Abstract
Drosophila use small-interfering RNA mechanisms to limit the amplification of viral genomes. However, it is unclear how small RNA interference components recognize and separate viral from cellular RNA. Dnmt2 enzymes are highly conserved RNA methyltransferases with substrate specificity towards cellular tRNAs. We report here that Dnmt2 is required for efficient innate immune responses in Drosophila. Dnmt2 mutant flies accumulate increasing levels of Drosophila C virus and show activated innate immune responses. Binding of Dnmt2 to DCV RNA suggests that Dnmt2 contributes to virus control directly, possibly by RNA methylation. These observations demonstrate a role for Dnmt2 in antiviral defence.
Keywords: innate immunity, DCV, Dnmt2, RNA, methylation
Introduction
Efficient immune responses are essential for the control and elimination of infections with viruses and the survival of the infected host. Recent work in Drosophila has revealed the importance of small RNA processing machineries for virus control [1, 2]. In particular, the activity of Dicer-2 (Dcr-2) is required for the production of virus-derived small-interfering RNAs (vsiRNAs), which are loaded into Argonaute-2 to target and cleave viral RNA genomes [3, 4]. While Dcr-2 accepts long double-stranded RNA substrates and dices these into vsiRNAs, it is unclear where and how Dcr-2 detects and sorts its substrate RNAs. Membrane compartments that are crucially important for viral entry and replication [5] have also been linked to the activation of innate immune responses [6, 7] and small-interfering RNA pathway activity [8]. These observations suggest that siRNA-mediated antiviral defence mechanisms evolve at the intersection of cellular and endosomal membrane systems.
Dnmt2 genes have been strongly conserved during evolution, suggesting a biological function that would justify long-term evolutionary selection [9, 10]. Although being annotated as DNA methyltransferases, recent findings have established that Dnmt2 enzymes are transfer RNA methyltransferases [11] and that this methylation becomes important under stress conditions [12, 13]. To analyse the biological function of Dnmt2 in adult flies, transcriptional profiling was performed. Several stress response genes, including components of the Drosophila innate immune system, were upregulated in Dnmt2 mutant flies. Accumulation of (+) RNA viruses was associated with increased immune responses in Dnmt2 mutants. Infection studies suggested that Dnmt2 has a role in the acute immune response to Drosophila C virus (DCV) infection by binding to and possibly methylation of viral RNA. Our data indicate that Dnmt2 proteins are required to control specific RNA viruses in Drosophila.
Results
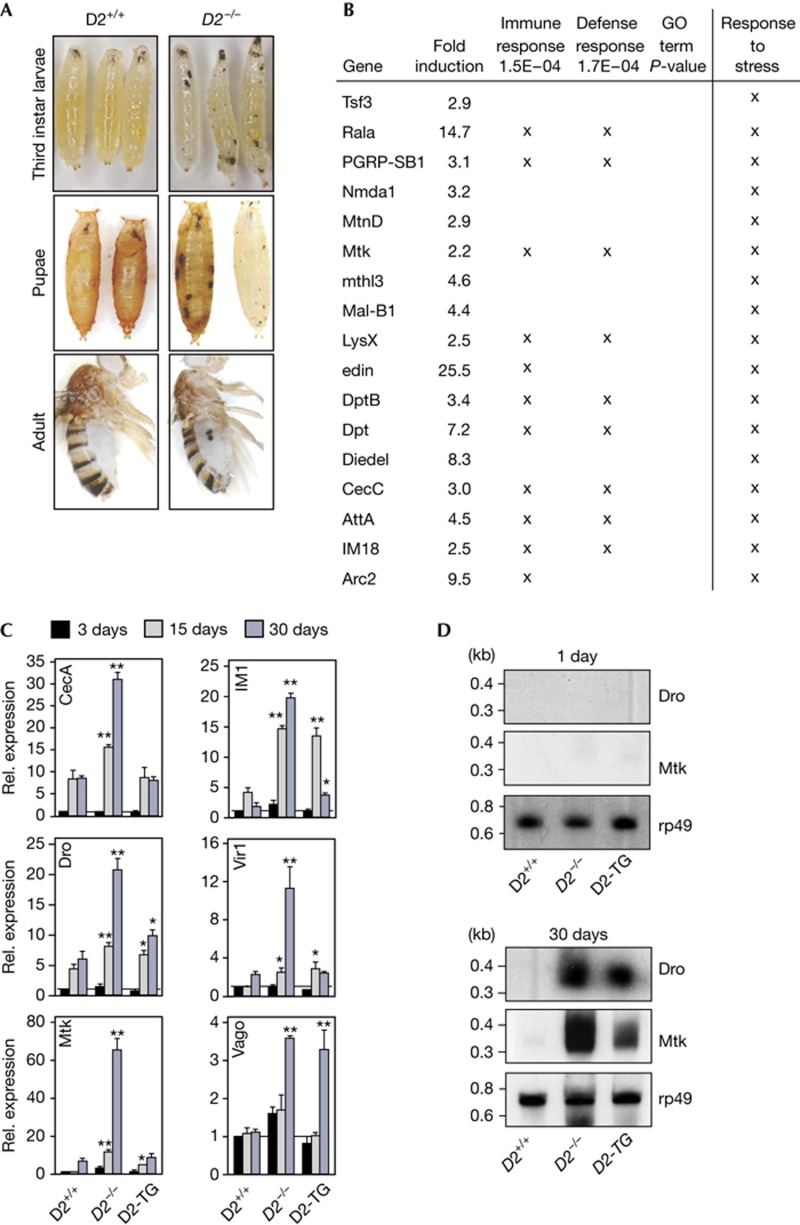
Adult flies with a null mutation in Dnmt2 are sensitive to heat and oxidative stress [12]. To better understand this increased sensitivity to heat stress, larvae were heat-shocked and the resulting adults were scored for phenotypes. A significant increase of black spots was detected in Dnmt2 null mutant flies (supplementary Fig S1A,B online). These spots resembled melanotic lesions, which have been associated with upregulated immune responses in flies [14, 15]. Also, when Dnmt2 mutants were cultured under sub-optimal conditions (for example, crowded population), they regularly developed melanotic lesions (Fig 1A). These observations indicate that stress-induced signals caused increased immune responses in Dnmt2 mutant animals.
Figure 1.
Increased stress and immune responses in Dnmt2 mutant flies. (A) Melanotic spots after keeping fly stocks under crowded growth conditions. Third instar larvae, pupae and adults of wild-type (D2+/+) and Dnmt2 mutants (D2−/−) flies are shown. (B) Enrichment analysis for ⩾2-fold upregulated genes in Dnmt2 mutants relative to w1118 males that can be linked to stress responses. Benjamani–Hochberg multiple hypothesis correction P-value is displayed for two stress-related GO classifications and (X) marks genes that are associated with these terms. Last column includes genes that are not associated with GO terms but can be linked to stress responses using annotation of the Flybase consortium ( http://flybase.org/). (C) Q-PCR analysis for immune response genes in wild-type (D2+/+), Dnmt2 mutant (D2−/−) and transgenic rescue (D2-TG) flies at various time points after hatching (3, 15 and 30 days). (D) Northern blotting for Drosocin and Metchnikowin from young (1 day) and aged (30 days) flies of the genotypes as in (C). Rp49 was probed as loading control. RNA expression of genes in wild-type was set to 1 and normalized to rp49 messenger RNA in individual experiments. Error bars represent s.d.’s from three biological replicates. P-values were determined by Student’s t-test (*P<0.05; **P<0.01). GO, gene ontology; Q-PCR, quantitative PCR; rel., relative.
To analyse the molecular effects of the Dnmt2 mutation, adult wild-type and Dnmt2 mutant flies were compared using gene expression micro-arrays. This analysis showed that 164 genes displayed a robust and significant (fold change ⩾2, P<0.05) upregulation, whereas 121 genes showed a robust and significant downregulation in Dnmt2 mutant flies (supplementary Table S1, S2 online). Enrichment analysis revealed clusters of genes associated with general stress response (for example, cytochrome and glutathione S-transferase genes, Table S3) as well as with immune response pathways (Fig 1B). Gene ontology (GO) term analysis indicated that various processes that were associated with the innate immune response and related stress pathways were misregulated in Dnmt2 mutants (supplementary Fig S2A,B online).
Quantitative PCR analysis was used to validate the micro-array data. Dnmt2 mutant flies showed significantly increased expression of genes in response pathways to microbes, fungi and viruses that became more pronounced with age (Fig 1C). The expression of anti-microbial peptide RNA in Dnmt2 mutants was also confirmed by northern blotting (Fig 1D). Importantly, a fly line that carries a transgenic Dnmt2–EGFP construct in the Dnmt2 mutant background (D2-TG, [16]) showed gene expression levels, which were similar to wild-type controls (Fig 1C, D) indicating the rescue of the Dnmt2 mutant phenotype. These results confirm the induction of innate immune responses in Dnmt2 mutants.
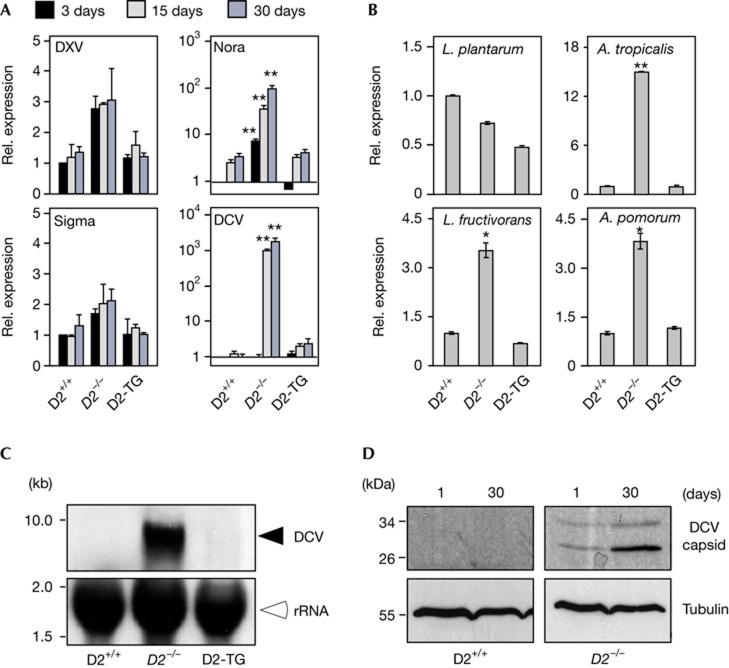
Elevated expression of virus-induced genes (Fig 1B, C) indicated that viruses were present in Dnmt2 mutant flies. As DNA viruses have not been detected in Drosophila laboratory strains and Dnmt2 is an RNA methyltransferase, we determined the levels of naturally occurring RNA viruses in Dnmt2 mutant flies. While the RNA levels of double-stranded (ds) Drosophila X virus and single-stranded (−) Sigma virus were low and not affected by the Dnmt2 mutation and age of the flies (Fig 2A), Dnmt2 mutants contained high RNA levels of single-stranded (+) RNA viruses. Compared with wild-type flies, Nora virus and DCV levels were increased between 100 and 3,000-fold, respectively, particularly in older Dnmt2 mutant animals (Fig 2A). Transgenic rescue flies (D2-TG) carried viral RNA levels that were similar to wild-type controls (Fig 2A). We also determined the bacterial load of adult Drosophila using diagnostic quantitative PCR. Wolbachia was not present in Dnmt2 mutant flies (data not shown). Further analysis showed comparably moderate differences for three out of four dominant gut bacteria [17] between the tested genotypes (Fig 2B). These results indicate that highly increased levels of (+) RNA viruses rather than gut bacteria levels caused the upregulation of innate immunity responses in Dnmt2 mutant flies.
Figure 2.
Increase of (+) RNA viruses and bacteria in Dnmt2 mutant flies. (A) Q-PCR analysis for dsRNA Drosophila X virus, (−) RNA (Sigma) virus and (+) RNA (Nora and DCV) viruses in wild-type (D2+/+), Dnmt2 mutant (D2−/−) and transgenic rescue (D2-TG) and at various time points after hatching (3, 15, 30 days). (B) Q-PCR analysis for Lactobacillus plantarum, Acetobacter tropicalis, Lactobacillus fructivorans and Acetobacter pomorum in genotypes as in (A) flies 30 days after hatching. DNA content of bacteria in wild-type was set to 1 and normalized to rp49 content in individual experiments. Error bars represent s.d.'s from three biological replicates. (C) Northern blotting for DCV genomes from aged (30 days) flies of the genotypes as in (A). Cross-hybridization of the DCV probe with ribosomal RNA was used as loading control. (D) Western blotting for DCV capsid from young (1 day) and aged (30 days) wild-type (D2+/+) and Dnmt2 mutant (D2−/−) flies. Tubulin was probed as loading control. RNA expression of genes in wild-type was set to 1 and normalized to rp49 mRNA in individual experiments. Error bars represent s.d.’s from three biological replicates. Student’s t-test P-values are indicated (*P<0.05; **P<0.01). DCV, Drosophila C virus; dsRNA, double-stranded RNA; Q-PCR, quantitative PCR; rel., relative.
DCV infection has been extensively studied in Drosophila [18]. Northern blotting confirmed full-length DCV RNA genomes in 30-day-old Dnmt2 mutant flies (Fig 2C), and western blotting revealed significant amounts of DCV capsid proteins in Dnmt2 mutants of this age (Fig 2D). These results indicate that Dnmt2 mutants were unable to control DCV levels and suggested a role for Dnmt2 in antiviral defence mechanisms.
The natural route for DCV infections is the digestive system. To establish a physiological infection system, fly carcasses of 30-day-old flies (donors) were fed for 24 h to freshly hatched (recipient) flies (supplementary Fig S3A online). In a first set of experiments, Dicer-2 mutants (Dcr-2L811fsX) served as recipients, because Dcr-2 has been shown to be important for the restriction of viruses in Drosophila [4]. Infection with DCV supplied by donors (supplementary Fig S3B online) was monitored using quantitative PCR at two time points (1 and 10 days after feeding). Dcr-2 mutant recipients showed slightly increased DCV RNA levels at day 1 after infection with wild-type or D2-TG donors (supplementary Fig S3C online). In contrast, Dnmt2 mutant donors caused a 250-fold increase of DCV RNA in Dcr-2 mutants (supplementary Fig S3C online). Importantly, at day 10 after infection with Dnmt2 mutant donors, Dcr-2 mutant recipients had accumulated 12,000 times more DCV RNA than control recipients (Fig S3C online). These results confirm that infection-by-feeding (oral infection) can be used to propagate DCV in flies with compromised antiviral defence mechanisms.
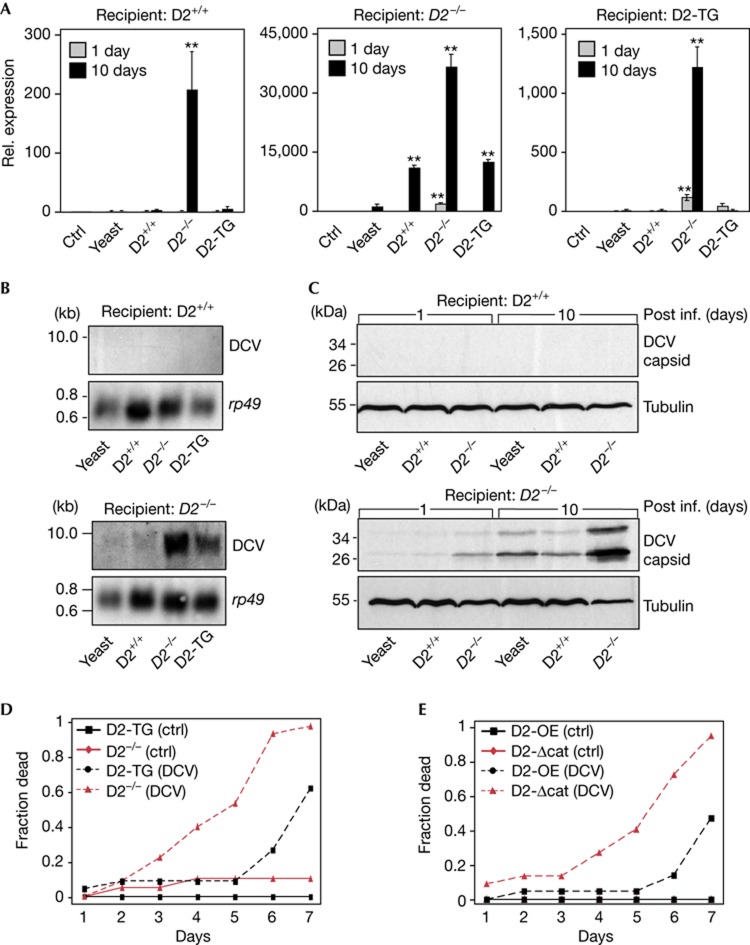
To analyse the function of Dnmt2 in DCV suppression, wild-type, Dnmt2 mutant and D2-TG recipients were fed with Dnmt2 mutant donors (supplementary Fig S3B online). DCV levels in recipient flies were comparable at the time of hatching (supplementary Fig S3D online). Wild-type recipients suppressed DCV levels efficiently with only residual virus detectable after feeding with Dnmt2 mutant donors (Fig 3A, left). In contrast, Dnmt2 mutant recipients showed significant amplification of DCV RNA (10,000 to 35,000-fold, Fig 3A, middle). DCV levels in D2-TG recipients were similar to wild-type controls, thus confirming that DCV suppression was Dnmt2-dependent (Fig 3A, right). Northern blotting (10 days after infection) showed full-length DCV RNA genomes in Dnmt2 mutant but not in wild-type recipients (Fig 3B). Western blotting also confirmed that infected Dnmt2 mutant recipients contained more viral capsid protein if compared with wild-type and D2-TG recipients (Fig 3C). To further demonstrate a role for Dnmt2 in the control of DCV infection, we used purified DCV to challenge young animals by intra-thoracic injection. Using this approach, we found that Dnmt2 mutants succumbed more rapidly to the infection than transgenic rescue controls (D2-TG, Fig 3D). These results clearly show that Dnmt2 is required for the efficient control of DCV in Drosophila.
Figure 3.
Dnmt2 is required to suppress DCV accumulation. (A) Q-PCR analysis for DCV in wild-type (D2+/+), Dnmt2 mutant (D2−/−) and transgenic rescue (D2-TG) recipients after feeding with yeast or donors (x axis) and two incubation periods (1 or 10 days). (B) Northern blotting for DCV in wild-type (D2+/+) and Dnmt2 mutant (D2−/−) recipients after feeding with yeast or donors as described in (A). Rp49 was probed as loading control. (C) Western blotting for DCV capsid from wild-type (D2+/+) and Dnmt2 mutant (D2−/−) recipients after feeding with donors and incubation periods as described in (A). Tubulin was probed as loading control. RNA expression of virus was set to 1 in non-infected (ctrl) and normalized to rp49 mRNA in individual experiments. Error bars represent s.d.’s from three biological replicates. P-values were determined by Student’s t-test (**P<0.01). (D) Lethality of wild-type (D2+/+) and Dnmt2 mutant (D2−/−) flies challenged with purified DCV (2 × 104 virus particles per μl) by intra-thoracic injection. A representative of three experiments is shown; D2−/− and D2-TG (ctrl) P=0.533; D2−/− and D2-TG (DCV) P=0.000034 by log-rank test. (E) Lethality of flies that overexpress Dnmt2 (D2-OE) and flies overexpressing a catalytically inactive Dnmt2 (D2-catΔ) challenged with purified DCV (2 × 104 virus particles per μl) by intra-thoracic injection. A representative of three experiments is shown; D2-OE and D2-catΔ (ctrl) P=1; D2-OE and D2-catΔ (DCV) P=0.005 by log-rank test. DCV, Drosophila C virus; Q-PCR, quantitative PCR; rel., relative.
To test whether Dnmt2 directly affects the accumulation of DCV, recipients that overexpressed Dnmt2 (supplementary Fig S4A online) were used in oral infection experiments. Expression of Dnmt2 under the ubiquitin promoter in the wild-type background (w1118; pUbq>>Dnmt2–EGFP; D2-OE) suppressed DCV RNA levels during the infection with Dnmt2 mutant donors (supplementary Fig S4B online). Importantly, recipients that expressed a catalytically inactive Dnmt2 (point mutation in motif IV) under the same promoter but in the Dnmt2 mutant background (Dnmt299; pUbq>>Dnmt2-catmutant-FLAG; D2-catΔ) also suppressed DCV but not as efficiently as catalytically active Dnmt2 (supplementary Fig S4C–E online). To further analyse the contribution of the catalytic activity of Dnmt2 to virus control, we challenged these transgenic flies with purified DCV by intra-thoracic injection. We found that D2-catΔ flies succumbed more rapidly to the infection than D2-OE flies (Fig 3E). These results show that Dnmt2 is necessary and sufficient to suppress DCV levels and indicate that the cytosine-5 methyltransferase activity of Dnmt2 contributes to Dnmt2-mediated DCV control.
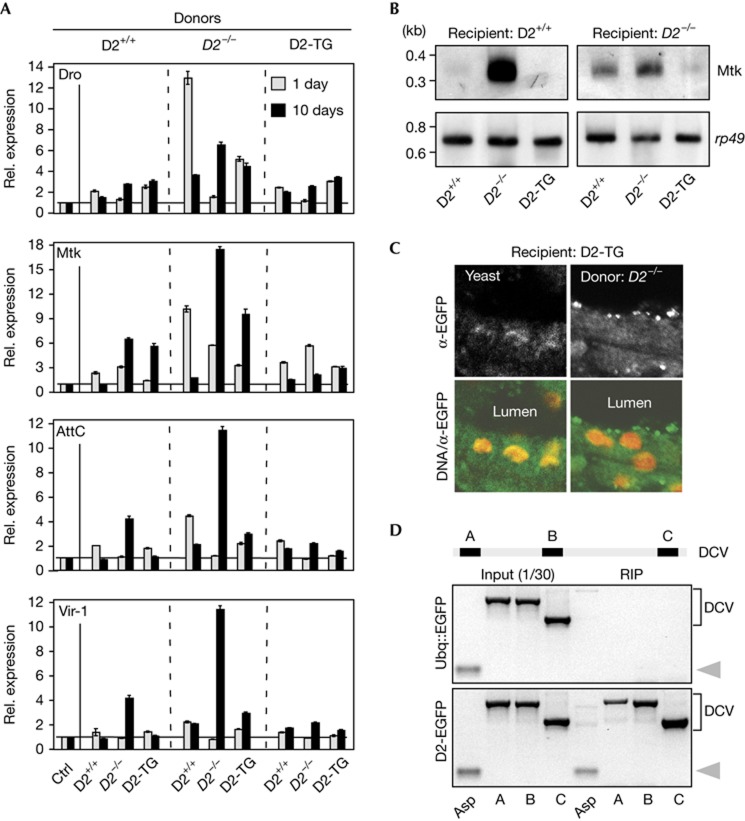
Gene expression analysis for anti-microbial genes and the DCV-induced gene, vir-1, after oral infection with Dnmt2 mutant donors showed rapid upregulation of immune response genes in control, but not in Dnmt2 mutant recipients (Fig 4A). Northern blotting also confirmed that wild-type but not Dnmt2 mutant recipients expressed high RNA levels of the immune-inducible peptide Metchnikowin at day 1 after infection (Fig 4B). In contrast, Dnmt2 mutant recipients showed significantly increased immune responses only at day 10 after oral infection (Fig 4A). Importantly, at this time, wild-type and D2-TG recipients had efficiently downregulated the initial immune response (Fig 4A). These results indicate that Dnmt2 function is important during the acute phase of immune system activation.
Figure 4.
Dnmt2 mutants show delayed immune responses and Dnmt2–EGFP relocalizes and interacts with DCV RNA during infection. (A) Q-PCR analysis for immune response genes in wild-type (D2+/+), Dnmt2 mutant (D2−/−) and transgenic rescue (D2-TG) recipient flies after feeding with donors and incubation periods as described in Fig 3A. (B) Northern blotting for Metchnikowin RNA in wild-type (D2+/+) and Dnmt2 mutant (D2−/−) recipients after feeding with donors as described in Fig 3A and an incubation period of 1 day. Rp49 was probed as loading control. (C) Immunofluorescence images of fixed Drosophila hindgut tissue from D2-TG recipients after feeding with yeast alone (yeast) or Dnmt2 mutant (D2−/−) donors followed by an incubation period of 1 day. The border between lumen and gut cells is shown. Dnmt2–EGFP fusion protein was visualized using antibodies against EGFP (white in upper panels, green in lower panels). DNA is shown in red. (D) Schematic illustration of the DCV RNA genome with three primer sets used for the analysis of RIPs. Agarose gels show the results of RIP from control (Ubq::EGFP) and D2-TG (D2-EGFP) recipients after continuous feeding (72 h) with Dnmt2 mutant donors. Both samples contained tRNA-AspGTC (grey arrowhead) and DCV in the input fraction. After RIP, only Dnmt2–EGFP complexes contained tRNA-AspGTC (grey arrowhead) and DCV. RNA expression was set to 1 in non-infected recipients (ctrl) and normalized to rp49 mRNA in individual experiments. Error bars represent s.d.’s from three biological replicates. DCV, Drosophila C virus; Q-PCR, quantitative PCR; RIP, RNA immunoprecipitation; rel., relative.
To further explore the function of Dnmt2 during acute infection, Dnmt2 localization was analysed in infected gut tissues. To this end, transgenic rescue flies (D2-TG) were used, which express Dnmt2–EGFP at comparable levels to wild-type flies (supplementary Fig S4E online). While Dnmt2–EGFP was ubiquitous in controls, feeding Dnmt2 mutant donors caused the accumulation of Dnmt2–EGFP into distinct cytoplasmic macromolecular structures (Fig 4C). This observation indicates that relocalization of Dnmt2 is important for its function during the response to infection.
Finally, the interaction of Dnmt2 with DCV was tested using RNA immunoprecipitation (RIP) with EGFP nanobodies after infection of D2-TG recipients with Dnmt2 mutant donors. Control RIP was performed on infected flies, which expressed EGFP from the ubiquitin promoter (Ubq::EGFP). The specificity of RIP was confirmed by analysing the precipitate for tRNA-AspGTC, which is a substrate of Dnmt2. The analysis showed that Dnmt2–EGFP complexes specifically interacted with tRNA-AspGTC and with at least three regions of the DCV RNA genome (Fig 4D). These results indicate that Dnmt2 is associated with DCV genomic RNA and suggest direct interactions of Dnmt2 with viral RNA.
Discussion
Eukaryotic cells induce the innate immune system after detecting parts of a pathogen such as RNA, DNA or specific surface molecules of microbial and viral stressors. Vertebrates have developed an intricate system of membrane-based receptors, which can distinguish and bind various polynucleotides on the basis of their termini and structure [19, 20, 21]. Invertebrates share such receptor-based detection but additionally use RNA-mediated interference-based mechanisms, which limit the amplification of viral RNA genomes and RNA-based intermediates of replicating virus. As pattern recognition receptors can also detect the modification status of nucleic acids [21, 22, 23], it is conceivable that host-induced modification of viral RNA genomes facilitates their detection and contributes to efficient antiviral mechanisms.
Our findings support this notion and implicate the RNA methyltransferase Dnmt2 in the activation of the innate immune system during (+) RNA virus infections. This conclusion is based upon the observation that immune peptides are induced in wild-type but not in Dnmt2 mutants after oral infection. Additionally, the relocalization of Dnmt2 into distinct macromolecular foci in gut cells suggests compartmentalized activity of Dnmt2 during this phase of infection. The observation that high levels of non-functional Dnmt2 are unable to efficiently suppress DCV after oral infection and that these flies are more sensitive to intra-thoracic infections using pure DCV suggests a role for RNA methylation in the process. An analysis of the RNA methylation status of the DCV genome is complicated by the suppression of viral genomes through Dnmt2 activity and needs to be addressed in future studies. It has also been reported that Dnmt2 can bind tightly to DNA without methylating it [24]. It is therefore also conceivable that Dnmt2 presents viral RNA to innate immunity receptors or RNA processing enzymes to control specific RNA viruses in Drosophila.
Methods
Fly husbandry and phenotypic analysis. Flies were usually maintained on standard medium at 25° C, 60% humidity, under a 12-h light–dark cycle. For culturing flies under crowded conditions, flies were allowed to hatch but were not removed from the vials. These flies reproduced and finally succumbed to the crowded conditions. Developing larvae were feeding not only on medium but also on carcasses of the parental generation. Larvae, pupae and adults arising from this generation were scored for melanotic inclusions.
Gene expression analysis. RNA was isolated from two biological replicates (50 male flies per replicate, non-stressed, 15 days after hatching) per genotype (w1118 and Dnmt299 [12]) and analysed for gene expression using the Drosophila Gene Expression Microarray 4 × 44K (Agilent Technologies). Sample preparation and hybridization were performed according to the manufacturer’s instructions. Gene expression array raw data were preprocessed and analysed using Chipster v1.4. Data were filtered for significance (Student’s t-test) and analysed using DAVID 6.7 (http://david.abcc.ncifcrf.gov/) and tools provided by the GO consortium (http://www.geneontology.org/).
Protein extraction and western blotting. Twenty adult flies were homogenized in 1 volume of RIPA buffer and incubated at 4 °C for 10 min. Cells were disrupted by 20 strokes in a douncer, followed by passing the extract 10 × through a syringe needle (G20), followed by centrifugation. Fifty micrograms of total protein extracts were analysed by 10% SDS–PAGE. Western blotting was performed using antibodies against DCV capsid (chicken; 1:25,000) and Tubulin (mouse DM1A; 1:10,000; Sigma).
Fly infection by feeding. Inoculation experiments were performed with crude extracts from 200 mg of w1118, Dnmt299, D2-TG flies. After homogenization in 500 μl H2O, carcasses were mixed with 500 μl yeast paste. This mix (donors) was used for feeding of freshly hatched flies (recipients). After 24 h, flies were transferred to standard Drosophila medium and RNA was isolated at specified time points.
Intra-thoracic injections. DCV was purified and used for intra-thoracic challenge of groups of 20 flies (4–7-day-old) as previously described [25]. Mortality was monitored as a function of time. A log-rank test was used to determine significance.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Nevcin Senturk for the establishment of RIPs. This work was supported by grants from the Deutsche Forschungsgemeinschaft to M.S. and F.L. (FOR1082). Z.D. is a fellow of the HBIGS graduate programme at the University of Heidelberg.
Author contributions: M.S. conceived the study. Z.D. and T.P. performed molecular work and phenotypic characterization of Drosophila. T.P. performed microarray expression analysis. Z.D. and K.H. performed validation of RNA expression analysis and northern blotting. Z.D. performed oral infection experiments. B.G. and S.C. performed intra-thoracic infections and Z.D. performed RIP assays. Z.D., S.C., F.L. and M.S. designed the experiments and interpreted the results. MS wrote the manuscript with contributions from Z.D., S.C. and F.L. as well as input from the other co-authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R (2006) The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 20: 2985–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW (2006) RNA interference directs innate immunity against viruses in adult Drosophila. Science 312: 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL (2006) Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat Immunol 7: 590–597 [DOI] [PubMed] [Google Scholar]
- Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, Dostert C, Antoniewski C, Hoffmann JA, Imler JL (2008) The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in Drosophila. Nat Immunol 9: 1425–1432 [DOI] [PubMed] [Google Scholar]
- Cherry S, Silverman N (2006) Host-pathogen interactions in Drosophila: new tricks from an old friend. Nat Immunol 7: 911–917 [DOI] [PubMed] [Google Scholar]
- Dalpke A, Helm M (2012) RNA mediated Toll-like receptor stimulation in health and disease. RNA Biol 9: 828–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HR, Chen ZJ, Kunes S, Chang GD, Maniatis T (2010) Endocytic pathway is required for Drosophila Toll innate immune signaling. Proc Natl Acad Sci USA 107: 8322–8327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS et al. (2009) Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol 11: 1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514 [DOI] [PubMed] [Google Scholar]
- Schaefer M, Lyko F (2010) Solving the Dnmt2 enigma. Chromosoma 119: 35–40 [DOI] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH (2006) Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311: 395–398 [DOI] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F (2010) RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24: 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiagarajan D, Dev RR, Khosla S (2011) The DNA methyltranferase Dnmt2 participates in RNA processing during cellular stress. Epigenetics 6: 103–113 [DOI] [PubMed] [Google Scholar]
- Rodriguez AJ, Zhou Z, Tang ML, Meller S, Chen J, Bellen H, Kimbrell DA (1996) Identification of immune system and response genes, and novel mutations causing melanotic tumor formation in Drosophila melanogaster. Genetics 143: 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson KL, Johnson TK, Denell RE (1991) Lethal(1) aberrant immune response mutations leading to melanotic tumor formation in Drosophila melanogaster. Dev Genet 12: 173–187 [DOI] [PubMed] [Google Scholar]
- Schaefer M, Steringer JP, Lyko F (2008) The Drosophila cytosine-5 methyltransferase Dnmt2 is associated with the nuclear matrix and can access DNA during mitosis. PLoS ONE 3: e1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ZS, Hedges LM, Brownlie JC, Johnson KN (2011) Wolbachia-mediated antibacterial protection and immune gene regulation in Drosophila. PLoS ONE 6: e25430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler JL (2005) The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat Immunol 6: 946–953 [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124: 783–801 [DOI] [PubMed] [Google Scholar]
- Pétrilli V, Dostert C, Muruve DA, Tschopp J (2007) The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol 19: 615–622 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Reis e Sousa C (2010) RIGorous detection: exposing virus through RNA sensing. Science 327: 284–286 [DOI] [PubMed] [Google Scholar]
- Gehrig S et al. (2012) Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J Exp Med 209: 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Akira S (2005) TLR ignores methylated RNA? Immunity 23: 111–113 [DOI] [PubMed] [Google Scholar]
- Dong A, Yoder JA, Zhang X, Zhou L, Bestor TH, Cheng X (2001) Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res 29: 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S, Perrimon N (2004) Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat Immunol 5: 81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.