dpp signalling represses Brk, allowing Myc levels to increase, thereby increasing the rates of ribosome biogenesis and cell growth.
METHODS
ChIP-Seq. Brk ChIP-Seq from wing imaginal discs was performed as previously described [24]. A total of 1,600 wing imaginal discs from Canton S third-instar larva were lysed in immunoprecipitation buffer (0.5% NP40, 150 mM NaCl, 200 mM Tris–HCl, pH 8, 20 mM EDTA, protease inhibitor) and 2 μg of Brk antibody used per ChIP. Immunoprecipitated and input samples were processed and sequenced following Solexa/Illumina protocols at the Ultrasequencing Unit of the CRG (Barcelona, Spain). An aliquot of 10 ng of each sample was used. Fragments sized 300–350 bp were selected before sequencing. Sequencing analysis was based on single reads of 36 nt aligned against the melanogaster genome (dm3 assembly, BDGP Release 5). After sequencing, 35,702,391 reads were obtained and 4,808,148 could be uniquely mapped to the genome. We ran PeakSeq [25] (READLENGTH=325, MAXGAP=40, MINFDR=0.05 and PVALTHRESH<10−10) to identify regions significantly enriched compared with normalized input control (published in the study by Perez-Lluch et al [24]). Brk ChIP-Seq profile and target regions were deposited at NCBI GEO as wiggle (WIG) and Browser Extensible Data (BED) files, respectively (accession GSE40957). To define Brk target genes we filtered for genes with Brk peaks <1,000 bp upstream of the transcription start site (TSS) or within introns.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
N.D. is part of the Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology (HBIGS), University of Heidelberg. We thank K. Weischenfeldt and the Ultrasequencing Unit of the CRG (Barcelona, Spain) for technical support. This work was supported by a Helmholtz Young Investigator Grant to A.A.T., and National Institutes of Health grant NIH R01 GM51186 to B.E. M.C. and E.B. acknowledge funding from the Consolider-Ingenio 2020 (CSD2007-00008), and M.R.-R. was supported by a Formación de Personal Investigador fellowship (BFU2006-07334), Ministerio de Ciencia e Innovación, Spain.
Author contributions: All authors designed the project and experiments, analysed results, and wrote the manuscript. N.D., M.R.-R., E.B. and A.A.T. performed the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Shingleton AW (2010) The regulation of organ size in Drosophila: physiology, plasticity, patterning and physical force. Organogenesis 6: 76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank G, Basler K (2010) Regulation of organ growth by morphogen gradients. Cold Spring Harb Perspect Biol 2: a001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA, Gallant P (2002) Control of growth and organ size in Drosophila. Bioessays 24: 54–64 [DOI] [PubMed] [Google Scholar]
- Goberdhan DC, Wilson C (2003) The functions of insulin signaling: size isn't everything, even in Drosophila. Differentiation 71: 375–397 [DOI] [PubMed] [Google Scholar]
- Edgar BA (2006) How flies get their size: genetics meets physiology. Nat Rev Genet 7: 907–916 [DOI] [PubMed] [Google Scholar]
- Mirth CK, Riddiford LM (2007) Size assessment and growth control: how adult size is determined in insects. Bioessays 29: 344–355 [DOI] [PubMed] [Google Scholar]
- Tumaneng K, Russell RC, Guan KL (2012) Organ size control by Hippo and TOR pathways. Curr Biol 22: R368–R379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL (2010) The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev 24: 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affolter M, Basler K (2007) The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat Rev Genet 8: 663–674 [DOI] [PubMed] [Google Scholar]
- Campbell G, Tomlinson A (1999) Transducing the Dpp morphogen gradient in the wing of Drosophila: regulation of Dpp targets by brinker. Cell 96: 553–562 [DOI] [PubMed] [Google Scholar]
- Minami M, Kinoshita N, Kamoshida Y, Tanimoto H, Tabata T (1999) Brinker is a target of Dpp in Drosophila that negatively regulates Dpp-dependent genes. Nature 398: 242–246 [DOI] [PubMed] [Google Scholar]
- Yao LC, Phin S, Cho J, Rushlow C, Arora K, Warrior R (2008) Multiple modular promoter elements drive graded brinker expression in response to the Dpp morphogen gradient. Development 135: 2183–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B, Hartmann B, Pyrowolakis G, Affolter M, Basler K (2003) Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell 113: 221–233 [DOI] [PubMed] [Google Scholar]
- Jazwinska A, Kirov N, Wieschaus E, Roth S, Rushlow C (1999) The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell 96: 563–573 [DOI] [PubMed] [Google Scholar]
- Sivasankaran R, Vigano MA, Muller B, Affolter M, Basler K (2000) Direct transcriptional control of the Dpp target omb by the DNA binding protein Brinker. EMBO J 19: 6162–6172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick H, Johnson K, Laughon A (2001) Repression of dpp targets by binding of brinker to mad sites. J Biol Chem 276: 18216–18222 [DOI] [PubMed] [Google Scholar]
- Weiss A, Charbonnier E, Ellertsdottir E, Tsirigos A, Wolf C, Schuh R, Pyrowolakis G, Affolter M (2010) A conserved activation element in BMP signaling during Drosophila development. Nat Struct Mol Biol 17: 69–76 [DOI] [PubMed] [Google Scholar]
- Wartlick O, Mumcu P, Kicheva A, Bittig T, Seum C, Julicher F, Gonzalez-Gaitan M (2011) Dynamics of Dpp signaling and proliferation control. Science 331: 1154–1159 [DOI] [PubMed] [Google Scholar]
- Schwank G, Yang SF, Restrepo S, Basler K (2012) Comment on ‘Dynamics of dpp signaling and proliferation control’. Science 335: 401. [DOI] [PubMed] [Google Scholar]
- Schwank G, Tauriello G, Yagi R, Kranz E, Koumoutsakos P, Basler K (2011) Antagonistic growth regulation by Dpp and Fat drives uniform cell proliferation. Dev Cell 20: 123–130 [DOI] [PubMed] [Google Scholar]
- Martin FA, Perez-Garijo A, Moreno E, Morata G (2004) The brinker gradient controls wing growth in Drosophila. Development 131: 4921–4930 [DOI] [PubMed] [Google Scholar]
- Schwank G, Restrepo S, Basler K (2008) Growth regulation by Dpp: an essential role for Brinker and a non-essential role for graded signaling levels. Development 135: 4003–4013 [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD (2011) Cooperative regulation of growth by Yorkie and Mad through bantam. Dev Cell 20: 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Lluch S, Blanco E, Carbonell A, Raha D, Snyder M, Serras F, Corominas M (2011) Genome-wide chromatin occupancy analysis reveals a role for ASH2 in transcriptional pausing. Nucleic Acids Res 39: 4628–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB (2009) PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol 27: 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio R, de Celis JF (2004) Regulation of spalt expression in the Drosophila wing blade in response to the Decapentaplegic signaling pathway. Proc Natl Acad Sci USA 101: 6021–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS (2002) Genome-wide analysis of clustered dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci USA 99: 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P (1999) Drosophila myc regulates cellular growth during development. Cell 98: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DC, Johnston LA (2010) Control of wing size and proportions by Drosophila myc. Genetics 184: 199–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto-Silva RM, de Beco S, Johnston LA (2010) Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell 19: 507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell NC, Johanson TM, Cranna NJ, Er AL, Richardson HE, Hannan RD, Quinn LM (2010) Hfp inhibits Drosophila myc transcription and cell growth in a TFIIH/Hay-dependent manner. Development 137: 2875–2884 [DOI] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM (2008) Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab 7: 21–32 [DOI] [PubMed] [Google Scholar]
- Prober DA, Edgar BA (2002) Interactions between Ras1, dMyc, and dPI3K signaling in the developing Drosophila wing. Genes Dev 16: 2286–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E, Basler K, Morata G (2002) Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature 416: 755–759 [DOI] [PubMed] [Google Scholar]
- Johnston LA (2009) Competitive interactions between cells: death, growth, and geography. Science 324: 1679–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.