Abstract
The mechanistic target of rapamycin is a protein kinase that, as part of the mechanistic target of rapamycin complex 1 (mTORC1), senses both local nutrients and, through insulin signalling, systemic nutrients to control a myriad of cellular processes. Although roles for mTORC1 in promoting protein synthesis and inhibiting autophagy in response to nutrients have been well established, it is emerging as a central regulator of lipid homeostasis. Here, we discuss the growing genetic and pharmacological evidence demonstrating the functional importance of its signalling in controlling mammalian lipid metabolism, including lipid synthesis, oxidation, transport, storage and lipolysis, as well as adipocyte differentiation and function. Defining the role of mTORC1 signalling in these metabolic processes is crucial to understanding the pathophysiology of obesity and its relationship to complex diseases, including diabetes and cancer.
Keywords: adipocytes, Akt, insulin, liver, mTOR
See Glossary for abbreviations used in this article.
Glossary.
- 4E-BP1/2
eIF4E-binding protein 1/2
- AMPK
adenosine monophosphate-activated protein kinase
- ATG5/7
autophagy-related 5/7
- ATGL
adipose triglyceride lipase
- C/EBP
CCAAT/enhancer-binding protein
- CPT1
carnitine palmitoyltransferase 1
- DAG
diacylglycerol
- eIF4E
eukaryotic translation initiation factor 4E
- GSK3
glycogen synthase kinase 3
- HSL
hormone-sensitive lipase
- IDL
intermediate density lipoprotein
- IGF1
insulin-like growth factor 1
- Insig
insulin-induced gene
- LDL
low density lipoprotein
- LDLR
LDL receptor
- lipin 1
phosphatidate phosphatase LPIN1
- LPL
lipoprotein lipase
- LST8
lethal with SEC13 protein 8
- MAG
monoacylglycerol
- MEF
mouse embryonic fibroblast
- N-CoR1
nuclear receptor co-repressor 1
- PCSK9
proprotein convertase subtilisin/kexin type 9
- PKA/C
protein kinase A/C
- PPARα/γ
peroxisome proliferator-activated receptor α/γ
- Raptor
regulatory-associated protein of mTOR
- Rictor
Raptor-independent companion of mTOR
- S6K1/2
ribosomal S6 kinase 1/2
- SCAP
SREBP cleavage-activating protein
- SCD
stearoyl-CoA desaturase
- SGK
serum and glucocorticoid regulated kinase
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- SRE
sterol response element
- SREBP
sterol regulatory element-binding protein
- TAG
triacylglycerol
- TBC1D7
TBC1 domain family, member 7
- TCA
tricarboxylic/citric acid
- TSC1/2
tuberous sclerosis complex 1/2
- VLDL
very low density lipoprotein
Introduction
Of the four main classes of biological macromolecule, our understanding of the molecular mechanisms by which cellular signalling pathways regulate lipid metabolism has lagged behind that of carbohydrates, proteins and nucleic acids. However, lipids are crucially important both structurally and functionally in all living organisms. An obvious reason for this dependence is the lipid makeup of the plasma membrane and many subcellular organelles. Moreover, lipids act as signalling molecules on both a cellular, for example phosphoinositides, and organismal, for example steroid hormones, scale. Lipids are also used for energy storage, primarily as triacylglycerides in adipocytes, and as an alternative to glucose for catabolic metabolism. Despite the dependence of living organisms on lipids, we know little about how lipid homeostasis is controlled by the intricate network of cellular signalling pathways that sense cellular growth conditions. As detailed in this review, the mechanistic target of rapamycin (mTOR) protein kinase has emerged as a crucial link between cellular and systemic growth signals and the regulation of lipid metabolism.
mTOR is an evolutionarily conserved serine/threonine kinase that exists within two functionally distinct protein complexes, the mechanistic target of rapamycin complexes 1 (mTORC1) and 2 (mTORC2). mTORC1 senses and integrates a diverse array of cellular signals, with mTOR kinase activity within the complex being influenced by a variety of nutrients—for example, amino acids, glucose and oxygen, cellular energy levels, such as ATP, and many secreted growth factors, cytokines and hormones, including insulin. All of these signals require the Ras-related small G protein Rheb, which on GTP-loading is an essential upstream activator of mTORC1 [1]. Many of the signals that regulate mTORC1 do so by altering the GTP-binding status of Rheb through activation or inhibition of a GTPase-activating protein complex, comprised of TSC1, TSC2 and TBC1D7—the TSC–TBC complex [2]. For instance, insulin, IGF1 and other growth factors inhibit the complex to activate Rheb and mTORC1 through Akt-mediated phosphorylation of TSC2 [3,4]. By contrast, a decrease in cellular ATP, such as the decrease that occurs during glucose depletion, activates the complex to inhibit Rheb and mTORC1, at least in part, through the action of AMPK (Fig 1; [5,6,7]). On activation, mTORC1 directly phosphorylates S6K1 and S6K2, 4E-BP1 and 4E-BP2, and a growing number of other downstream targets [8]. Whilst the overall effects of mTORC1 signalling differ in cells and tissues, it has an evolutionarily conserved role in promoting anabolic cell growth and inhibiting the catabolic process of autophagy. On the other hand, mTORC2 seems to be regulated primarily by growth factor signalling and phosphorylates a conserved hydrophobic motif in the protein kinases Akt, SGK and some isoforms of PKC, thereby increasing their kinase activity [9]. Through these targets, and probably through others, mTORC2 signalling is believed to promote cell survival, proliferation, metabolism and changes in the actin cytoskeleton. The two mTOR complexes can be distinguished from one another by their differential sensitivity to rapamycin, an allosteric and partial inhibitor of mTOR (Sidebar A).
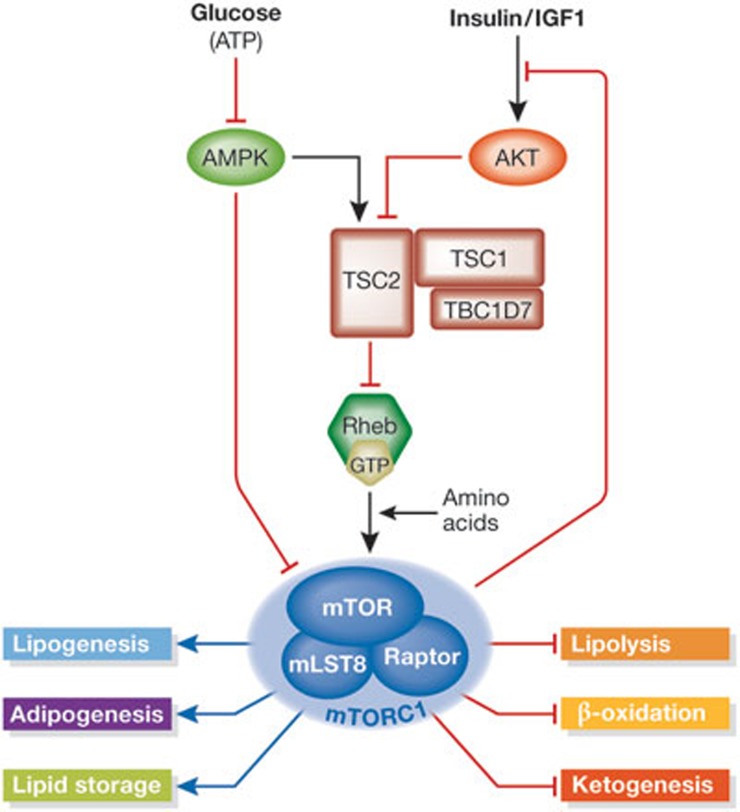
Figure 1.
Upstream regulation from the mTORC1 and its downstream functions related to lipid metabolism. The presence of amino acids is required for the activation of mTORC1 by GTP-bound Rheb. Upstream from Rheb, the TSC–TBC complex receives signals about systemic and local nutrient and energy availability, in part through AMPK and Akt. These signals either activate or inhibit the ability of the TSC–TBC complex to act as a GAP for Rheb, thereby inhibiting or activating mTORC1, respectively. Activated mTORC1 leads to enhanced phosphorylation of IRS1, which serves as negative feedback to dampen the insulin response. mTORC1 has many roles in regulating lipid metabolism, including the promotion of lipid synthesis and storage and inhibition of lipid release and consumption, which are detailed in the text. AMPK, adenosine monophosphate-activated protein kinase; GAP, GTPase-activating protein; IRS1, insulin receptor substrate 1; IGF1, insulin-like growth factor 1; mTORC1, mechanistic target of rapamycin complex 1; Raptor, regulatory-associated protein of mTOR; TSC, tuberous sclerosis complex; TBC, Tre-2/Bub2/Cdc16 domain-containing protein.
Sidebar A | mTORC1 versus mTORC2 and the differential effects of mTOR inhibitors.
In studying the mechanistic target of rapamycin (mTOR) signalling network, or interpreting the mTOR literature, it is crucial to understand some of the basic complexities of mTOR signalling and inhibition. The mechanistic target of rapamycin complex 1 (mTORC1) is composed of the core essential components mTOR, mTOR-associated protein, LST8 homologue (mLST8) and the regulatory-associated protein of mTOR (Raptor), whereas mTORC2 is composed of mTOR, mLST8, SAPK-interacting protein 1 (SIN1) and the Raptor-independent companion of mTOR (Rictor). Although these complexes are functionally distinct, they can have an influence on each other's activity. For instance, as mTORC2 stimulates an increase in Akt activity [84], it might influence its downstream signalling from mTORC1. On the other hand, several negative feedback mechanisms are triggered by mTORC1 activation, which influences mTORC2 activity, including one leading to direct phosphorylation of Rictor within mTORC2 by ribosomal S6 kinase 1 (S6K1) downstream from mTORC1 [85,86]. Regarding mTOR inhibitors, the widely used rapamycin and its many analogues, which on interaction with the ubiquitous protein FK506 binding protein of 12 kDa (FKBP12) binds to an allosteric site amino terminal to the mTOR kinase domain—the FKBP12-rapamycin binding domain—only has access to mTOR within mTORC1. However, it is evident that in both cell culture and mice, prolonged exposure to rapamycin can block the assembly of mTORC2 by sequestering uncomplexed mTOR [82,87]. Therefore, although rapamycin is specific to mTORC1 for acute inhibition and generally leads to an increase in upstream signalling from mTORC2 and Akt by blocking negative feedback mechanisms, one must consider that the observed effects of long-term rapamycin treatment might be due to loss of mTORC2 in some experimental systems, which affects the many processes downstream from Akt. Also, the development of mTOR kinase domain inhibitors, which completely block mTOR within both complexes, has revealed that rapamycin only partly inhibits mTORC1 activity. Whilst the nature of this differential sensitivity is unknown, rapamycin strongly affects the phosphorylation of some mTORC1 targets (for example, S6K1) but only modestly inhibits other targets (for example, eIF4E-binding protein 1; [88]).
Many studies in cell and mouse models, combined with preclinical and clinical data on mTOR inhibitors, have revealed a pivotal role for mTOR—particularly within mTORC1—in controlling lipid homeostasis in many settings, both physiological and pathological. We review this evidence below, with a focus on the key aspects of lipid synthesis, storage and mobilization. The emerging picture is that, through a variety of molecular mechanisms, mTORC1 signalling promotes processes to synthesize and store lipids, whilst inhibiting those leading to lipid consumption (Fig 1).
Lipogenesis
The regulation of de novo sterol and fatty acid synthesis by signalling pathways, especially insulin signalling, has garnered intense interest. Unlike most terminally differentiated cells, hepatocytes and adipocytes synthesize significant amounts of lipid de novo through pathways in which cytosolic acetyl-CoA, derived from glucose or amino acid catabolism, is used to form the hydrophobic carbon backbone of lipids. Acetyl-CoA is either committed to sterol and isoprenoid biosynthesis through the action of HMG-CoA synthase or to fatty acid biosynthesis through acetyl-CoA carboxylase. Both the sterol and fatty acid synthesis branches comprise many steps requiring many specific enzymes. Importantly, the SREBPs are transcription factors that stimulate the expression of genes encoding nearly all of these lipogenic enzymes [10]. The three SREBP isoforms, encoded by two genes, are produced as inactive transmembrane proteins at the endoplasmic reticulum (Fig 2). Under conditions of abundant sterols, full-length SREBP, through its sterol-sensing binding partner SCAP, is retained in the endoplasmic reticulum by the INSIG proteins [11]. Depletion of intracellular sterols results in release of the SREBP–SCAP complex from Insig and their transport to the Golgi apparatus, in which two proteolytic cleavage events by the site-specific proteases S1P and S2P liberate the active amino-terminus of SREBP. This fragment then enters the nucleus and induces transcription from SREs within target genes. SREBP1a and 1c are products of alternative splicing of the SREBF1 gene and have been primarily implicated in the control of genes involved in fatty acid synthesis, although SREBP1a is thought to activate most SRE-containing genes [12]. SREBP2 is encoded by SREBF2 and is believed to have a more important role in the transcription of steroidogenic genes, including those involved in cholesterol synthesis in the liver [13,14]. Although the SREBPs preferentially activate transcription of different sets of genes, there is substantial overlap between the targets of the SREBP isoforms and the tissue specificity of these preferences, which has not been fully established. Importantly, independent studies have identified the SREBPs as major transcriptional effectors of mTORC1 signalling and have demonstrated that mTORC1 activation promotes lipogenesis through this family of transcription factors [15,16].
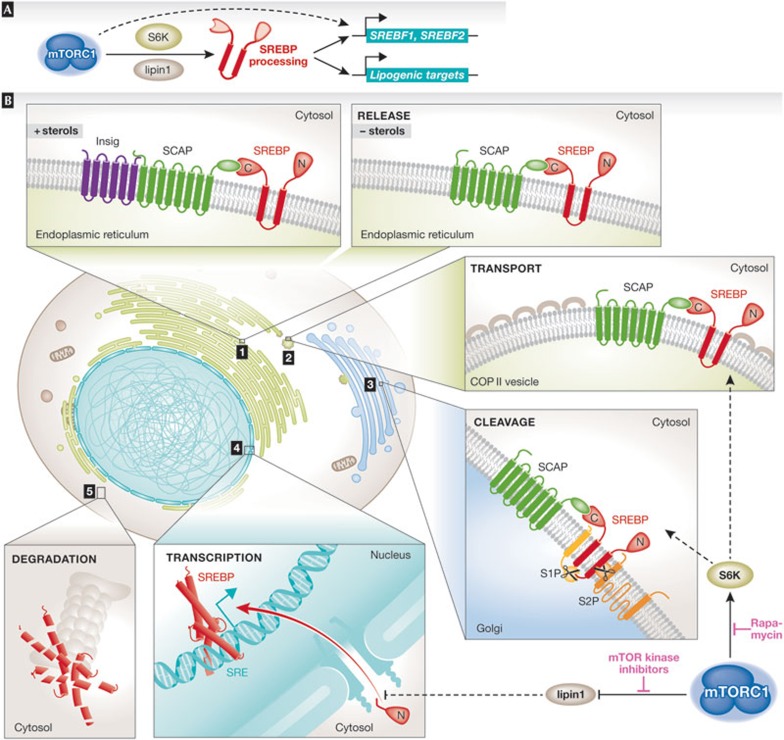
Figure 2.
The complex steps leading to SREBP activation and input from mTORC1 signalling. (A) SREBP processing and activation is regulated by mTORC1 through S6K and lipin 1 leading to the transcriptional induction of the SREBF1 and SREBF2 genes, encoding SREBP1 and SREBP2, respectively, and genes encoding many lipogenic enzymes involved in both fatty acid and sterol synthesis. The mTORC1-mediated transcriptional activation of SREBF1 could result from either autoregulation by SREBP1 or from an unknown parallel pathway downstream from mTORC1. (B) In the presence of sterols, SREBP resides in the endoplasmic reticulum bound to SCAP and the Insig proteins. When sterols become scarce SCAP undergoes a conformational change, which releases the SCAP–SREBP complex from the Insig, allowing its transport from the endoplasmic reticulum to the Golgi apparatus through COPII vesicles. Once in the Golgi, SREBP comes into contact with two site-specific proteases. S1P cleaves the luminal loop of SREBP and S2P cleaves the amino-terminal transmembrane region of SREBP, which releases the N-terminal region of SREBP containing the DNA-binding and -transactivating domains. The NLS-containing processed form of SREBP enters the nucleus to activate transcription of genes containing SREs in their promoters. Finally, the processed form of SREBP is unstable and subject to proteasome-mediated degradation. In some settings, SREBP processing has been found to require S6K1 downstream from mTORC1 and is therefore sensitive to rapamycin. However, the nuclear shuttling of SREBP has been found to require lipin 1 downstream from mTORC1, the phosphorylation of which is largely resistant to rapamycin but sensitive to mTOR kinase domain inhibitors (Sidebar A). The precise molecular mechanisms by which either of these two mTORC1 targets regulates SREBP activation are unknown. COPII, coatamer protein II; Insig, insulin-induced gene; lipin 1, phosphatidate phosphatase LPIN1; mTORC1, mechanistic target of rapamycin complex 1; NLS, nuclear localization signal; S1/2P, site 1/2 protease; S6K1, ribosomal S6 kinase 1; SCAP, SREBP cleavage-activating protein; SRE, sterol response element; SREBP1/2, sterol regulatory element-binding protein 1/2.
mTORC1 signalling promotes SREBP activation and lipogenesis in response to both physiological and genetic stimuli. In primary rodent hepatocytes and the intact liver, insulin or feeding has been shown to increase the expression of the major liver isoform of SREBP (SREBP1c) and its targets, and to promote de novo lipid synthesis in a manner that is sensitive to rapamycin [17,18,19]. Insulin activates mTORC1 through a pathway involving the Akt-mediated phosphorylation and inhibition of TSC2, within a complex with TSC1 and TBC1D7 ]2,3,4]. Expression of constitutively active Akt or loss of either TSC1 or TSC2, both of which result in insulin-independent activation of mTORC1 signalling, stimulates the global expression of SREBP1 and SREBP2 targets and drives lipogenesis through mTORC1 [15,16]. These latter studies found that mTORC1 signalling promotes accumulation of the processed, mature form of SREBP1, which resides in the nucleus to induce its own expression and that of genes involved in both steroid and fatty acid biosynthesis. In exploring the molecular mechanism of this regulation, it was found that S6K1 is required downstream from mTORC1 to stimulate the increase in levels of active SREBP1, expression of SREBP1 and SREBP2 targets, and de novo lipogenesis in TSC2-deficient cells [15]. SREBP1 regulation in this setting is independent of the effects on the proteasomal degradation of its active form, suggesting that S6K1 promotes the processing of SREBP1. Consistent with these findings, S6K1 has been found to promote the activation of hepatic SREBP1c by having an effect on its processing [20,21], and to affect the processing of SREBP2 in a hepatocellular carcinoma cell line [22]. mTORC1 signalling has also been suggested to increase SREBP1 activation in an S6K1-dependent manner in cultured myotubes [23].
Genetic mouse models have demonstrated that mTORC1 activation is essential, but not sufficient, to stimulate hepatic SREBP1c and its lipogenic targets in response to feeding [18,24]. Mice lacking mTORC1 in their liver, through liver-specific Raptor knockout, fail to induce SREBP1c and lipogenesis [24], and have reduced levels of both liver triglycerides and circulating cholesterol on a ‘Western’ diet [25]. However, characterization of mice with a liver-specific knockout of Tsc1 (LTsc1KO) and constitutive activation of mTORC1, which is independent of insulin and feeding, revealed that mTORC1 signalling, although essential, is not capable of activating SREBP1c and hepatic lipid synthesis on its own [18]. In fact, these mice were found on two independent strain backgrounds to be resistant to the development of both age- and diet-induced hepatic steatosis due to decreased SREBP1c activation [18,26]. These seemingly paradoxical findings are the result of a strong feedback attenuation of Akt signalling that accompanies loss of function of the TSC1–TSC2 complex in all settings [27]. A crucial role for Akt signalling in the induction of SREBP1c and lipogenesis in the liver has been established through rodent models [28,29,30], and this has been extended by using mice with liver-specific Rictor knockout, which results in the loss of mTORC2 activity and its activating phosphorylation of Akt [31]. Consistent with the essential nature of Akt signalling to hepatic SREBP1c, a restoration of Akt activity in LTsc1KO hepatocytes restores SREBP1c activation and lipogenesis [18]. Whilst many mTORC1-independent pathways might function in parallel downstream from Akt to help to promote the activation of hepatic SREBP1c, including GSK3 inhibition [32], data from the LTsc1KO mice suggest that one pathway involves the repression of an isoform of the SREBP inhibitor Insig, Insig2a, which is only expressed in the liver [18]. A liver-specific mechanism is also consistent with the fact that mTORC1 activation alone is sufficient to promote SREBP activation and lipogenesis in other settings, even in the absence of Akt signalling [15].
The molecular mechanism by which S6K1 promotes SREBP processing is unknown, and it is clear from additional studies that S6K1 is not the only direct target downstream from mTORC1 involved in SREBP isoform regulation, which might vary by cellular context. For instance, siRNA knockdown of the mRNA cap-binding protein eIF4E, which is normally activated by mTORC1 signalling through the phosphorylation and release of its inhibitory binding partner 4E-BP1, decreases overall levels of SREBP1 and its canonical target SCD in breast cancer cell lines [33]. The potential involvement of 4E-BP1 regulation by mTORC1 in some cells might explain the resistance of SREBP1 or SREBP2 activation to rapamycin in specific settings [22,34]. The resistance of some mTORC1 targets to rapamycin (Sidebar A) is an important consideration when examining the role of mTORC1 signalling in any aspect of lipid metabolism. Another direct target of mTORC1 that, as with 4E-BP1, is partly resistant to rapamycin for its regulation is the phosphatidic acid phosphatase lipin 1, which has also been implicated in SREBP regulation [25,35]. Lipin 1 seems to have a role in the remodelling of the nuclear lamina, which is inhibited by mTORC1-mediated phosphorylation of many residues on this enzyme. Lipin 1 phosphorylation also coincides with an increase in the levels of processed, nuclear SREBP1 and SREBP2, and the expression of SREBP targets. Although the phosphatidic acid phosphatase activity of lipin 1 was shown to be important for its inhibitory effect on nuclear SREBP levels [35], the molecular mechanism and tissue specificity of this regulation, as with S6K1 and 4E-BP1, remains unknown. Finally, it is clear that mTORC1 signalling also increases the transcript levels of SREBP1 and SREBP2 in cell culture models [15], and SREBP1c in both rodent hepatocytes and the intact liver in response to insulin or feeding [18,19,20,21]. This mTORC1-dependent transcriptional response leads to an increase in full-length SREBP isoforms that accompany the increased processing and activation of SREBP. However, it remains unclear whether this transcriptional effect is simply a result of autoregulation by processed SREBPs at the SREBF1 or SREBF2 promoter or a parallel pathway independent from the effects of mTORC1 on SREBP processing (Fig 2). Both SREBF1 and SREBF2 contain a characterized SRE in their promoters [36,37]. In cell culture models, exogenous expression of processed SREBP1a stimulates the expression of endogenous SREBP1 and SREBP2 transcripts in a manner that is no longer sensitive to rapamycin, suggesting that the transcriptional effects of mTORC1 signalling on SREBP expression are upstream from processed SREBP [15]. However, elegant studies with a transgenic version of SREBP1c in rats suggest that the role of mTORC1 in SREBP1c processing and gene expression is separable [21]. More studies are needed to understand the many inputs of mTORC1 signalling, especially in vivo, into the regulation of SREBP isoforms.
Adipogenesis
Adipocytes are specialized mesenchymal cells that either store lipids as energy reserves (white adipose tissue) or burn lipids through oxidation to generate heat (brown adipose tissue). Pharmacological and genetic studies have demonstrated that the differentiation of mesenchymal stem cells into mature adipocytes—adipogenesis—requires mTOR signalling (Fig 3). Rapamycin treatment has been reported to reduce adipogenesis in a variety of cell culture models. Rapamycin seems to block the early determination step in brown adipocyte differentiation, in which a mesenchymal stem cell commits to becoming a preadipocyte [38]. Similarly, rapamycin treatment or shRNA-mediated knockdown of S6K1 in embryoid bodies hinders their commitment to preadipocytes [39]. However, much of our knowledge of adipogenesis comes from cell culture models of preadipocytes after lineage commitment and also from MEFs, and has therefore been focused on the later steps of white adipose differentiation. Treatment of preadipocytes with rapamycin leads to a marked decrease in adipocyte differentiation [40,41,42,43,44]. mTOR has been implicated in hormonal induction of clonal expansion, which is an initial step of differentiation that occurs through the action of two C/EBP family transcription factors, C/EBP-β and -δ. Overall levels of C/EBP-β have been found to decrease on rapamycin treatment, which corresponds with a repression of clonal expansion of preadipocytes [41]. However, rapamycin has also been shown to inhibit preadipocyte differentiation after clonal expansion, thereby ruling out the anti-proliferative effects of rapamycin as its primary mode of inhibiting adipogenesis [42,43,44].
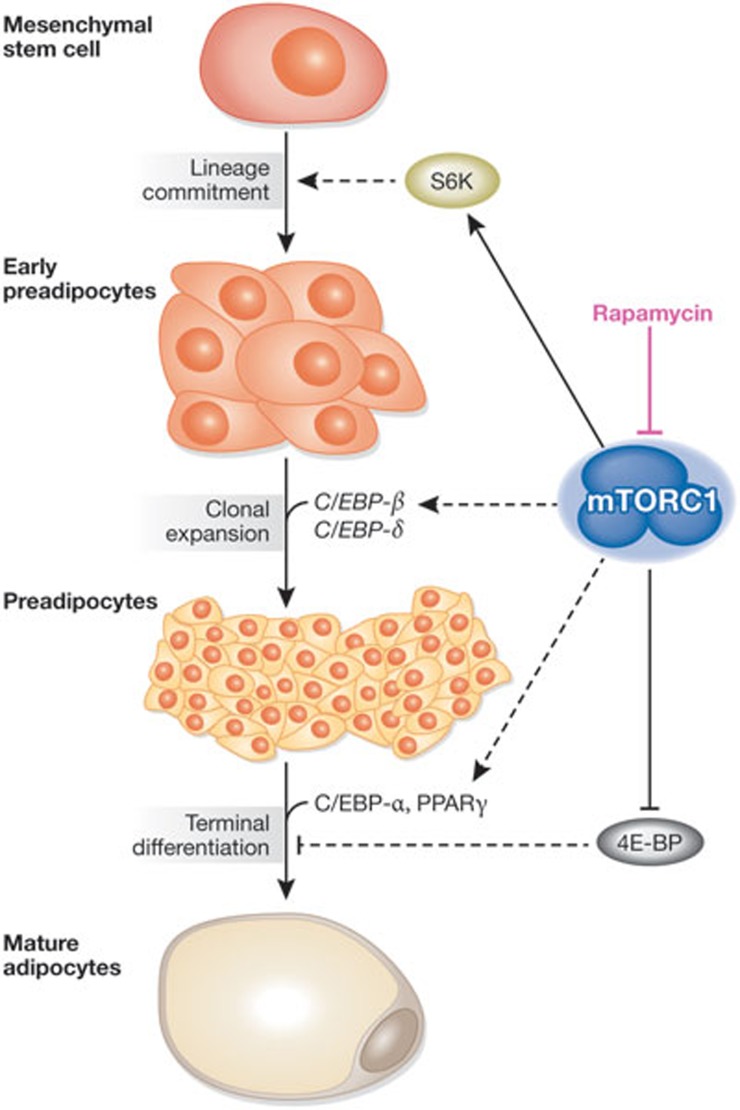
Figure 3.
mTORC1 signalling has been implicated in promoting the three main steps of adipogenesis. Adipogenesis consists of the differentiation of a mesenchymal stem cell to a mature adipocyte, which makes up a significant part of adipose tissue in which energy is stored as lipids. The commitment of the mesenchymal stem cells to the adipocyte lineage is the first step of adipogenesis and is facilitated by S6K1 activity. C/EBP-β and -δ are the primary drivers of clonal expansion, which is crucial for preadipocyte maturation, and the former has been suggested to be activated by mTORC1 signalling. The terminal differentiation of preadipocytes to mature adipocytes is mediated by PPARγ and C/EBP-α. mTORC1 promotes this final step through both its inhibition of 4E-BP and its activation of PPARγ through a poorly understood mechanism. Although the precise molecular mechanisms have yet to be defined, rapamycin blocks adipogenesis. 4E-BP, eIF4E-binding protein; C/EBP-α/β/δ, CCAAT/enhancer-binding protein-α/β/δ; mTORC1, mechanistic target of rapamycin complex 1; PPARγ, peroxisome proliferator-activated receptor γ; S6K1, ribosomal S6 kinase 1.
Several genetic models have further supported a crucial role for mTORC1 activation in terminal adipocyte differentiation, in which it seems to be both necessary and sufficient. For instance, MEFs lacking TSC1 or TSC2, which have sustained, insulin-independent activation of mTORC1 signalling, have an mTORC1-dependent enhanced capacity to differentiate into adipocytes despite these cells being severely resistant to insulin, a major adipogenic factor [45]. Reciprocally, TSC2-deficient MEFs that express a phosphorylation site mutant of TSC2, which blocks the ability of mTORC1 to be activated by insulin and Akt signalling, show reduced adipogenesis [45]. The enhanced adipogenesis in mesenchymal cells lacking the TSC tumour suppressors probably explains the common development of adipocyte-rich renal angiomyolipomas in patients with TSC [46]. Consistent with an essential role for mTORC1, RNA interference knockdown of Raptor also blocks adipogenesis in preadipocytes [47]. Downstream from mTORC1, genetic evidence suggests a role for both S6K and 4E-BP in the control of adipogenesis. The involvement of S6K in the commitment of stem cells to preadipocytes was reinforced by the reduced size of this progenitor cell population in S6K1 knockout mice and a defect in the capacity of embryonic stem cells from these mice to commit to the adipocyte lineage [39]. Reciprocally, 4E-BP1/2 double-knockout MEFs show enhanced differentiation towards adipocytes [48], suggesting that the ability of mTORC1 to both activate S6K and inhibit 4E-BP contributes to its role in promoting adipogenesis. Interestingly, the S6K1 knockout mice have a lean phenotype on both normal and high-fat diets [39,49], whereas the 4E-BP1/2 double-knockout mice are more sensitive to diet-induced obesity than their wild-type counterparts [48]. However, the differences in adiposity in these systemic mouse models probably reflect many effects of mTORC1 signalling on lipid synthesis and mobilization, discussed elsewhere in this review, in addition to its role in promoting the development of adipose deposits.
The molecular mechanisms by which mTORC1 and its downstream targets stimulate adipocyte differentiation have yet to be fully defined. The temporal activation of two transcription factors, C/EBP-α and PPARγ—the master regulator of terminal adipocyte differentiation—is responsible for inducing the final stages of differentiation [50]. mTORC1 signalling has been shown to increase PPARγ transcript and protein levels, as well as its transactivating activity [45,47,51,52], albeit through unknown mechanisms. Cell culture experiments have suggested that regulation of the final differentiation steps is primarily independent of S6K and is probably dependent on 4E-BP inhibition downstream from mTORC1 [40,48]. However, a study has indicated that PPARγ activation can also be suppressed by hyperactive mTORC1 signalling through its negative feedback effects on insulin signalling [53]. These findings indicate that there are probably mTORC1-dependent and -independent inputs into PPARγ activation and adipocyte differentiation downstream from insulin signalling, with more in vivo experiments needed.
Lipolysis
In addition to its role in stimulating lipogenesis through SREBP, mTORC1 signalling is believed to promote the storage of fatty acids in lipid stores by inhibiting lipolysis. Neutral lipids, in the form of MAG, DAG and TAG inside the cell are subject to lipolysis to mobilize free fatty acids for energy production or remodelling into new lipid species, including specific membrane and signalling lipids. Patients treated with rapamycin frequently have dyslipidaemia, one facet of which is elevated levels of plasma free fatty acids, which could reflect an increase in lipolysis in adipose tissue [54,55]. Mice treated with rapamycin show a reduction in adipocyte size and overall adiposity, and rapamycin stimulates lipolysis in cultured adipocytes [56,57,58]. Genetic manipulations of mTORC1 signalling in several mouse models have reinforced the link between mTORC1 activation and an inhibition of lipolysis. The adipose tissue of 4E-BP1/2 double-knockout mice shows decreased lipolysis [48], and S6K1 knockout mice are leaner with elevated rates of lipolysis [49]. However, mice with adipose-specific Raptor knockout, whilst also lean with reduced adiposity, do not show an obvious increase in lipolysis [47]. This suggests that the lipolysis phenotypes observed in the whole-body 4E-BP and S6K1 knockout models could be due to systemic effects rather than those intrinsic to the adipocyte. Interestingly, adipose-specific Atg7 knockout mice that have a defect in autophagy, show decreased adipocyte lipolysis [59], suggesting that the inhibitory effects of mTORC1 on lipolysis could be, at least in part, through its attenuation of autophagy.
Although the molecular mechanisms of lipolytic regulation by mTOR are not fully understood, mTORC1 signalling has been found to influence three distinct lipases: ATGL, HSL and LPL [60]. In adipocytes, ATGL catalyses the lipolysis of TAGs to DAGs within lipid droplets. HSL then converts the DAGs to MAGs. In 3T3-L1 adipocytes, mTORC1 suppression increases the transcription of ATGL, which parallels the enhanced lipolysis induced by rapamycin or siRNA knockdown of Raptor [57]. The phosphorylation of HSL at Ser 563, an established PKA site, is associated with an increase in its lipase activity. A decrease in HSL phosphorylation correlates with mTORC1 activation and the diminished release of free fatty acids [58]. However, as with ATGL transcriptional suppression, how mTORC1 signalling negatively affects HSL phosphorylation on this PKA site is unknown. Similarly to mTORC1 inhibition, adipocyte-specific Rictor knockout also leads to the phosphorylation of HSL at Ser 563 [61]. In addition to adipocyte lipolysis, mTORC1 has been implicated in the control of the extracellular lipase LPL. LPL is a water-soluble lipase present in plasma, as well as on the surface of endothelial cells, primarily in muscle and adipose tissue. It hydrolyses TAG in circulating VLDL to promote conversion to IDL and LDL, which facilitates the uptake of lipoprotein into tissues [62]. Systemic rapamycin treatment has been found to decrease LPL activity in mouse adipose tissue, and mouse and human plasma, albeit through an unknown mechanism [63,64]. The collective studies in patients treated with rapamycin and a variety of cell and mouse models suggest that mTORC1 activation, which occurs in metabolic tissues after feeding, promotes the synthesis and storage of lipids. By contrast, mTORC1 inhibition, such as during fasting, stimulates lipolysis and the release of free fatty acids into the circulation.
β-oxidation and ketogenesis
Consistent with the inhibition of mTORC1 signalling promoting fatty acid release and consumption, there is growing evidence that mTORC1 suppresses the β-oxidation of fatty acids for energy or ketogenesis. Rapamycin has been found to increase β-oxidation in rat hepatocytes and this has been attributed to increased expression of β-oxidation enzymes, including long-chain acyl-CoA dehydrogenase and carnitine acyltransferase [17,65]. This effect of rapamycin could be due to the induction of autophagy, which seems to promote the β-oxidation of fatty acids from TAGs in hepatocytes [66]. However, genetic evidence suggests that autophagy has inhibitory effects on β-oxidation in adipose tissue [59,67]. Mice with whole-body knockout of S6K1 seem to have enhanced β-oxidation, as evidenced by increased levels of CPT1 transcript in isolated adipocytes [49]. Consistent with mTORC1 signalling attenuating β-oxidation, myoblasts isolated from S6K1/S6K2 double-knockout mice also show enhanced β-oxidation of fatty acids [68]. However, this phenotype was attributed to indirect effects from energy stress and AMPK activation in this setting. As with the S6K1 knockout and the S6K1/S6K2 double-knockout mice, mice with adipose-specific Raptor knockout are lean with adipocytes that show increased mitochondrial uncoupling, which could allow them to burn lipids rapidly without generating ATP [47,49,68]. Paradoxically, mTORC1 activation has also been linked to increased mitochondrial biogenesis in some settings [69]. This could explain the decrease in oxidative capacity of muscle [69,70,71] and Jurkat T cells [72] after the inhibition or complete loss of mTORC1 signalling. However, further studies are needed to determine how the observed changes in mitochondrial gene expression and oxygen consumption in these settings influence the β-oxidation of fatty acids. The collective data suggest that mTORC1 signalling inhibits fatty acid oxidation, whilst also promoting mitochondrial biogenesis in some settings.
The acetyl-CoA released from β-oxidation can either enter the TCA cycle or, under fasting conditions in the liver, be converted to ketone bodies. Genetic evidence suggests that mTORC1 signalling in the liver, which is respectively inhibited and activated by fasting and feeding, suppresses ketogenesis [73]. Mice with LTsc1KO that show sustained mTORC1 signalling under fasting have a defect in ketogenesis, whereas mice with liver-specific Raptor knockout show an increase in fasting-induced ketogenesis. mTORC1 seems to suppress the expression of ketogenic enzymes through its regulation of N-CoR1 and PPARα [73], by a mechanism probably dependent on S6K2 [74]. These inhibitory effects on PPARα and its transcriptional targets could also explain the negative regulation of fatty acid oxidation by mTORC1. The repression of β-oxidation and ketogenesis by mTORC1 probably acts together with its stimulation of lipogenesis, further promoting the flux of acetyl-CoA towards lipid synthesis and storage.
Lipid transport
Several lines of evidence suggest a role for mTORC1 signalling in the control of lipid mobilization and transport. As stated above, patients treated with mTORC1 inhibitors suffer frequently from a dyslipidaemia consisting of hypertriglyceridaemia and hypercholesterolaemia, as well as increased levels of plasma free fatty acids [55]. The source of the elevated circulating lipids in these patients is unknown. However, TAG and cholesterol transport out of the liver involves their packaging into apolipoprotein complexes, and plasma levels of both apolipoprotein B-100 and apolipoprotein C-III have been found to be increased in patients treated with rapamycin [54]. A study in guinea pigs revealed that the increase in circulating TAGs observed in the response to rapamycin correlates with an increase in VLDL, the primary mode of TAG export from the liver [75]. In cultured hepatocytes, the ability of insulin to repress the expression of both apolipoprotein B and apolipoprotein A-5 is sensitive to rapamycin, suggesting that the increase in apolipoproteins observed on rapamycin treatment in vivo might be due to direct effects on hepatocytes [76,77]. How mTORC1 negatively regulates the expression or protein levels of specific apolipoproteins is unknown and could be secondary to changes in apolipoprotein uptake or degradation. Conversely, mTORC1 signalling seems to upregulate LDLR, which facilitates the uptake of cholesterol-rich LDL from the plasma into the liver and peripheral tissues. LDLR gene expression is controlled by SREBP [78] and would, therefore, be predicted to be stimulated by insulin in an mTORC1-dependent manner. In addition, mTORC1 signalling downstream from the insulin receptor in the liver has been found to repress the expression of PCSK9, a known negative regulator of LDLR protein levels [79]. Consequently, rapamycin treatment decreases LDLR levels in a PCSK9-dependent manner, thereby reducing LDL uptake and increasing its circulating levels. Combined with the rapamycin-stimulated increase in lipolysis and apolipoprotein levels, these effects on the LDLR suggest a mechanistic basis for the dyslipidaemia observed in patients treated with mTORC1 inhibitors.
mTORC1 in physiology, obesity and diabetes
The global effects of the mTORC1-mediated regulation of lipid metabolism detailed above are predicted to promote the systemic flux of carbon into lipids and their storage as TAGs within adipose tissue (Fig 4). The postprandial increase in both glucose and insulin stimulates the acute activation of mTORC1 within metabolic tissues, in which mTORC1 has contextual roles in controlling lipid metabolism. In the liver, and probably in adipose tissue, mTORC1 activation induces lipogenesis. At the same time, mTORC1 probably blocks the β-oxidation of fatty acids in the liver, adipose, and perhaps muscle, instead promoting the use and storage of glucose in these tissues. TAGs and cholesterol produced in the liver facilitate the packaging and release of VLDL into circulation. mTORC1 signalling might enhance uptake of lipids by peripheral tissues through the activation of LPL, which hydrolyses VLDL to IDL, and an increase in the levels of LDLR. In adipose tissue, the insulin-stimulated activation of mTORC1 is predicted to contribute to the inhibition of lipolysis, further promoting the storage of TAGs, either mobilized from the liver or produced de novo within the adipocytes.
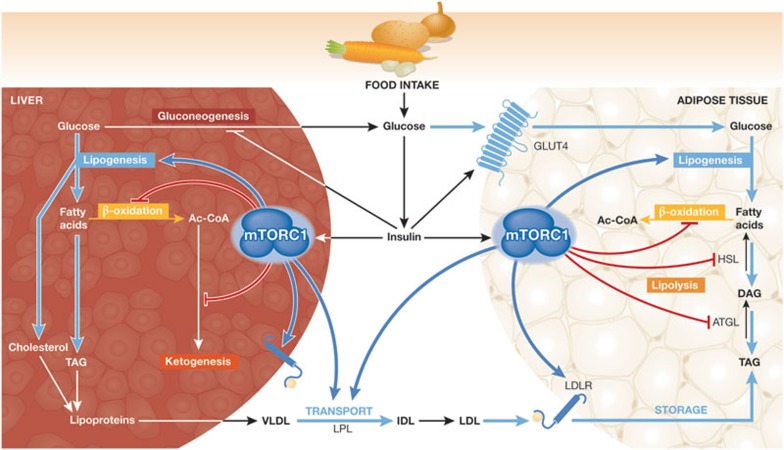
Figure 4.
The increase in insulin levels after a meal alters hepatic and adipose lipid metabolism, at least in part, through mTORC1 signalling (a working model). In the liver, mTORC1 promotes lipid synthesis through SREBP1c activation. In addition, mTORC1 signalling blocks lipid catabolism by blocking β-oxidation and ketogenesis in the liver. Consequently, mTORC1 activation in the liver promotes the synthesis of TAGs and perhaps cholesterol, which are incorporated into VLDL for transport to peripheral tissues. Evidence suggests that mTORC1 signalling positively influences LPL activity, which promotes lipid delivery to peripheral tissues by hydrolysing VLDL to IDL, which is then converted to LDL. Lipoprotein-bound TAGs are taken up by tissues, including adipocytes, through the LDLR. Both the expression and stability of LDLR, at least in the liver, are probably promoted by mTORC1 activation. In response to insulin, mTORC1 has been suggested to inhibit lipolysis in adipocytes by downregulating ATGL and HSL. Therefore, the systemic effects of postprandial mTORC1 activation are to promote the flux of carbon from glucose towards TAG storage in adipose tissue. See text for details regarding the evidence underlying this model. Ac-COA, acetyl-CoA; ATGL, adipose triglyceride lipase; DAG, diacylglycerol; GLUT4, glucose transporter type 4; HSL, hormone-sensitive lipase; IDL, intermediate density lipoprotein; LDL, low density lipoprotein; LDLR, LDL receptor; LPL, lipoprotein lipase; mTORC1, mechanistic target of rapamycin complex 1; SREBP1c, sterol regulatory element-binding protein 1c; TAG, triacylglycerol; VLDL, very low density lipoprotein.
Whilst mTORC1 is activated transiently within metabolic tissues by normal feeding, conditions of nutrient overload and obesity can lead to chronically elevated mTORC1 signalling in these tissues [49,80]. The mechanism by which obesity leads to hyperactivation of mTORC1 is unknown but happens probably through a combination of hyperglycaemia and hyperinsulinaemia under these conditions. Furthermore, evidence suggests that increased circulating levels of branch-chain amino acids, which are known to activate mTORC1, correlates with the development of obesity and insulin resistance [81]. In addition to potentially exacerbating obesity by further promoting lipid storage in adipose depots, chronic mTORC1 activation under such conditions is believed to contribute to the development of insulin resistance, which frequently accompanies obesity. Increased mTORC1 signalling can trigger several distinct feedback mechanisms, which in a cell-autonomous manner, dampens the cellular response to insulin. The in vivo contribution of these feedback mechanisms to insulin resistance is well illustrated by loss- and gain-of-function mouse models of mTORC1 signalling. For instance, S6K1 knockout mice have enhanced peripheral insulin sensitivity [49], whereas mice with LTsc1KO show hepatic insulin resistance with greatly reduced Akt signalling [18]. Therefore, under conditions of obesity, mTORC1 activation in metabolic tissues probably both perpetuates obesity and promotes insulin resistance, thereby expediting the progression to type II diabetes.
The fundamental role of mTORC1 in regulating whole-body lipid homeostasis, paired with its frequent upregulation in obesity and type 2 diabetes, suggests that mTOR inhibitors might offer some therapeutic benefit in metabolic diseases. In theory, mTORC1-specific inhibitors should suppress lipid synthesis and promote lipolysis and lipid catabolism, in addition to blocking mTORC1-dependent feedback mechanisms to resensitize tissues to insulin. However, important caveats arise from the use of mTORC1 inhibitors to combat obesity and diabetes. First, prolonged treatment with rapamycin disrupts mTORC2 and therefore Akt activation downstream from the insulin receptor, further exacerbating the insulin-resistant phenotype (Sidebar A; [82]). Second, patients treated with rapamycin frequently have increased levels of circulating TAGs, cholesterol and free fatty acids [55]. Therefore, whilst rapamycin treatment might help mobilize lipids and deplete fat stores, lipid clearance offers an additional pathological challenge. Targeting mTORC1 signalling indirectly might offer a more promising avenue. AMPK is a potent negative regulator of mTORC1, blocking its function through phosphorylation of both the TSC–TBC complex ]2,5] and Raptor [6]. Therefore, mTORC1 signalling is blocked on activation of AMPK, which is stimulated by a large variety of natural and synthetic compounds, including metformin, resveratrol and aspirin [83]. Importantly, metformin is the most widely prescribed anti-diabetes drug in the world. Whether any of the beneficial metabolic effects of metformin are attributed to its inhibition of mTORC1 signalling is one of several important outstanding questions (Sidebar B).
Sidebar B | In need of answers.
What are the molecular mechanisms by which mTORC1 regulates SREBP1 and SREBP2?
Which lipid species are most influenced by the activation state of mTORC1 signalling?
Does mTORC1 stimulate the synthesis of membrane lipids in addition to storage lipids?
How do lipids influence mTORC1 signalling?
How does mTORC1 become dysregulated under conditions of obesity?
Does mTORC1 inhibition contribute to the effects of AMPK-activating compounds on cellular and systemic metabolism?
What is the role of mTORC1 activation in the common lipogenic phenotype of cancer cells?
How is lipid metabolism differentially regulated by mTORC1 in different tissues?
Acknowledgments
We apologize to our colleagues whose work we were not able to cover in this review due to space constraints. Research in the Manning laboratory related to the subject of this review was supported by a predoctoral training grant DGE-1144152 from the National Science Foundation (S.J.H.R.) and by National Institutes of Health grants R01-CA122617 and P01-CA120964, Department of Defense grants TS093033 and TS110065, a Sanofi Innovation Award and grants from the American Diabetes Association and Ellison Medical Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Huang J, Manning BD (2008) The TSC1–TSC2 complex: a molecular switchboard controlling cell growth. Biochem J 412: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC et al. (2012) TBC1D7 is a third subunit of the TSC1–TSC2 complex upstream of mTORC1. Mol Cell 47: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL (2002) TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4: 648–657 [DOI] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC (2002) Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell 10: 151–162 [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL (2003) TSC2 mediates cellular energy response to control cell growth and survival. Cell 115: 577–590 [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC (2004) The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6: 91–99 [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM (2012) mTOR signaling in growth control and disease. Cell 149: 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks CA, Guertin DA (2010) Targeting mTOR: prospects for mTOR complex 2 inhibitors in cancer therapy. Oncogene 29: 3733–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109: 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon TI, Osborne TF (2011) SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab 23: 65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL (1997) Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest 99: 846–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shimomura I, Brown MS, Hammer RE, Goldstein JL, Shimano H (1998) Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest 101: 2331–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA 100: 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel K et al. (2010) Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39: 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A (2008) SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab 8: 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NF, Stefanovic-Racic M, Sipula IJ, Perdomo G (2007) The mammalian target of rapamycin regulates lipid metabolism in primary cultures of rat hepatocytes. Metabolism 56: 1500–1507 [DOI] [PubMed] [Google Scholar]
- Yecies JL et al. (2011) Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab 14: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Brown MS, Goldstein JL (2010) Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci USA 107: 3441–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ogawa W, Emi A, Hayashi K, Senga Y, Nomura K, Hara K, Yu D, Kasuga M (2011) Role of S6K1 in regulation of SREBP1c expression in the liver. Biochem Biophys Res Commun 412: 197–202 [DOI] [PubMed] [Google Scholar]
- Owen JL, Zhang Y, Bae SH, Farooqi MS, Liang G, Hammer RE, Goldstein JL, Brown MS (2012) Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc Natl Acad Sci USA 109: 16184–16189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BT, Ducker GS, Barczak AJ, Barbeau R, Erle DJ, Shokat KM (2011) The mammalian target of rapamycin regulates cholesterol biosynthetic gene expression and exhibits a rapamycin-resistant transcriptional profile. Proc Natl Acad Sci USA 108: 15201–15206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yuan H, Niu Y, Niu W, Fu L (2012) The role of AMPK/mTOR/S6K1 signaling axis in mediating the physiological process of exercise-induced insulin sensitization in skeletal muscle of C57BL/6 mice. Biochim Biophys Acta 1822: 1716–1726 [DOI] [PubMed] [Google Scholar]
- Wan M, Leavens KF, Saleh D, Easton RM, Guertin DA, Peterson TR, Kaestner KH, Sabatini DM, Birnbaum MJ (2011) Postprandial hepatic lipid metabolism requires signaling through Akt2 independent of the transcription factors FoxA2, FoxO1, and SREBP1c. Cell Metab 14: 516–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR et al. (2011) mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146: 408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenerson HL, Yeh MM, Yeung RS (2011) Tuberous sclerosis complex-1 deficiency attenuates diet-induced hepatic lipid accumulation. PLoS ONE 6: e18075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD (2009) A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans 37: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann M, Iynedjian PB (2000) Regulation of sterol regulatory-element binding protein 1 gene expression in liver: role of insulin and protein kinase B/cAkt. Biochem J 349: 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavens KF, Easton RM, Shulman GI, Previs SF, Birnbaum MJ (2009) Akt2 is required for hepatic lipid accumulation in models of insulin resistance. Cell Metab 10: 405–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono H et al. (2003) Hepatic Akt activation induces marked hypoglycemia, hepatomegaly, and hypertriglyceridemia with sterol regulatory element binding protein involvement. Diabetes 52: 2905–2913 [DOI] [PubMed] [Google Scholar]
- Hagiwara A et al. (2012) Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab 15: 725–738 [DOI] [PubMed] [Google Scholar]
- Bengoechea-Alonso MT, Ericsson J (2009) A phosphorylation cascade controls the degradation of active SREBP1. J Biol Chem 284: 5885–5895 [DOI] [PubMed] [Google Scholar]
- Luyimbazi D et al. (2010) Rapamycin regulates stearoyl CoA desaturase 1 expression in breast cancer. Mol Cancer Ther 9: 2770–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D et al. (2009) EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci Signal 2: ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman TA, Mothe-Satney I, Lawrence JC Jr (2002) Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proc Natl Acad Sci USA 99: 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato R, Inoue J, Kawabe Y, Kodama T, Takano T, Maeda M (1996) Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J Biol Chem 271: 26461–26464 [DOI] [PubMed] [Google Scholar]
- Amemiya-Kudo M et al. (2000) Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem 275: 31078–31085 [DOI] [PubMed] [Google Scholar]
- Vila-Bedmar R, Lorenzo M, Fernández-Veledo S (2010) Adenosine 5′-monophosphate-activated protein kinase-mammalian target of rapamycin cross talk regulates brown adipocyte differentiation. Endocrinology 151: 980–992 [DOI] [PubMed] [Google Scholar]
- Carnevalli LS et al. (2010) S6K1 plays a critical role in early adipocyte differentiation. Dev Cell 18: 763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Chaâr D, Gagnon A, Sorisky A (2004) Inhibition of insulin signaling and adipogenesis by rapamycin: effect on phosphorylation of p70 S6 kinase vs eIF4E-BP1. Int J Obes Relat Metab Disord 28: 191–198 [DOI] [PubMed] [Google Scholar]
- Yeh WC, Bierer BE, McKnight SL (1995) Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc Natl Acad Sci USA 92: 11086–11090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A, Grunder L, Sorisky A (2000) Rapamycin inhibits human adipocyte differentiation in primary culture. Obes Res 8: 249–254 [DOI] [PubMed] [Google Scholar]
- Gagnon A, Lau S, Sorisky A (2001) Rapamycin-sensitive phase of 3T3-L1 preadipocyte differentiation after clonal expansion. J Cell Physiol 189: 14–22 [DOI] [PubMed] [Google Scholar]
- Cho HJ, Park J, Lee HW, Lee YS, Kim JB (2004) Regulation of adipocyte differentiation and insulin action with rapamycin. Biochem Biophys Res Commun 321: 942–948 [DOI] [PubMed] [Google Scholar]
- Zhang HH, Huang J, Düvel K, Boback B, Wu S, Squillace RM, Wu CL, Manning BD (2009) Insulin stimulates adipogenesis through the Akt–TSC2–mTORC1 pathway. PLoS ONE 4: e6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crino PB, Nathanson KL, Henske EP (2006) The tuberous sclerosis complex. N Engl J Med 355: 1345–1356 [DOI] [PubMed] [Google Scholar]
- Polak P, Cybulski N, Feige JN, Auwerx J, Rüegg MA, Hall MN (2008) Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab 8: 399–410 [DOI] [PubMed] [Google Scholar]
- Le Bacquer O, Petroulakis E, Paglialunga S, Poulin F, Richard D, Cianflone K, Sonenberg N (2007) Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest 117: 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um SH et al. (2004) Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431: 200–205 [DOI] [PubMed] [Google Scholar]
- Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM (2002) C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev 16: 22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Chen J (2004) Regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes 53: 2748–2756 [DOI] [PubMed] [Google Scholar]
- Yu W et al. (2008) Critical role of phosphoinositide 3-kinase cascade in adipogenesis of human mesenchymal stem cells. Mol Cell Biochem 310: 11–18 [DOI] [PubMed] [Google Scholar]
- Laplante M, Horvat S, Festuccia WT, Birsoy K, Prevorsek Z, Efeyan A, Sabatini DM (2012) DEPTOR cell-autonomously promotes adipogenesis, and its expression is associated with obesity. Cell Metab 16: 202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisett JD et al. (2002) Effects of sirolimus on plasma lipids, lipoprotein levels, and fatty acid metabolism in renal transplant patients. J Lipid Res 43: 1170–1180 [PubMed] [Google Scholar]
- Kasiske BL, de Mattos A, Flechner SM, Gallon L, Meier-Kriesche HU, Weir MR, Wilkinson A (2008) Mammalian target of rapamycin inhibitor dyslipidemia in kidney transplant recipients. Am J Transplant 8: 1384–1392 [DOI] [PubMed] [Google Scholar]
- Zhang C, Yoon MS, Chen J (2009) Amino acid-sensing mTOR signaling is involved in modulation of lipolysis by chronic insulin treatment in adipocytes. Am J Physiol Endocrinol Metab 296: E862–E868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti P, English T, Shi J, Smas CM, Kandror KV (2010) Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes 59: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman GA, Acosta-Jaquez HA, Fingar DC (2010) mTORC1 inhibition via rapamycin promotes triacylglycerol lipolysis and release of free fatty acids in 3T3-L1 adipocytes. Lipids 45: 1089–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S (2009) Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci USA 106: 19860–19865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F (2012) FAT SIGNALS—lipases and lipolysis in lipid metabolism and signaling. Cell Metab 15: 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Lawrence JC Jr, Jung DY, Ko HJ, Keller SR, Kim JK, Magnuson MA, Harris TE (2010) Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes 59: 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Eckel RH (2009) Lipoprotein lipase: from gene to obesity. Am J Physiol Endocrinol Metab 297: E271–E288 [DOI] [PubMed] [Google Scholar]
- Tory R, Sachs-Barrable K, Hill JS, Wasan KM (2008) Cyclosporine A and rapamycin induce in vitro cholesteryl ester transfer protein activity, and suppress lipoprotein lipase activity in human plasma. Int J Pharm 358: 219–223 [DOI] [PubMed] [Google Scholar]
- Blanchard PG et al. (2012) Major involvement of mTOR in the PPARγ-induced stimulation of adipose tissue lipid uptake and fat accretion. J Lipid Res 53: 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Golub TR, Sabatini DM (2002) The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol Cell Biol 22: 5575–5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ (2009) Autophagy regulates lipid metabolism. Nature 458: 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R et al. (2009) Autophagy regulates adipose mass and differentiation in mice. J Clin Invest 119: 3329–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar V et al. (2007) S6 kinase deletion suppresses muscle growth adaptations to nutrient availability by activating AMP kinase. Cell Metab 5: 476–487 [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P (2007) mTOR controls mitochondrial oxidative function through a YY1–PGC-1alpha transcriptional complex. Nature 450: 736–740 [DOI] [PubMed] [Google Scholar]
- Risson V et al. (2009) Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J Cell Biol 187: 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger CF et al. (2008) Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab 8: 411–424 [DOI] [PubMed] [Google Scholar]
- Schieke SM, Phillips D, McCoy JP, Aponte AM, Shen RF, Balaban RS, Finkel T (2006) The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem 281: 27643–27652 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM (2010) mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468: 1100–1104 [DOI] [PubMed] [Google Scholar]
- Kim KK, Pyo S, Um SH (2012) S6 kinase 2 deficiency enhances ketone body production and increases peroxisome proliferator-activated receptor alpha activity in the liver. Hepatology 55: 1727–1737 [DOI] [PubMed] [Google Scholar]
- Aggarwal D, Fernandez ML, Soliman GA (2006) Rapamycin, an mTOR inhibitor, disrupts triglyceride metabolism in guinea pigs. Metabolism 55: 794–802 [DOI] [PubMed] [Google Scholar]
- Sidiropoulos KG, Meshkani R, Avramoglu-Kohen R, Adeli K (2007) Insulin inhibition of apolipoprotein B mRNA translation is mediated via the PI-3 kinase/mTOR signaling cascade but does not involve internal ribosomal entry site (IRES) initiation. Arch Biochem Biophys 465: 380–388 [DOI] [PubMed] [Google Scholar]
- Nowak M et al. (2005) Insulin-mediated down-regulation of apolipoprotein A5 gene expression through the phosphatidylinositol 3-kinase pathway: role of upstream stimulatory factor. Mol Cell Biol 25: 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicher R, Kotzka J, Müller-Wieland D, Siemeister G, Munck M, Avci H, Krone W (1996) SREBP-1 mediates activation of the low density lipoprotein receptor promoter by insulin and insulin-like growth factor-I. J Biol Chem 271: 7128–7133 [DOI] [PubMed] [Google Scholar]
- Ai D et al. (2012) Regulation of hepatic LDL receptors by mTORC1 and PCSK9 in mice. J Clin Invest 122: 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamzina L, Veilleux A, Bergeron S, Marette A (2005) Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats: possible involvement in obesity-linked insulin resistance. Endocrinology 146: 1473–1481 [DOI] [PubMed] [Google Scholar]
- Newgard CB et al. (2009) A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9: 311–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW et al. (2012) Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335: 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA (2012) AMP-activated protein kinase: a target for drugs both ancient and modern. Chem Biol 19: 1222–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307: 1098–1101 [DOI] [PubMed] [Google Scholar]
- Dibble CC, Asara JM, Manning BD (2009) Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol Cell Biol 29: 5657–5670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien LA, Carriere A, Moreau J, Roux PP (2010) mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. Mol Cell Biol 30: 908–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 22: 159–168 [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM (2009) The pharmacology of mTOR inhibition. Sci Signal 2: pe24. [DOI] [PubMed] [Google Scholar]



 Brendan D Manning & Stéphane J H Ricoult
Brendan D Manning & Stéphane J H Ricoult