Abstract
Autophagosome formation is a dynamic process that is strictly controlled by autophagy-related (Atg) proteins. However, how these Atg proteins are recruited to the autophagosome formation site or autophagic membranes remains poorly understood. Here, we found that FIP200, which is involved in proximal events, directly interacts with Atg16L1, one of the downstream Atg factors, in an Atg14- and phosphatidylinositol 3-kinase-independent manner. Atg16L1 deletion mutants, which lack the FIP200-interacting domain, are defective in proper membrane targeting. Thus, FIP200 regulates not only early events but also late events of autophagosome formation through direct interaction with Atg16L1.
Keywords: autophagy, FIP200, ULK1, Atg16L1
INTRODUCTION
Macroautophagy, hereafter referred to simply as autophagy, is an intracellular degradation system mediated by the autophagosome. Cytoplasmic materials, such as proteins, lipids, glycogens and organelles, are engulfed by an isolation membrane/phagophore, leading to the formation of a double-membrane autophagosome. Next, the autophagosome fuses with a lysosome, and the sequestered contents and the inner membranes are degraded [1]. So far, more than 30 autophagy-related (ATG) genes have been identified in yeast [2, 3]. These include Atg1–10, 12–14, 16–18, 29 and 31, most of which are conserved in higher eukaryotes and are essential for the formation of the autophagosome [1, 3].
Following the comprehensive hierarchical analysis of yeast Atg proteins [4], the genetic hierarchy of mammalian Atg proteins has also been determined. The ULK1–Atg13–FIP200–Atg101 complex and Atg9A are independently recruited to the autophagosome formation site, and both are required for recruitment of the class III phosphatidylinositol 3-kinase (PtdIns3K) complex (Beclin1–Atg14(L)–Vps15–Vps34) [5,6,7]. PtdIns3K activity is required for further recruitment of the Atg2–WIPI and Atg12—Atg5–Atg16L1 complexes (— indicates a covalent attachment) [5, 8,9,10]. Finally, LC3 is recruited in an Atg12—Atg5–Atg16L1-dependent manner [11].
Whereas WIPI proteins have a phosphoinositide-binding motif for recruitment to the autophagic membrane, Atg5, Atg12 and Atg16L1 do not have such typical membrane-binding motifs. Thus, it remains unknown how the Atg12—Atg5–Atg16L1 complex is recruited during starvation-induced autophagy. Furthermore, conflicting results have also been reported with regard to selective autophagy. In Salmonella xenophagy, the recruitment of LC3 and Atg5 to the Salmonella-containing vacuole is independent of PtdIns3K, FIP200 and Atg9A [12]. In Parkin-dependent mitophagy, LC3 can also be recruited to depolarized mitochondria even in FIP200 knockout (KO) mouse embryonic fibroblasts (MEFs) and Atg9A KO MEFs [6]. Therefore, the recruitment of Atg12—Atg5–Atg16L1 and LC3 to the autophagosome formation site appears to be complicated.
Here, we report the physical interaction between the Atg12—Atg5–Atg16L1 complex and the ULK1 complex, and describe a new mode of Atg16L1 targeting to the isolation membrane. Furthermore, we discuss the role of the carboxy-terminal WD-repeat domain of Atg16L1, which is absent in yeast Atg16.
RESULTS
Identification of FIP200 as an Atg16L1-interacting protein
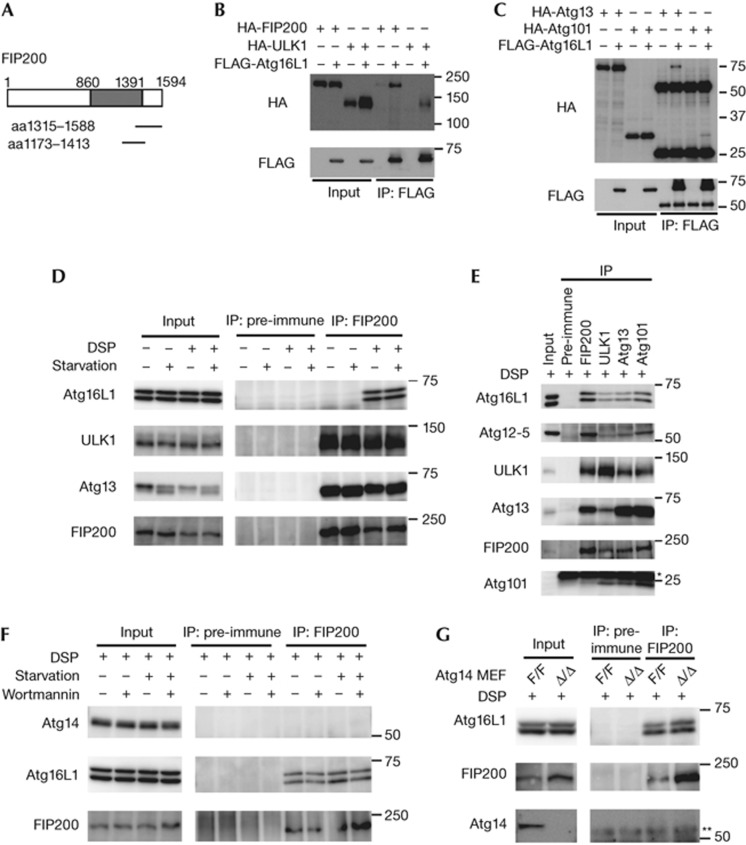
We screened a mouse brain complementary DNA yeast two-hybrid library with mouse Atg16L1 as ‘bait’ and isolated clones that all encoded partial sequences of FIP200 (Fig 1A). To confirm the interaction between Atg16L1 and FIP200, we first performed transient transfection experiments in HEK293T cells. Immunoprecipitation (IP) analysis revealed that FLAG–Atg16L1 interacts with HA–FIP200 (Fig 1B). As FIP200 forms a stable complex with ULK1, Atg13 and Atg101 [13,14,15], we examined the interaction between Atg16L1 and other components of the ULK1 complex. Interactions between FLAG–Atg16L1 and HA–ULK1, HA–Atg13, and HA–Atg101 were detected, but were weaker than the interaction between FLAG–Atg16L1 and HA–FIP200 (Fig 1B,C). Furthermore, endogenous Atg16L1 was co-immunoprecipitated with endogenous FIP200 in the presence, but not the absence, of the chemical crosslinker dithiobis(succinimidyl propionate) (DSP) (Fig 1D). Endogenous interactions between Atg16L1 and other components of the ULK1–Atg13–FIP200–Atg101 complex were also detected (Fig 1E). In addition, the immunoprecipitates using antibodies against FIP200, ULK1, Atg13 and Atg101 included not only Atg16L1 but also the Atg12—Atg5 conjugate (Fig 1E), and Atg12—Atg5 and Atg16L1 were included in a large (∼3 MDa) protein complex dependent on FIP200 (supplementary Fig S1 online). The interaction between Atg16L1 and FIP200 was not significantly changed by nutrient conditions (Fig 1D,F), and by deletion of ULK1 and ULK2 (supplementary Fig S2 online). Collectively, these results indicate that the Atg12—Atg5–Atg16L1 complex weakly interacts with the ULK1–Atg13–FIP200–Atg101 complex via direct interaction with Atg16L1–FIP200.
Figure 1.
Atg16L1 interacts with the ULK1–Atg13–FIP200–Atg101 complex. (A) Yeast two-hybrid screening for Atg16L1-interacting proteins. The Atg16L1-interacting FIP200 clones are indicated. (B, C) HEK293T cells were co-transfected with the indicated constructs. Cell lysates were subjected to IP using anti-FLAG antibodies. The resulting precipitates were examined by immunoblot analysis with anti-FLAG and anti-HA antibodies. (D) HEK293T cells were cultured in regular or starvation medium for 2 h. Cells were harvested and treated with DSP. (E) HEK293T cells were harvested and treated with DSP. (F) HEK293T cells were cultured in regular DMEM or starvation medium in the presence or absence of 200 nM wortmannin for 2 h. After treatment with DSP, cell lysates were subjected to IP analysis. (G) Atg14F/F (undeleted) or Atg14Δ/Δ (deleted) MEFs were harvested and treated with DSP. * (E) and ** (G) indicate the positions of the immunoglobulin light and heavy chains, respectively. aa, amino acid; DSP, dithiobis(succinimidyl propionate); IP, immunoprecipitation; MEFs, mouse embryonic fibroblasts.
Atg14 is not essential for the Atg16L1–FIP200 interaction
As PtdIns(3)P is required for the recruitment of the Atg12—Atg5–Atg16L1 complex to the isolation membrane [1, 5], we examined the involvement of PtdIns3K in the interaction with Atg16L1–FIP200. First, we found that the Atg16L1–FIP200 interaction was insensitive to the PtdIns3K inhibitor wortmannin (Fig 1F). As Atg14 is a specific subunit of the autophagy-specific class III PtdIns3K complex [1, 16], we directly investigated the involvement of Atg14 in the Atg16L1–FIP200 interaction. IP analysis revealed that Atg14 was not included in the FIP200 complex (Fig 1F). In addition, the Atg16L1–FIP200 interaction was still observed in cell lysates derived from Atg14-deficient MEFs (Fig 1G). These data suggest that the Atg16L1–FIP200 interaction is not dependent on Atg14 and PtdIns(3)P. PtdIns(3)P might be important for concentration of Atg16L1 to the puncta or stabilization of Atg16L1 on the membranes, rather than for membrane targeting.
Atg12—5–16L1 stabilizes the ULK1 complex
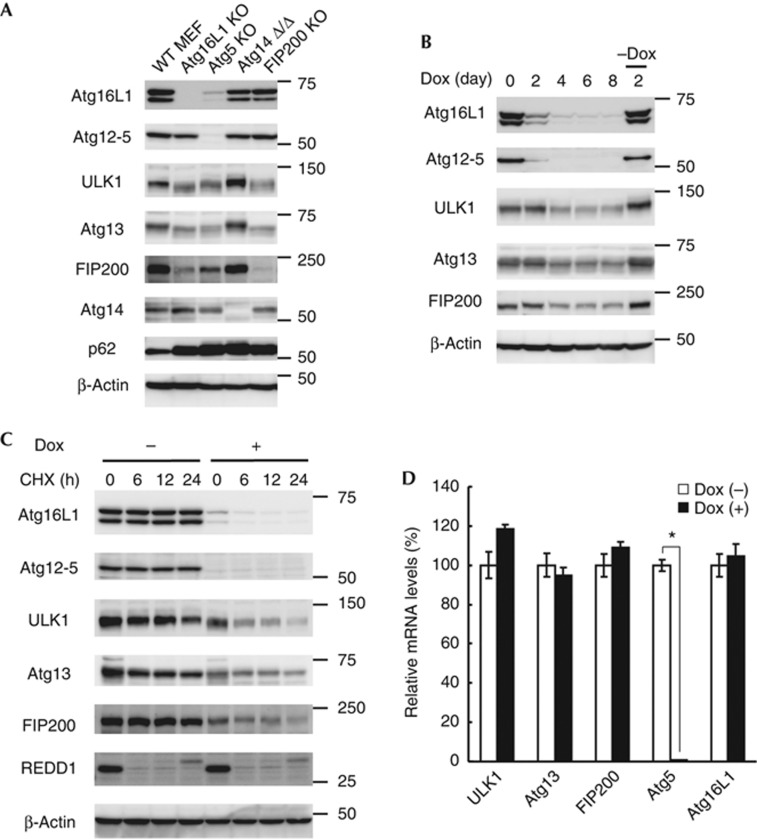
To further validate the interaction between the Atg12—Atg5–Atg16L1 and ULK1–Atg13–FIP200–Atg101 complexes in vivo, we determined expression levels of these proteins in various cells lacking Atg proteins. In FIP200 KO cells, the expression levels of Atg12—Atg5 and Atg16L1 were not significantly affected (Fig 2A). On the other hand, expression levels of ULK1, Atg13 and FIP200 were reduced in Atg5 KO and Atg16L1 KO MEFs (Fig 2A), although the ULK1–Atg13–FIP200 complex could be formed in Atg5 KO MEFs (supplementary Fig S3 online). Atg16L1 expression was also significantly reduced in Atg5 KO MEFs (Fig 2A). To exclude the possibility of clonal variation among cell lines, we used m5-7 cells, in which Atg5 expression can be regulated with doxycycline (Dox) [17]. As reported previously, expression of Atg5 was suppressed following addition of Dox (Fig 2B). Concomitantly, there was a decrease in Atg16L1 expression by day 4. After day 4, the expression levels of ULK1, Atg13 and FIP200 were also reduced. A further 2-day incubation after washing out Dox could reverse these effects, suggesting that deficiency of either Atg5 or Atg16L1 destabilizes ULK1, Atg13 and FIP200. Indeed, degradation of the ULK1 complex components was accelerated in the absence of Atg5 (Fig 2C), without significant changes in their messenger RNA levels (Fig 2D). On the other hand, Atg14 deficiency, which causes accumulation of the autophagy substrate p62, had no effect on these components (Fig 2A). Collectively, these results indicate that the Atg12—Atg5–Atg16L1 complex interacts with a large portion of the ULK1–Atg13–FIP200–Atg101 complex and stabilizes these components in the cell, even though the interaction between these two complexes was weak in the IP experiments. These studies reveal substantial and important interactions between protein complexes that mediate early and late steps in the autophagy pathway.
Figure 2.
The Atg12—Atg5–Atg16L1 complex regulates the stability of the ULK1–Atg13–FIP200 complex. (A) Immunoblot analysis of Atg proteins in the indicated MEFs. (B) m5-7 cells were cultured in regular medium containing 10 ng/ml Dox for 8 days, and then cultured in the absence of Dox for a further 2 days. (C,D) m5-7 cells were cultured in regular medium with or without 10 ng/ml Dox for 4 days, and then cultured in the presence of 50 μg/ml CHX for the indicated time periods (C). mRNA levels of the indicated genes were measured by quantitative PCR following the 4-day Dox treatment. Data represent mean±s.e. (*P<0.05) (D). CHX, cycloheximide; Dox, doxycycline; MEFs, mouse embryonic fibroblasts; mRNA, messenger RNA; WT, wild-type.
Identification of the FIP200-interacting domain in Atg16L1
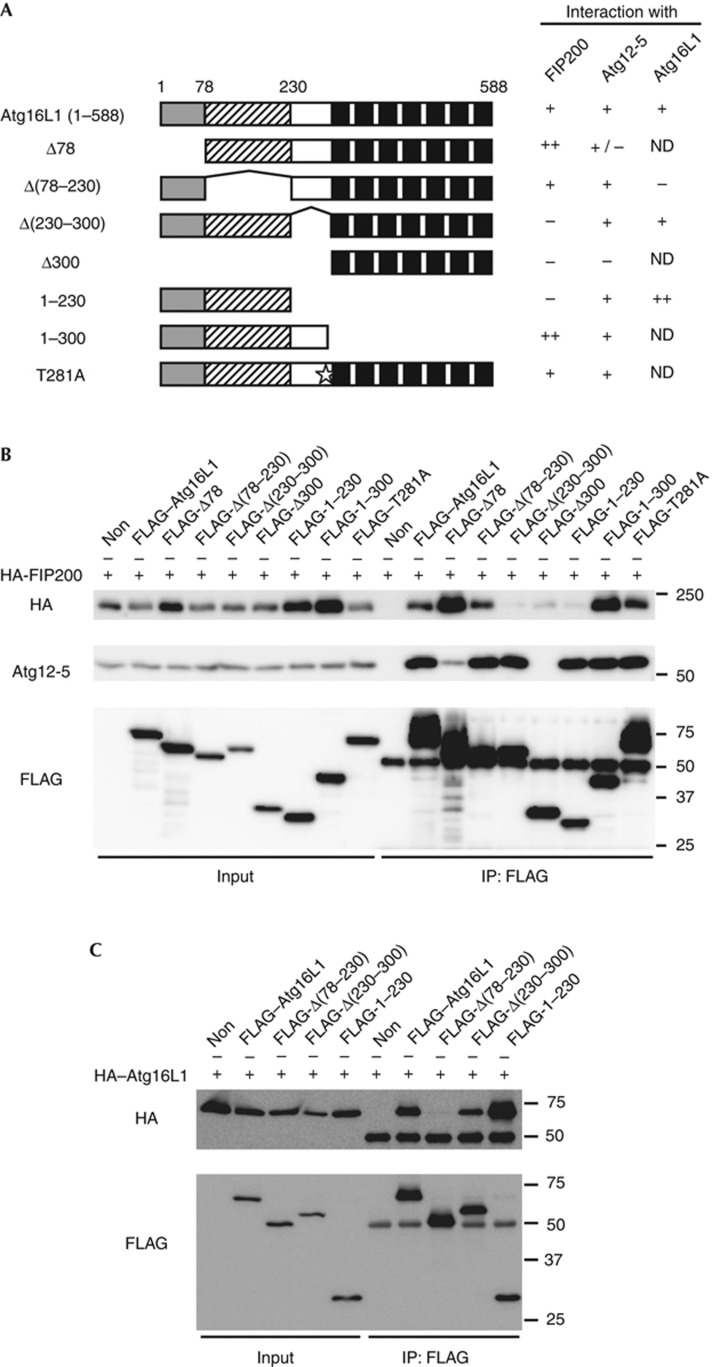
IP analysis revealed that the Atg16L1 mutants Δ(230–300), Δ300 and 1–230 did not interact with FIP200, suggesting that the middle region of Atg16L1α (amino acids 230–300) was essential for interaction with FIP200 (Fig 3A,B). Introduction of the Crohn’s disease-associated mutation T281A (corresponding to T300A in human Atg16L1β) did not significantly affect the Atg16L1–FIP200 interaction (Fig 3A,B). While the interaction between the Atg16L1Δ78 mutant and Atg5 was reduced as reported previously [18], two FIP200-interacting domain deletion mutants, Atg16L1Δ(230–300) and Atg16L1(1–230), could interact with endogenous Atg5 (Fig 3A,B). We further narrowed down the interaction domain to two regions in Atg16L1, 230–250 and 288–300, which were involved in the interaction with FIP200 (supplementary Fig S4A,B online). These regions are highly conserved among species but not in human Atg16L2, which cannot localize to the isolation membrane [19] (supplementary Fig S4C online). The FIP200-interacting domain deletion mutants Atg16L1Δ(230–300) and Atg16L1(1–230) could still form heterodimers with full-length Atg16L1, whereas the coiled–coil deletion mutant Atg16L1Δ(78–230) could not (Fig 3A,C). These results indicate that the middle region of Atg16L1 is required for its interaction with FIP200, but not with Atg5 or Atg16L1.
Figure 3.
The middle region of Atg16L1 is required for interaction with FIP200. (A) Schematic representations of the wild-type Atg16L1 and its mutants. Human Atg16L1α (NP_060444) has an amino-terminal Atg5-interacting domain (grey box), a coiled–coil domain (shaded box) and seven WD repeats (black boxes). Star indicates the position of the Crohn’s disease-associated T281A mutation. (B,C) HEK293T cells were co-transfected with the indicated constructs. Cell lysates were analysed by IP. IP, immunoprecipitation; ND, not determined.
Although it was reported that Rab33B interacts with the middle region of Atg16L1 [20], Rab33B was not immunoprecipitated with FIP200 and overexpression of Rab33B or OATL1, a GAP for Rab33B, did not significantly affect the Atg16L1–FIP200 interaction (supplementary Fig S5 online), suggesting that Rab33B is not involved in this interaction.
FIP200 regulates membrane targeting of Atg16L1
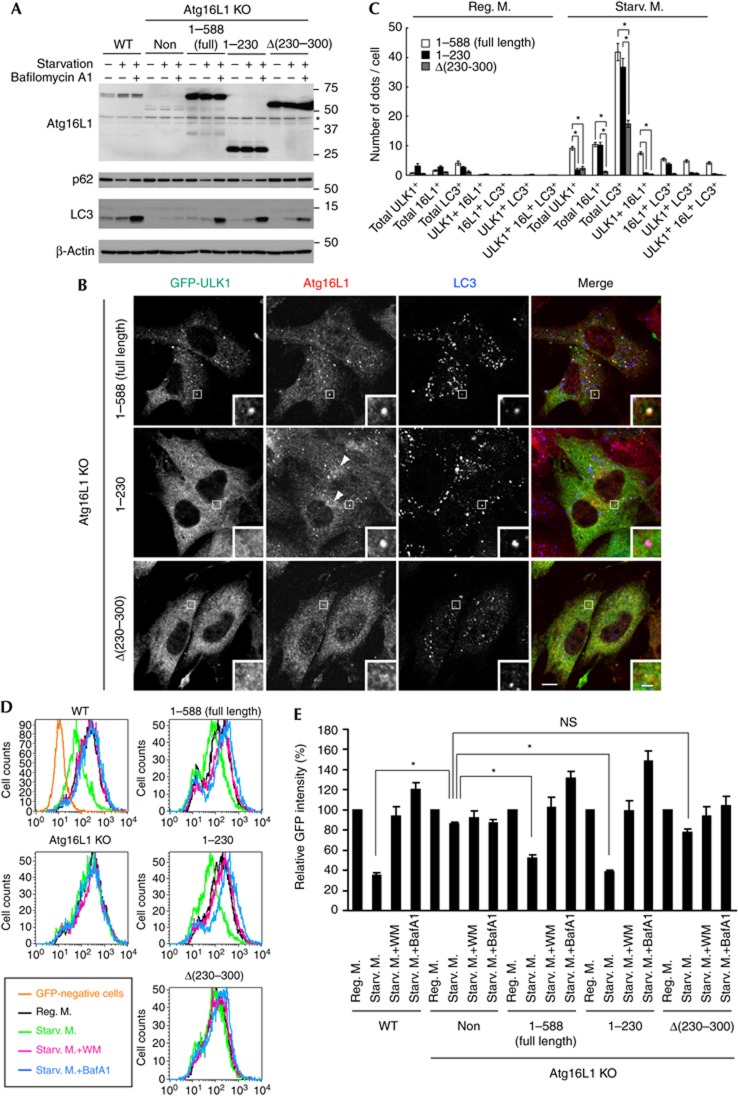
To investigate the role of the Atg16L1–FIP200 interaction in autophagy, we performed an autophagic flux assay using Atg16L1 KO MEFs. LC3 conjugation was blocked in Atg16L1 KO MEFs, and was restored by expression of non-tagged full-length Atg16L1α (Fig 4A). Bafilomycin A1-induced accumulation of LC3-II, which represents autophagic flux, was also restored. However, the expression of FIP200-binding-deficient Atg16L1Δ(230–300) only partially restored the autophagic flux, compared with that of full-length Atg16L1α (Fig 4A). The several LC3 puncta generated in response to starvation was also significantly reduced in Atg16L1 KO MEFs expressing the Atg16L1Δ(230–300) mutant (Fig 4B,C). In line with this, lysosomal degradation of p62 (Fig 4A) and GFP–LC3 (Fig 4D,E), both of which are known to be through autophagy, was induced by starvation in Atg16L1 KO MEFs expressing full-length Atg16L1, but not Atg16L1Δ(230–300). Moreover, the Atg16L1Δ(230–300) mutant did not show a clear punctate distribution pattern under starvation conditions (Fig 4B,C). Whereas there were about 10 dots per cell for full-length Atg16L1α, there was only about 1 dot per cell for the Atg16L1Δ(230–300) mutant (Fig 4B,C). Similar results were obtained in HeLa cells using FLAG-tagged Atg16L1Δ(230–300) (supplementary Fig S6 online).
Figure 4.
Atg16L1Δ(230–300) mutant is impaired in isolation membrane targeting and partially restores the autophagic defects of Atg16L1 KO MEFs. (A) WT MEFs, Atg16L1 KO MEFs or Atg16L1 KO MEFs stably expressing either full-length Atg16L1(1–588), Atg16L1(1–230) or Atg16L1Δ(230–300) were cultured in regular DMEM or starvation medium in the presence or absence of 100 nM BafA1 for 2 h. * indicates nonspecific immunoreactive bands. (B,C) Atg16L1 KO MEFs stably expressing GFP–ULK1 and either full-length Atg16L1(1–588), Atg16L1(1–230) or Atg16L1Δ(230–300) were cultured in regular DMEM (C) or starvation medium (B,C) for 1 h. Cells were fixed and analysed by immunofluorescence microscopy using anti-GFP, anti-Atg16L1 and anti-LC3 antibodies. Arrowheads indicate the perinuclear localization of Atg16L1(1–230). The number of dots was quantified from more than 30 randomly selected cells from three independent samples as described in the Methods. Data represent mean±s.e. (*P<0.001, analysis of variance followed by Bonferroni/Dunn post hoc test). Scale bar, 10 μm, and 2 μm in inset. (D, E) Cells stably expressing GFP–LC3 were cultured in regular DMEM, starvation medium or starvation medium in the presence of 0.2 μM WM or 100 nM BafA1 for 6 h. Total cellular GFP–LC3 signals were analysed by flow cytometry. Representative FACS data were shown (D). The geometric mean of fluorescence intensity was determined. Values are expressed as a percentage of the mean of control cells cultured in regular DMEM. Data represent mean±s.e. (*P<0.05) (E). BafA1, bafilomycin A1; GFP, green fluorescent protein; KO, knockout; MEFs, mouse embryonic fibroblasts; NS, not significant; WM, wortmannin; WT, wild-type.
By contrast, the Atg16L1(1–230) mutant, which also lacks the FIP200-binding domain, fully restored the autophagic flux defects of Atg16L1 KO MEFs (Fig 4A,D). The several puncta of Atg16L1(1–230) observed under starvation conditions was similar to that of full-length Atg16L1 (Fig 4B,C). However, the colocalization of Atg16L1(1–230) puncta with GFP–ULK1, which associates with the isolation membrane, was decreased significantly. Although more than 70% of puncta of full-length Atg16L1 were colocalized with GFP–ULK1 under starvation conditions, Ag16L1(1–230) puncta were mostly GFP–ULK1 negative (Fig 4B,C; supplementary Fig S6 online). We noticed that the several ULK1 puncta was significantly reduced in the Atg16L1 KO MEFs expressing the FIP200-interacting mutant compared with those expressing full-length Atg16L1 (Fig 4B,C). Treatment with wortmannin recovered formation of the ULK1 puncta in these cells (supplementary Fig S7 online), suggesting that the reduction of ULK1 puncta is caused by enhanced dissociation and/or reduced expression of ULK1, rather than by impaired recruitment.
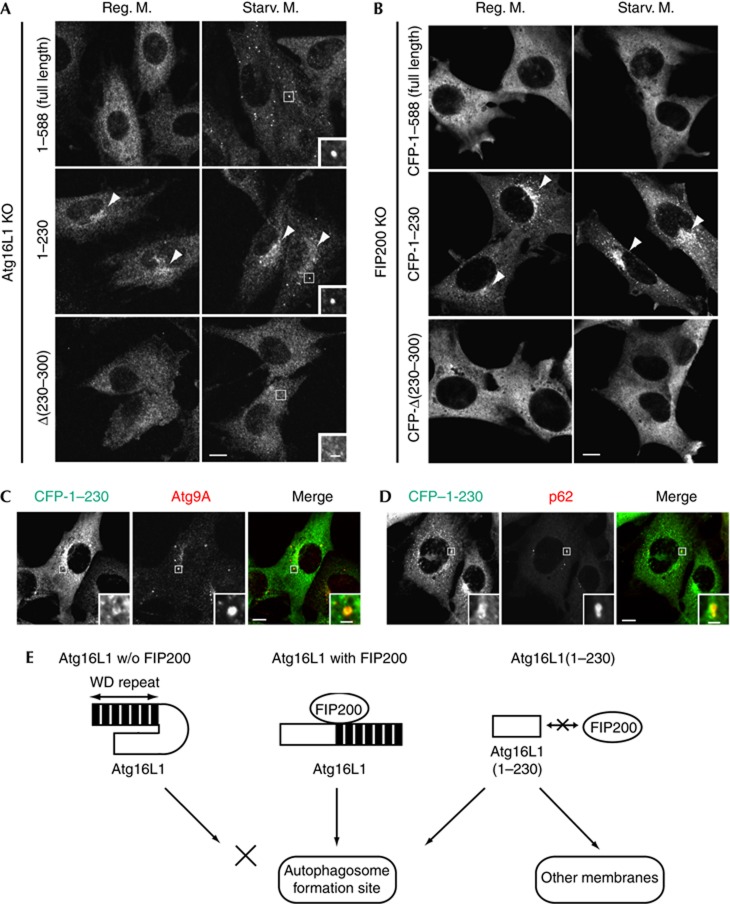
We also found that, in addition to the punctate structures, the Atg16L1(1–230) mutant localized to the perinuclear region both in nutrient-rich and in starvation condition (Figs 4B, 5A, arrowheads). It is interesting that this perinuclear localization of Atg16L1(1–230) was observed even in the absence of FIP200 (Fig 5B, arrowheads), indicating that the recruitment of the 1–230 mutant can be independent of FIP200. A small proportion of Atg16L1(1–230) also colocalized with Atg9A and p62 in FIP200 KO MEFs (Fig 5C,D), suggesting that Atg16L1(1–230) itself has some affinity for the autophagic membrane. However, Atg16L1(1–230) also targeted to the unknown perinuclear compartments, to which Atg9A and p62 did not localize, suggesting that the membrane targeting of Atg16L1(1–230) is deregulated. Collectively, these findings suggest that the Atg16L1–FIP200 interaction is required for the proper targeting of Atg16L1 to the isolation membrane.
Figure 5.
Atg16L1(1–230) localizes to aberrant membranes in a FIP200-independent manner. (A) Atg16L1 KO MEFs stably expressing either full-length Atg16L1(1–588), Atg16L1(1–230) or Atg16L1Δ(230–300) were cultured in regular DMEM or starvation medium for 1 h. Cells were fixed and subjected to immunofluorescence microscopy using anti-Atg16L1 antibody. Note that full-length Atg16L1 and Atg16L1(1–230), but not Atg16L1Δ(230–300), showed punctate structures under starvation conditions (inset). (B) FIP200 KO MEFs stably expressing either CFP–Atg16L1(1–588), CFP–Atg16L1(1–230) or CFP–Atg16L1Δ(230–300) were cultured in regular DMEM or starvation medium for 1 h. Cells were fixed and analysed by immunofluorescence microscopy using anti-GFP antibody. Arrowheads indicate the perinuclear localization of Atg16L1(1–230). (C,D) FIP200 KO MEFs stably expressing CFP–Atg16L1(1–230) were cultured in regular DMEM. Scale bar, 10 μm, and 2 μm in inset. (E) Hypothetical model of membrane targeting of Atg16L1. Interaction of Atg16L1 with FIP200 leads to proper targeting of this complex to the autophagosome formation site (middle). Without FIP200, puncta formation of Atg16L1 is impaired probably owing to a self-inhibitory role of the C-terminal WD-repeat domain (left). If the WD-repeat domain is deleted, the N-terminal half of Atg16L1 localizes to aberrant membranes as well as the autophagosome formation site (right). KO, knockout; MEFs, mouse embryonic fibroblasts.
DISCUSSION
We have shown that Atg16L1 directly interacts with FIP200 (Fig 1) and that this interaction is important for the proper targeting of Atg16L1 to the isolation membrane (Fig 5). During revision of this manuscript, the Atg16L1–FIP200 interaction and its importance in autophagy induction were also reported by Gammoh et al [21]. Considering that the Atg12—Atg5–Atg16L1 complex interacts with the ULK1–Atg13–FIP200–Atg101 complex independently of nutrient condition, it is reasonable to assume that these two units form a large complex in the cytoplasm, which then targets to the membrane as one complex. This suggests that the sequential action of components of the autophagy machinery could reflect the actions of different components of a large complex rather than the sequential recruitment of distinct and non-interacting protein complexes. These data are consistent with our findings of live imaging analysis, which shows that ULK1 and Atg5 are recruited to the same compartment with similar kinetics (Koyama-Honda et al, unpublished work).
Nonetheless, our results indicate that the 231–588 region of Atg16L1, including the FIP200-interacting domain and WD-repeat domain, is not essential for autophagy. As yeast Atg16 lacks this region, Atg16L1 might have acquired a new regulatory function in multicellular eukaryotes. To determine the precise function of the 231–588 region, we demonstrated that the Atg16L1(1–230) mutant localizes to punctate structures as well as a perinuclear structure in FIP200 KO MEFs, whereas full-length Atg16L1 shows a diffuse cytoplasmic pattern in the absence of FIP200 (Fig 5). This result suggests the presence of autoinhibitory regulation (Fig 5E). We propose that the 231–588 region of Atg16L1 inhibits the membrane targeting of Atg16L1 and, following interaction of Atg16L1 with FIP200 via its 230–300 region, Atg16L1 can target to the membrane by removal of autoinhibition. FIP200 could also serve as a recruiter and ensure the specific targeting of Atg16L1 to the isolation membrane. Structural analysis of Atg16L1 will be required for further understanding of its precise molecular function.
METHODS
The experimental procedures are described in detail in the supplementary information online.
Cell culture. Atg14flox/flox MEFs were obtained from Atg14flox/flox mouse embryos [12], and immortalized with SV40 large T antigen using pEF321-T. Other cell lines are described in the supplementary information online.
Chemical crosslinking. Cells were harvested, resuspended in ice-cold phosphate-buffered saline containing 1 mM DSP and incubated on ice for 30 min.
Statistical analysis. Differences were statistically analysed using unpaired t-test or analysis of variance with Bonferroni/Dunn post hoc test.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Jun-Lin Guan for providing FIP200 KO MEFs, Sharon A. Tooze for providing ULK1/2 DKO MEFs, Shoji Yamaoka for the retroviral vectors, Atsushi Miyawaki for the plasmid-coding SECFP, Teruhito Yasui for the pCG-VSV-G and pCG-gag-pol plasmids, Mitsunori Fukuda for FLAG-Rab33B and T7-OATL1 plasmids, Nao Hosokawa for preparing the HA–Atg101 plasmid, Takeshi Yagi for the pCre-Pac plasmid and Sumio Sugano for the pEF321-T plasmid. This work was supported in part by the Funding Program for Next Generation World-Leading Researchers and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to N.M.), and by the Naito Foundation (to T.N.).
Author contributions: T.N., T.K., H.W.V. and N.M. designed the experiments; T.S. and S.A. generated Atg14flox/flox MEFs; T.N., T.K., K.C. and M.H.S. performed the experiments; T.N., T.K., K.C., M.H.S., H.W.V. and N.M. analysed the data; and T.N., T.K., K.C., H.W.V. and N.M. wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27: 107–132 [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12: 209–218 [DOI] [PubMed] [Google Scholar]
- Itakura E, Mizushima N (2010) Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6: 764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Koyama-Honda I, Mizushima N (2012) Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J Cell Sci 125: 1488–1499 [DOI] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley H, Robinson D, Weston A, Collinson L, Tooze S (2012) Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, is required for autophagy. Mol Biol Cell 23: 1860–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA (2010) Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6: 506–522 [DOI] [PubMed] [Google Scholar]
- Lu Q et al. (2011) The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell 21: 343–357 [DOI] [PubMed] [Google Scholar]
- Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N (2012) Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell 23: 896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T (2008) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19: 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama S, Omori H, Saitoh T, Sone T, Guan JL, Akira S, Imamoto F, Noda T, Yoshimori T (2011) The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell 22: 2290–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N et al. (2009) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N (2009) Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5: 973–979 [DOI] [PubMed] [Google Scholar]
- He C, Levine B (2010) The Beclin 1 interactome. Curr Opin Cell Biol 22: 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara Y, Mizushima N (2006) Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett 580: 2623–2629 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T (2003) Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci 116: 1679–1688 [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Fujita N, Kanno E, Omori H, Yoshimori T, Itoh T, Fukuda M (2011) Atg16L2, a novel isoform of mammalian Atg16L that is not essential for canonical autophagy despite forming an Atg12-5-16L2 complex. Autophagy 7: 1500–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M (2008) Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell 19: 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammoh N, Florey O, Overholtzer M, Jiang X (2012) Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat Struct Mol Biol [Epub ahead of print] doi:; DOI: 10.1038/nsmb.2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.