SUMMARY
The study of 5-hydroxylmethylcytosines (5hmC) has been hampered by the lack of a method to map it at single-base resolution on a genome-wide scale. Affinity purification-based methods cannot precisely locate 5hmC nor accurately determine its relative abundance at each modified site. We here present a genome-wide approach, Tet-assisted Bisulfite Sequencing (TAB-Seq), for mapping 5hmC at base resolution and quantifying the relative abundance of 5hmC as well as 5mC when combined with traditional bisulfite sequencing. Application of this method to embryonic stem cells not only confirms widespread distribution of 5hmC in the mammalian genome, but also reveals sequence bias and strand asymmetry at 5hmC sites. We observe high levels of 5hmC and reciprocally low levels of 5mC near but not on transcription factor binding sites. Additionally, the relative abundance of 5hmC varies significantly among distinct functional sequence elements, suggesting different mechanisms for 5hmC deposition and maintenance.
INTRODUCTION
5-methylcytosine (5mC) in mammalian genomic DNA is essential for normal development and impacts a variety of biological functions. In 2009, 5-hydroxymethylcytosine (5hmC) was discovered as another relatively abundant form of cytosine modification in embryonic stem cells (ESCs) and Purkinje neurons (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). The TET proteins, which are responsible for conversion of 5mC to 5hmC, have been shown to function in ESC regulation, myelopoiesis and zygote development (Dawlaty et al., 2011; Gu et al., 2011; Iqbal et al., 2011; Ito et al., 2010; Ko et al., 2010; Koh et al., 2011; Wossidlo et al., 2011). 5hmC was found to be widespread in many tissues and cell types, although with diverse levels of abundance (Globisch et al., 2010; Munzel et al., 2010; Song et al., 2011; Szwagierczak et al., 2010). Proteins that can recognize 5hmC-containing DNA have also been investigated (Frauer et al., 2011; Yildirim et al., 2011). In addition, 5hmC can be further oxidized to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) by TET proteins (He et al., 2011; Ito et al., 2011; Pfaffeneder et al., 2011), and demethylation pathways through these modified cytosines have been shown (Cortellino et al., 2011; Guo et al., 2011; He et al., 2011; Maiti and Drohat, 2011; Zhang et al., 2012). Together, these studies provide an emerging paradigm in which 5mC oxidation plays important roles in sculpting a cell’s epigenetic landscape and developmental potential through the regulation of dynamic DNA methylation states.
Strategies to label and/or enrich 5hmC in genomic DNA have been developed to investigate the distribution and function of 5hmC in the genome (Ficz et al., 2011; Pastor et al., 2011; Robertson et al., 2012; Robertson et al., 2011; Song et al., 2011; Stroud et al., 2011; Williams et al., 2011; Wu et al., 2011; Xu et al., 2011). While 5hmC is more enriched in gene bodies than transcription starting sites in mouse cerebellum (Song et al., 2011; Szulwach et al., 2011b), all genome-wide maps of 5hmC in human and mouse embryonic stem cells indicate that 5hmC tends to exist in gene bodies, promoters, and enhancers (Ficz et al., 2011; Pastor et al., 2011; Stroud et al., 2011; Szulwach et al., 2011a; Williams et al., 2011; Wu et al., 2011; Xu et al., 2011). However, in all cases, the resolution of these maps was restricted by the size of the immunoprecipitated or chemically captured DNA, which varied from several hundred to over a thousand bases.
The study of 5mC has been facilitated by the development of whole genome bisulfite sequencing methods that can resolve the genomic location of methylcytosine at single-base resolution (Cokus et al., 2008; Lister et al., 2008; Lister et al., 2009). However, current bisulfite sequencing methods cannot distinguish between 5mC and 5hmC (Huang et al., 2010; Jin et al., 2010). Therefore, the genome-wide bisulfite sequencing maps generated in recent years may not accurately capture the true abundance of 5mC at each base in the genome. A more detailed understanding of the function of 5hmC as well as 5mC has, therefore, been hampered by the lack of a single-base resolution sequencing technology capable of detecting the relative abundance of 5hmC per cytosine.
Here we present a Tet-assisted bisulfite sequencing (TAB-Seq) strategy, which provides a method for single-base resolution detection of 5hmC amenable to both genome-wide and loci-specific sequencing. Applying this new method, we have generated the first genome-wide, single-base resolution maps of 5hmC in ESCs. Distinct classes of functional elements exhibit variable abundance of 5hmC, with promoter-distal regulatory elements harboring the highest levels of 5hmC. High levels of 5hmC and reciprocally low levels of 5mC can be found near binding sites of transcription factors. In contrast to 5mC, 5hmC sites display strand asymmetry and sequence bias. Finally, the base-resolution maps of 5hmC provide more accurate estimates of both 5hmC and 5mC levels at each modified cytosine than previous whole genome bisulfite sequencing approaches. Our results support a dynamic DNA methylation process at distal regulatory elements, and suggest that different mechanisms of DNA modification may be involved at distinct classes of functional sequences in the genome.
RESULTS
TAB-Seq of Model DNA and Specific Loci
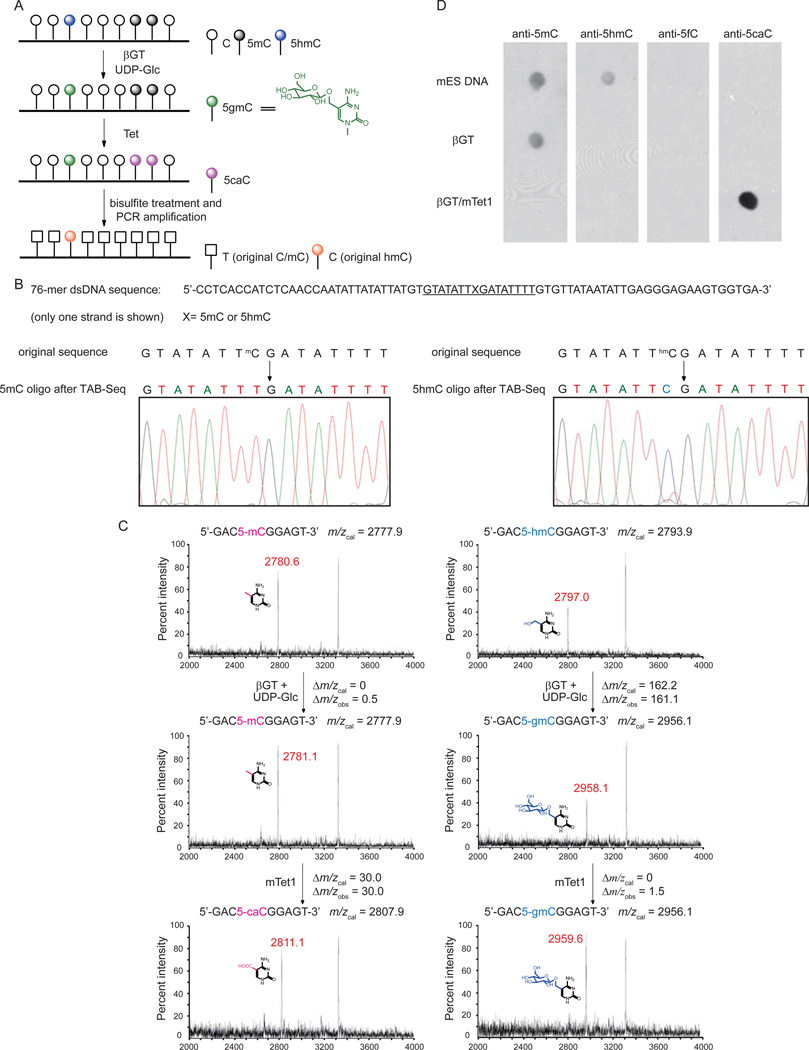
Traditional bisulfite sequencing cannot discriminate 5mC from 5hmC because both resist deamination by bisulfite treatment (Huang et al., 2010; Jin et al., 2010). We have recently found that TET proteins not only oxidize 5mC to 5hmC, but also further oxidize 5hmC to 5caC, and that 5caC exhibits similar behavior as unmodified cytosine after bisulfite treatment (He et al., 2011; Ito et al., 2011). This deamination difference between 5caC and 5mC/5hmC under standard bisulfite conditions inspired us to explore TAB-Seq. In this approach, we introduce a glucose onto 5hmC using β-glucosyltransferase (βGT), generating β-glucosyl-5-hydroxymethylcytosine (5gmC) to protect 5hmC from further TET oxidation. After blocking of 5hmC, all 5mC is converted to 5caC by oxidation with excess of recombinant Tet1 protein. Bisulfite treatment of the resulting DNA then converts all C and 5caC (derived from 5mC) to uracil or 5caU, respectively, while the original 5hmC bases remain protected as 5gmC. Thus, subsequent sequencing will reveal 5hmC as C, providing an accurate assessment of abundance of this modification at each cytosine when combined with traditional bisulfite sequencing (Figure 1A). We first confirmed that 5gmC is read as C in traditional bisulfite sequencing (data not shown). We cloned and expressed the catalytic domain of mouse Tet1 (mTet1) (Figure S1A), as previously reported (Ito et al., 2010). We tested a synthetic double-stranded DNA with site-specifically incorporated 5mC or 5hmC modification (Figure 1B). Application of our method with Sanger sequencing of the PCR amplified products showed that the original 5mC was completely converted into T after treatment, indicating efficient oxidation of 5mC to 5caC by mTET1 (Figure 1B). However, the original 5hmC was sequenced as C, confirming that the protected 5gmC is resistent to deamination under bisulfite treatment (Figure 1B). The products of each step were confirmed by MALDI-TOF/TOF using a shorter model duplex DNA (Figure 1C). Full conversion of 5mC in the context of genomic DNA was also confirmed by conventional bisulfite, PCR, and both Sanger and semiconductor sequencing (Figure S1B–C). Additionally, application to genomic DNA confirmed conversion of 5mC to 5caC and protection of 5hmC, and that 5fC is undetectable by immunoblot on the final reaction products (Figure 1D). Thus, coupling βGT-mediated transfer of glucose to 5hmC with mTet1-catalyzed oxidation of 5mC to 5caC enables the distinction of 5hmC from both C and 5mC after sodium bisulfite treatment.
Figure 1. TAB-Seq Strategy and Validation.
(A)Schematic diagram of TAB-Seq. 5 hmCs in genomic DNA are protected by glucosylation, and then 5mCs are converted to 5caCs by Tet-mediated oxidation. After bisulfite treatment, both 5caC (generated from 5mC) and C display as T while 5gmC (generated from original 5hmC) displays as C.
(B) TAB-Seq of 76-mer dsDNA with 5mC or 5hmC. The 76-mer dsDNA with 5mC (left) or 5hmC (right) modification was subject to TAB-Seq as described in Figure 1A. Sanger sequencing results showed that 5mC was completely converted to T (left) and 5hmC still read as C (right).
(C) Mass spectrometry characterization of the products from TAB-Seq with a model DNA. The dsDNA contains a 5mC (left) or 5hmC (right) on a 9mer strand annealed to a 11mer complementary strand. The DNA was subject to βGT-mediated glucosylation and mTet1-mediated oxidation. The reactions were monitored by MALDI-TOF/TOF with the calculated and observed molecular weight indicated.
(D)Validation of 5mC and 5hmC conversion in genomic DNA (mouse ES) with western blotting. The untreated DNA, βGT-treated DNA, and βGT/mTet1-treated DNA were tested with dot blot assays using antibodies against 5mC, 5hmC, 5fC and 5caC, respectively. No 5hmC could be observed after glucosylation. Almost all 5mCs were converted into 5caCs after the mTet1-mediated oxidation.
See also Figure S1.
The ability to distinguish 5hmC at base resolution offers a significant opportunity to further parse DNA methylation/hydroxymethylation states at specific genomic loci. We applied traditional bisulfite sequencing and TAB-Seq to known 5hmC-enriched loci in mouse cerebellum that were identified previously (Song et al., 2011; Szulwach et al., 2011b). Comparing the sequencing results, we were able to identify genuine 5hmC and 5mC sites (Figure S1D).
Generation of Base-Resolution Maps of 5hmC in Embryonic Stem Cells
We next applied TAB-Seq to genomic DNA from H1 human ES cells and E14Tg2a mouse ES cells, and sequenced to an average depth of 26.5X and 17X per cytosine, respectively. Successful detection of 5hmC is governed by three key parameters: 1) efficient conversion of unmodified cytosine to uracil; 2) efficient conversion of 5mC to 5caU/U; and 3) efficient protection of 5hmC. To directly assess these conversion rates in the context of genomic DNA, sequenced samples were spiked in with fragments of lambda DNA amplified by PCR to contain three distinct domains having either unmodified cytosine, 5mC, or 5hmC. We observe low non-conversion rates for unmodified cytosine (0.38%) and 5mC (2.21%), contrasted to a high non-conversion rate of 5hmC (84.4%) (Figure S2B). Further analysis indicates that this latter value is an underestimate of the true 5hmC protection rate in H1, which is closer to 87.0% (Figure S2D–E). These data further confirm the capability of TAB-Seq for robust distinction of 5hmC from 5mC and unmodified cytosine in the context of genomic DNA.
We next focused our analysis on the map of H1 human ES cells. To confidently identify 5hmC-modified bases we took advantage of the highly annotated H1 methylome generated using methylC-Seq, which identifies both 5mC as well as 5hmC. Accordingly, we restricted our search for 5hmC to the subset of methylated bases previously identified by methylC-Seq (Lister et al., 2009). The probability that a cytosine can be confidently identified as 5hmC is governed by the sequencing depth at the cytosine and abundance of the modification (Figure S2C). Modeling this probabilistic event with a binomial distribution (Lister et al., 2009) with N as the depth of sequencing at the cytosine and p as the 5mC non-conversion rate, we identified a total of 691,414 5hmCs with a false discovery rate of 5% (Figure S2F, see Extended Experimental Procedures). Given an average sequencing depth of 26.5, our assay can on average resolve 5hmC having an abundance of 20% or higher (Figure S2C).
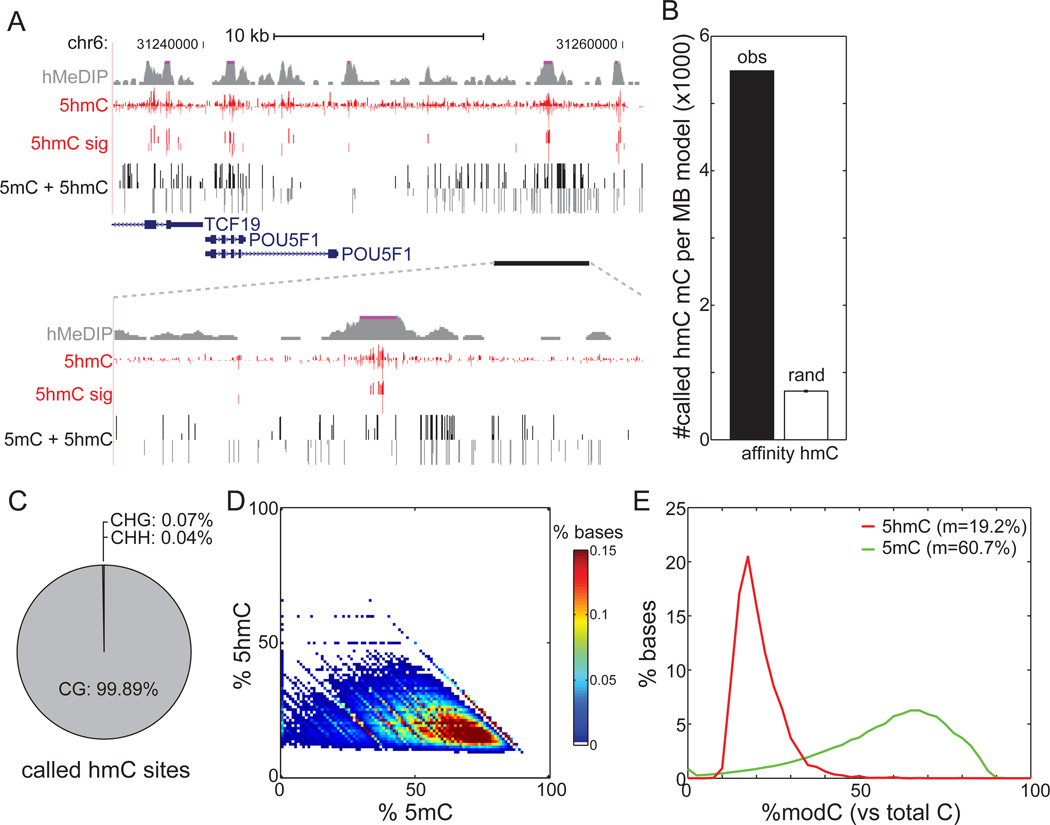
Genomic profiles of absolute 5hmC levels are comparable to a map previously generated using an affinity-based approach (Szulwach et al., 2011a) (Figure 2A). As sequenced fragments are equally distributed among the population of cells, TAB-Seq provides a steady-state glimpse of 5hmC in the entire population. This is in contrast to affinity-based approaches, which bias sequencing towards 5hmC-enriched DNA fragments. By TAB-Seq, identified 5hmCs are highly clustered, unlike 5mCs (Figure S3A), and track well with peaks of 5hmC enrichment previously identified by affinity sequencing (Figure 2A). There are 7.6 times as many 5hmCs overlapping affinity-identified regions as expected by chance (Figure 2B, Z-score = 1,579). Furthermore, 81.5% of these 82,221 affinity-identified regions were recovered by at least one 5hmC. In contrast, only 35.6% of 5hmCs are recovered by affinity-based approaches, suggesting an increased sensitivity of TAB-Seq. Using semiconductor sequencing, we verified the presence/absence of 5hmC at 57 out of 59 individual cytosines (9 out of 11 hydroxymethylated CpGs, with depth ≥ 30) within regions that previously escaped detection by 5hmC affinity capture (Figure S2A), underscoring the sensitivity and specificity of our approach.
Figure 2. Generation of Genome-wide Base-Resolution Maps of 5hmC.
(A)Snapshot of base-resolution 5hmC maps (red) compared to affinity-based 5hmC maps (grey) in H1 cells near the POU5F1 gene. Also shown are base-resolution maps of traditional bisulfite sequencing in H1 cells (black). Positive values (darker shades) indicate cytosines on the Watson strand, whereas negative values indicate cytosines on the Crick strand. For 5hmC, the vertical axis limits are −50% to +50%. For traditional bisulfite sequencing, the limits are −100% to +100%. Only cytosines sequenced to depth ≥5 are shown.
(B) Overlap of 5hmC with 82,221 genomic regions previously identified as enriched with 5hmC by affinity mapping (black), in comparison to randomly chosen 5mC (white) (see Extended Experimental Procedures).
(C) Sequence context of 5hmC sites compared to the reference human genome.
(D)Heatmap of estimated abundances of 5hmC and 5mC for modified cytosines significantly enriched with 5hmC. 5mC was estimated as the rate from traditional bisulfite sequencing (5hmC + 5mC) minus the measured 5hmC rate.
(E) The distribution of estimated abundances of 5hmC (red) and 5mC (green) at 5hmC sites. m: median.
See also Figure S2.
Application of TAB-Seq to mouse ESCs resulted in 2,057,636 high-confidence 5hmCs. This larger number of sites is likely attributable to higher level expression of both Tet1 and Tet2 in mouse ESCs as revealed by RNA-Seq analysis (Lister et al., 2011; Myers et al., 2011) (B.R., unpublished data). Like H1, these 5hmCs are also significantly enriched at genomic loci recovered by affinity sequencing (Figure S2J). In addition, these 5hmC sites are significantly enriched for previously mapped binding sites of Tet1 (Williams et al., 2011; Wu et al., 2011), confirming the TAB-Seq approach.
Base Composition and Genomic Distribution of 5-Hydroxymethylcytosine
DNA methylation of cytosines can exist in several contexts: CpG (denoted CG), CHG, and CHH (H = A, C, or T). While it has been suggested that mouse ESCs may harbor 5hmC in non-CG content (Ficz et al., 2011) and while non-CG methylation is present in human and mouse ESCs (Lister et al., 2009; Stadler et al., 2011), we found that nearly all (99.89%) of H1 5hmCs exist in the CG context (Figure 2C). Similarly, this figure is 98.7% in mouse ESCs (Figure S2G).
The combination of traditional methylC-Seq and TAB-Seq maps allows us to estimate the true abundance of both 5hmC and 5mC. We observe that, in a steady-state population of cells, 5mC and 5hmC often coexist at the same cytosine (Figure 2D). The median observed abundance of 5hmC at 5hmC-rich cytosines is 19.2%, compared to 60.7% for 5mC as estimated from traditional bisulfite sequencing (Figure 2E). Adjusting for the 87.0% protection rate of 5hmC by TAB-Seq, we estimate the corrected median 5hmC and 5mC abundance to be 22.1% and 57.8%, respectively. These results suggest that, at the base level, the abundance of 5hmC is lower than 5mC. This observation is corroborated in mouse ESCs (Figure S2H–I), and is consistent with a previous estimate of global 5hmC levels in embryonic stem cells (Tahiliani et al., 2009).
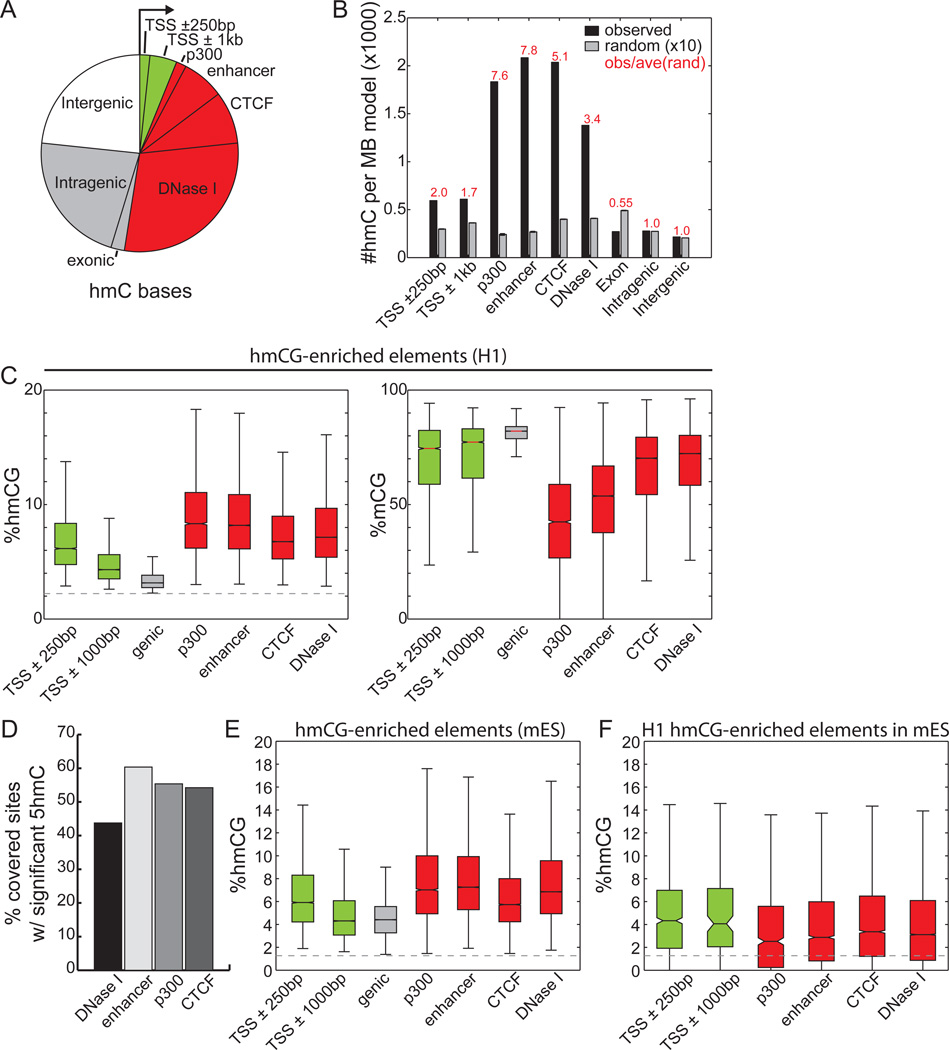
Previous studies using affinity-based approaches have demonstrated that 5hmC is enriched at promoters, enhancers, CTCF binding sites, exons, and gene bodies (Ficz et al., 2011; Pastor et al., 2011; Stroud et al., 2011; Szulwach et al., 2011a; Williams et al., 2011; Wu et al., 2011; Xu et al., 2011), suggesting an extensive role for this modification in gene regulation. Supporting a functional role of 5hmC, we observe a trend of increasing sequence conservation for increasing abundance of 5hmC (Figure S3B). However, the absolute abundance of 5hmC cannot be assessed from affinity-based detection methods, therefore precluding further quantitative analysis of 5hmC’s role at each class of regulatory elements. In H1, we found that almost half (46.4%) of the 5hmCs reside in distal regulatory elements mapped by ChIP-Seq and DNase-Seq (Figure 3A). Assessing relative enrichment of 5hmC at each class of regulatory element by normalizing with genomic coverage, H1 distal regulatory elements including p300 binding sites (observed/expected = 7.6), predicted enhancers (o/e =7.8), CTCF binding sites (o/e = 5.1), and DNase I hypersensitive sites (o/e = 3.4) are more enriched with 5hmC than other genic regions (Figure 3B). Intriguingly, the subset of cytosines showing nearly equal levels of 5mC and 5hmC are more enriched in distal regulatory elements and less enriched at promoters and genic features (Figure S3E), suggesting that active demethylation is strongest outside of genes. In support of this observation, promoter-distal ChIP-Seq peaks for OCT4, SOX2, NANOG, KLF4, and TAFII are also more enriched with 5hmC than genic features (Figure S3D). Finally, we observe that increasing DNase I hypersensitivity signal correlates well with increased 5hmC and decreased 5mC enrichment (Figure S3C). These results are also supported by observations in mouse ESCs (Figure S3F–G), though we observe an increase in intragenic 5hmC occupancy.
Figure 3. Genomic Distribution of 5hmC Sites.
(A)Overlap of H1 5hmC with genomic elements. Genic features were extracted from the UCSC Known Genes database (Hsu et al., 2006). Promoter-distal regulatory elements (>5kb from TSS) reflect those experimentally mapped in H1 cells from ChIP-Seq and DNase-Seq experiments. Each 5hmC base is counted once: the overlap of a genomic element excludes all previously overlapped cytosines counterclockwise to the arrow. Green: promoter-proximal; red: promoter-distal regulatory elements; grey: genic regions; white: intergenic regions.
(B) The relative enrichment of H1 5hmC (black) and random sites (grey) at genomic elements, normalized to the total coverage of the element type. Random consists of 10 random samplings of 5mC (see Extended Experimental Procedures).
(C) The levels of 5hmCG (left) and 5mCG (right) for several classes of genomic elements significantly enriched with 5hmCG in H1 (p = 0.01, binomial). The dotted line indicates the 5mC non-conversion rate. Colors as in (A).
(D)The percentage of distal regulatory elements significantly enriched with 5hmCG in H1.
(E) In mouse ES cells, the absolute level of 5hmCG for several classes of genomic elements significantly enriched with 5hmCG (p = 0.01, Fisher’s exact test). Colors as in (A).
(F) For genomic elements significantly enriched with 5hmCG in H1 ES cells and conserved in mouse, the distribution of 5hmCG in mouse ES cells. Colors as in (A).
In all panels, definitions of enhancers, p300, CTCF, and DNase I sites are promoter-distal (>5-kb from TSS).
See also Figure S3.
Examining only those genomic elements having significant 5hmC enrichment, we found that the absolute levels of 5hmC at all classes of distal regulatory elements are significantly higher than promoter-proximal elements (Figure 3C). In contrast, gene bodies with significant levels of 5hmC show statistically lower levels of 5hmC. Furthermore, examining the estimated level of 5mC at these loci, we observed an inverse relationship between 5mC and 5hmC (Figure 3C). Distal regulatory elements have the lowest levels of 5mC, with p300 and enhancers having median abundances of 42.2% and 53.7%, respectively. This suggests that highly demethylated elements such as p300 contain more cytosines in a non-5mC/5hmC form, implicating stronger demethylation at these regulatory elements.
In combination with the observations that: 1) between 44% and 74% of distal regulatory elements are significantly enriched with 5hmC in human and mouse ESCs (Figure 3D, S3H); 2) the same class of elements are also enriched in mouse ESCs (Figure 3E, S3G); and 3) the sequence-conserved distal-regulatory elements in H1 are conserved for 5hmC in mouse ESCs (Figure 3F), our data suggests that the marking of functional regulatory elements with 5hmC is an evolutionarily conserved phenomenon with potential functional consequences. Together, these data show that 5hmC is most abundant at promoter-distal regulatory elements, and particularly enriched in distal regulatory elements.
Besides distal regulatory elements, we observe significant enrichment of 5hmC at genes of all tiers, but lowly expressed genes are more enriched than highly expressed genes (Figure S3I), consistent with previous studies (Pastor et al., 2011). In contrast to the abundant 5hmC found at regulatory elements in H1, the vast majority of repetitive elements are highly enriched with 5mC, but not 5hmC (Figure S3J). Between 3.5 and 7.5% of repetitive elements are significantly enriched with 5hmC, with LTRs being the highest (Figure S3K). At these significant loci, the absolute abundance of 5hmC is on par with promoters, but less than distal-regulatory elements (Figure S3L).
Profiles of Hydroxymethylcytosine at Distal Regulatory Elements
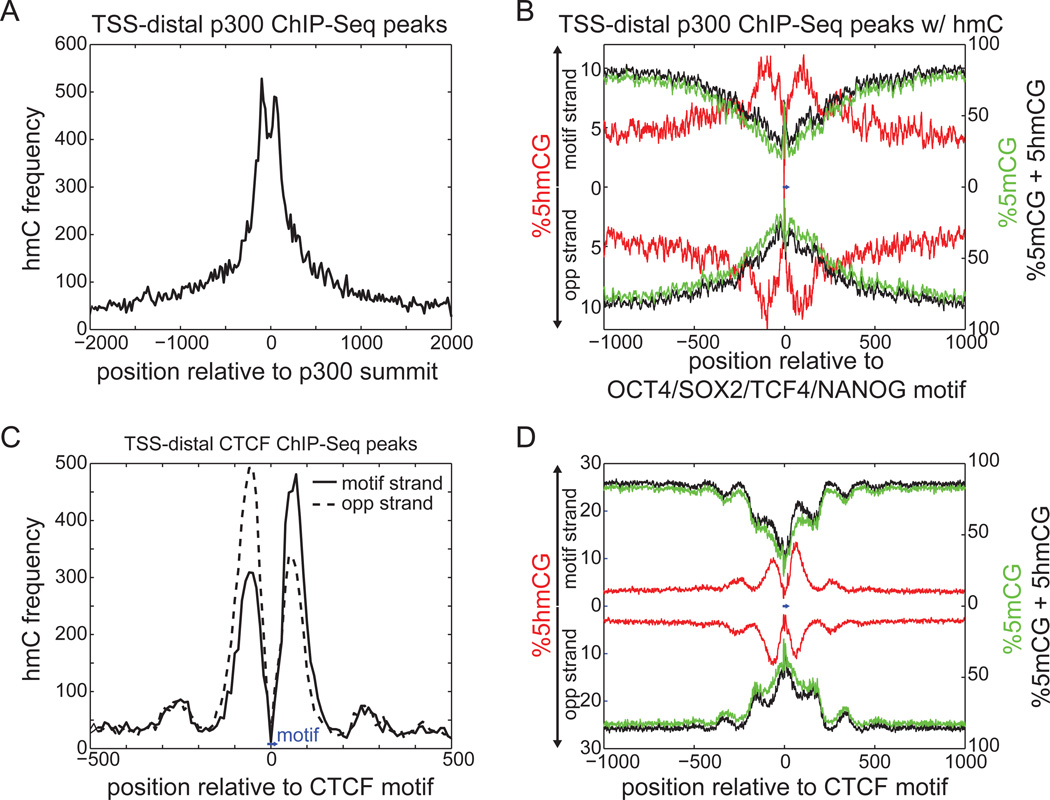
5mC is thought to confer specificity to gene regulation by influencing transcription factor binding or serving as a substrate of recognition for chromatin regulators (Bird, 2011; Chen and Riggs, 2011; Jaenisch and Bird, 2003; Quenneville et al., 2011). Similarly, it has been suggested that 5hmC offers a different platform upon which transcription factors may bind or 5mC specific binding proteins may be excluded (Hashimoto et al., 2012; Kriaucionis and Heintz, 2009; Valinluck et al., 2004; Yildirim et al., 2011). Since 5hmC is enriched near enhancers, one possibility is that this modified base is specifically recognized by transcription factors as a core base in binding motifs. But as sequence motifs are typically shorter than 20bp, the resolution of affinity-based approaches is not sufficient to resolve whether 5hmC is actually present within or outside of the binding site. We observed that while 5hmC is abundant within 500bp of distal p300 binding sites, there is a local depletion near the expected TF binding site (Figure 4A, Figure S4A). To increase resolution, we anchored p300 binding with the OCT4/SOX2/TCF4/NANOG consensus motif (Lister et al., 2009). Total DNA methylation (5mC+5hmC) decreases towards the motif, in agreement with a recent study (Stadler et al., 2011), while 5hmC displays a bimodal peak of enrichment centered at the motif with a maximum average abundance of 12.3% (Figure 4B).
Figure 4. Profiles of 5hmC at Distal Regulatory Elements.
(A)Frequency of 5hmC around distal p300 binding sites.
(B) Absolute levels of 5hmCG (red) and 5mCG+5hmCG (black) around the distal p300 binding sites containing an OCT4/SOX2/TCF4/NANOG motif (blue bar, center; consensus: ATTTGCATAACAATG). 5mC (green) was estimated as the rate from traditional bisulfite sequencing (5hmC + 5mC) minus the measured 5hmC rate. The top half indicates enrichment on the strand containing the motif, with the bottom half indicating the opposite strand.
(C) Frequency of 5hmC around distal CTCF binding sites, relative to the CTCF motif (blue bar, bottom). The different lines represent different strands, oriented with respect to the CTCF motif (consensus: ATAGTGCCACCTGGTGGCCA). Opp, opposite.
(D) Absolute levels of 5hmCG, 5mCG, and 5mCG+5hmCG around distal CTCF binding sites anchored at the CTCF motif (blue bar, center). Colors as in (B).
See also Figure S4.
Similarly for CTCF binding sites, we observed a bimodal enrichment profile of 5hmC abundance ~150bp around the motif, with almost no 5hmC within the motif itself (Figure 4C). 5hmC increases to a maximum abundance of 13.4%, coinciding with a dramatic depletion of 5mC from an average high of 86.2% to a low of 21.0% (Figure 3D). We also observed similar results for NANOG binding sites (Figure S4B–C). Together, these data suggest that 5hmC is typically not observed within potential binding sites of transcription factors, but rather is most enriched in regions immediately adjacent to sequence motifs. The reciprocal profiles of 5hmC and 5mC is consistent with a model of dynamic DNA methylation associated with DNA-binding transcription factors, and provides additional evidence supporting a role for 5hmC in the locally reduced levels of 5mC at distal regulatory elements (Stadler et al., 2011).
Asymmetric Hydroxymethylation at CG Sequences
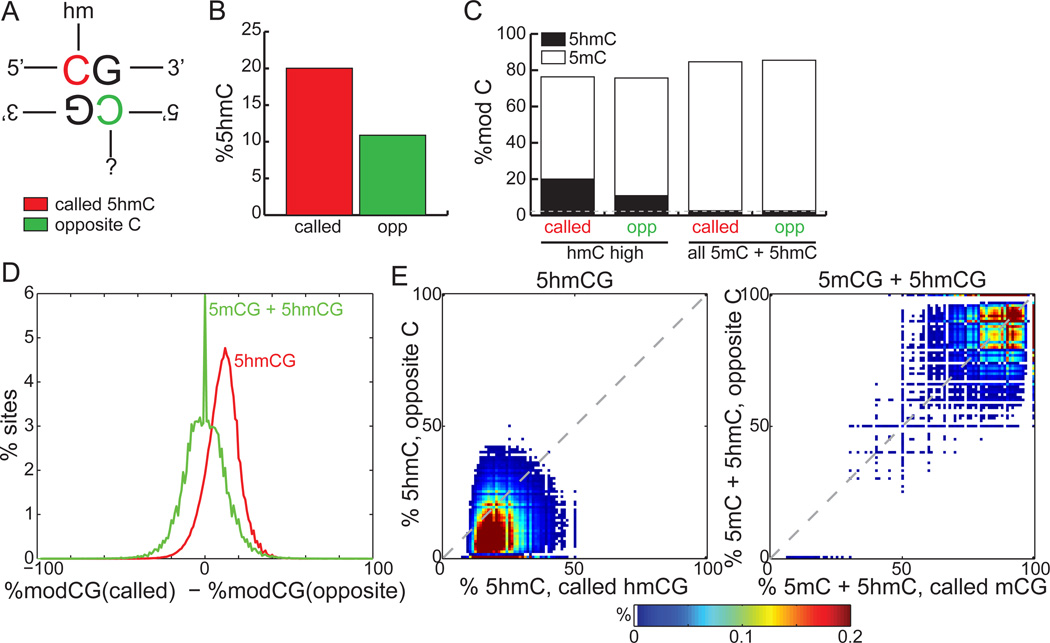
Cytosine methylation in CG context is symmetric, and the maintenance methyltransferase DNMT1 ensures efficient propagation of symmetric 5mCG during cell division, thus providing one of the central modes of epigenetic inheritance (Bird, 2011; Chen and Riggs, 2011; Goll and Bestor, 2005; Jaenisch and Bird, 2003; Wigler et al., 1981). Our observation that the bimodal distribution of 5hmC around CTCF is strand-asymmetric (Figure 4C–D) prompted us to examine if 5hmC is strand-biased in H1. While 91.8% of 5mCs are symmetrically modified, we found that only 21.0% of 5hmCs are symmetric. However, since the abundance of 5hmC is rare at any given cytosine (median 19.2%, Figure 2E), it is possible that sequencing depth was not sufficient to identify all 5hmCs, making this an under-estimate. To address this issue, we compared the pool of all called 5hmCs with the pooled 5hmC content on the opposite cytosine (Figure 5A). The average abundance of 5hmC is 20.0% at called 5hmCs, compared to 10.9% at the opposite cytosine, which corresponds to an 83.8% enrichment of 5hmC (Figure 5B, p < 1E-15, binomial). As a control, the base-line 5hmC content of all methylated cytosines in CG context is symmetric and comparable to the methylcytosine non-conversion rate (Figure 5C). At promoters and within gene bodies, we found that strand bias is not dependent on the orientation of the transcript (Figure S5A) (ppromoter = 0.0339, pgene body = 0.0719).
Figure 5. Asymmetry around 5hmCG.
(A) A schematic of nomenclature. The cytosine with 5hmC (red) designated as “called”, while the cytosine on the opposite strand (green) is designated as “opposite”.
(B) The average 5hmC abundance of called 5hmCG residues (red) compared to the opposite cytosine residues (green). called: called cytosine; opp: opposite cytosine.
(C) The average 5hmC (black) and 5mC (white) abundance at called and opposite cytosines, for called cytosines having 5hmC (left) or 5mC+5hmC (right). 5mC (white excluding black) was estimated as the rate from traditional bisulfite sequencing (5hmC + 5mC) minus the measured 5hmC rate. Grey line: 5mC non-conversion rate.
(D)The distribution of differences in 5hmCG (red) between called and opposite cytosines, in comparison to differences observed from traditional bisulfite sequencing (green, 5mCG + 5hmCG). Called and opposite cytosines are each sequenced to at least depth 10.
(E) For 5hmC-called sites, a heatmap of 5hmCG abundance at called and opposite cytosine pairs (left). For the 5mC-called sites from traditional bisulfite sequencing, a heatmap of 5mCG + 5hmCG abundance at called and opposite cytosine pairs (right).
See also Figure S5.
To confirm the asymmetry of 5hmCG, we examined the difference in methylation state of called 5hmCs and the cytosines located at the opposite strands. From traditional bisulfite sequencing, the median difference in total methylation (5mCG+5hmCG) between called and opposite cytosines is 0%. In contrast, TAB-Seq reveals a shifted distribution with a median of 10.9% less hydroxymethylation on the opposite cytosine (Figure 5D, p <1E-15, Wilcoxon). Simultaneous examination of the absolute levels of 5hmC on both called and opposite cytosines showed that the shift in hydroxymethylation state towards the called cytosine is evident, in contrast to DNA methylation levels that remain symmetric (Figure 5E). Our analysis of the spike-in lambda DNA showed no strand nor sequence bias of the TAB-Seq method (Figure S5B, Figure S2D). This conclusion was further supported by analyzing the βGT-catalyzed glucosylation efficiency of a fully-hydroxymethylated model dsDNA, which is over 90% (Figure S5C).
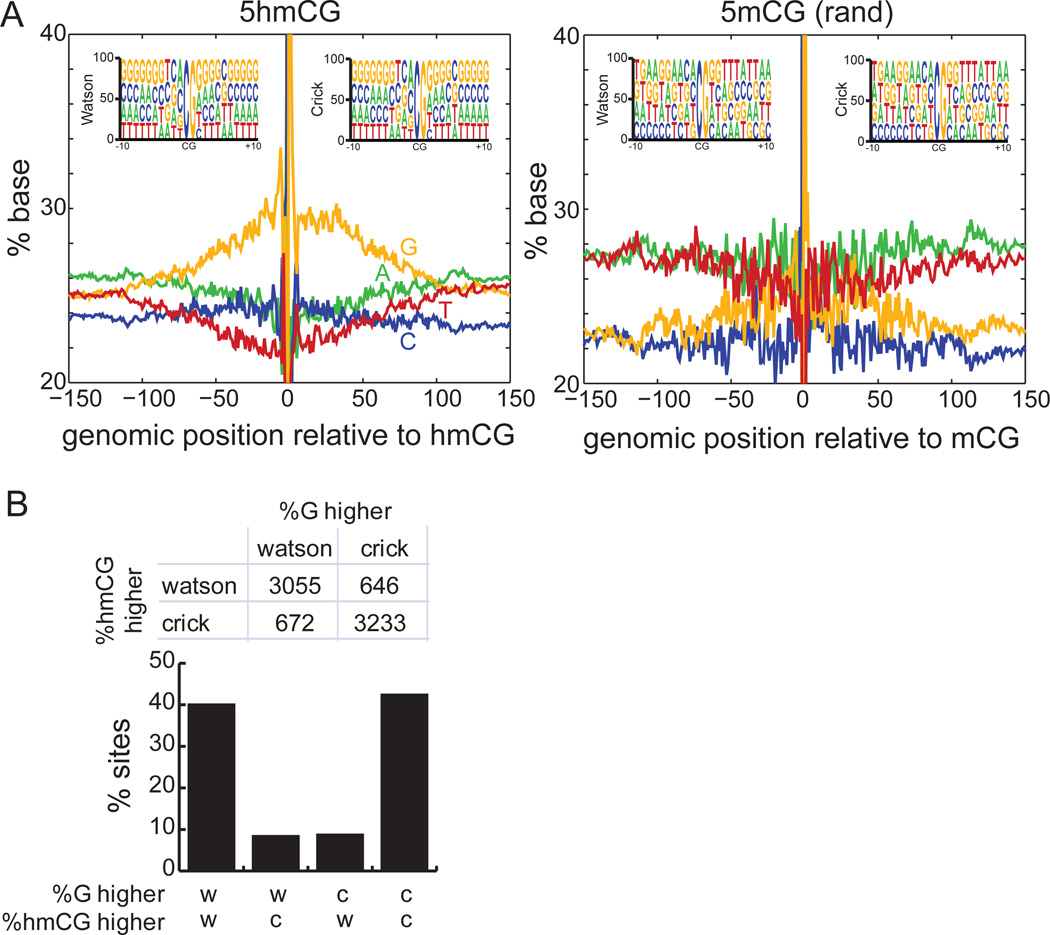
5hmC is Strand-Biased towards G-rich Sequences
The asymmetry of 5hmC in H1 suggests that, on a population average, one strand is more likely to be hydroxymethylated than the other strand. One possible explanation for this phenomenon is a sequence preference of 5hmC for one strand compared to the other. To examine this systematically, we aligned all 5hmCs in CG context and examined base composition (Figure 6A). On the strand containing 5hmC, we observed a modest increase in local guanine abundance with depletion of adenine and thymine content. Within a window of 100bp around 5hmCs, the local sequence content of guanine increases to an average of 29.9%, significantly higher than the 25.6% observed for randomly sampled methylated cytosines (Figure S6A, p < 1E-15, Wilcoxon). These observations are not a function of regulatory element class, as similar trends hold for subsets of 5hmC found at promoters, distal regulatory elements, and genic regions (Figure S6C). Furthermore, similar trends are observed in mouse ESCs (Figure S6D), and analysis of the spike-in lambda DNA shows that this observation is not a systematic bias of the TAB-Seq method (Figure S6B).
Figure 6. Local Sequence Context around 5hmCG.
(A)Sequence context ±150bp around 5hmCG sites (left), compared to the same number of randomly chosen mCG sites (right). Shown sequences are on the same strand as 5hmC. Inset: sequence context ±10bp around 5hmCG sites that are on the Watson or Crick strands. Positive coordinates indicate the 3’ direction.
(B) For cytosines showing significant difference in 5hmCG between Watson and Crick strands (p = 0.01, Fisher’s exact test), and for which the abundance of guanine ±50bp around the site showing significant strand bias (p = 0.01, Fisher’s exact test), shown is the frequency at which these two events co-occur.
See also Figure S6.
Our observations suggest that 5hmC deposition is biased towards the strand with a higher local density of guanine. To test this hypothesis, we developed a predictive algorithm: given that a strand-biased hydroxymethylation event exists at a particular CG (p-value = 0.01, Fisher’s exact test) and that one strand has local guanine content significantly different from the other strand (p-value = 0.01, Fisher’s exact test), we predict the strand with higher guanine content to have the hydroxymethylation event. This model correctly predicts the hydroxymethylated strand with 82.7% accuracy, significantly better than the 50% expected by chance (Figure 6B, p < 1E-15, binomial), confirming that local sequence content plays a role in strand-specific hydroxymethylation. However, while both human and mouse ESCs exhibit a bias of 5hmCG to occur on the strand with more guanine content (Figure S6C–D), the effect is weaker in mouse ESCs (Figure S6E–F), which is one potential reason that guanine content does not predict 5hmC in mouse ES cells. One possible explanation is the large difference in the expression levels of TET1 and TET2 in human and mouse ESCs.
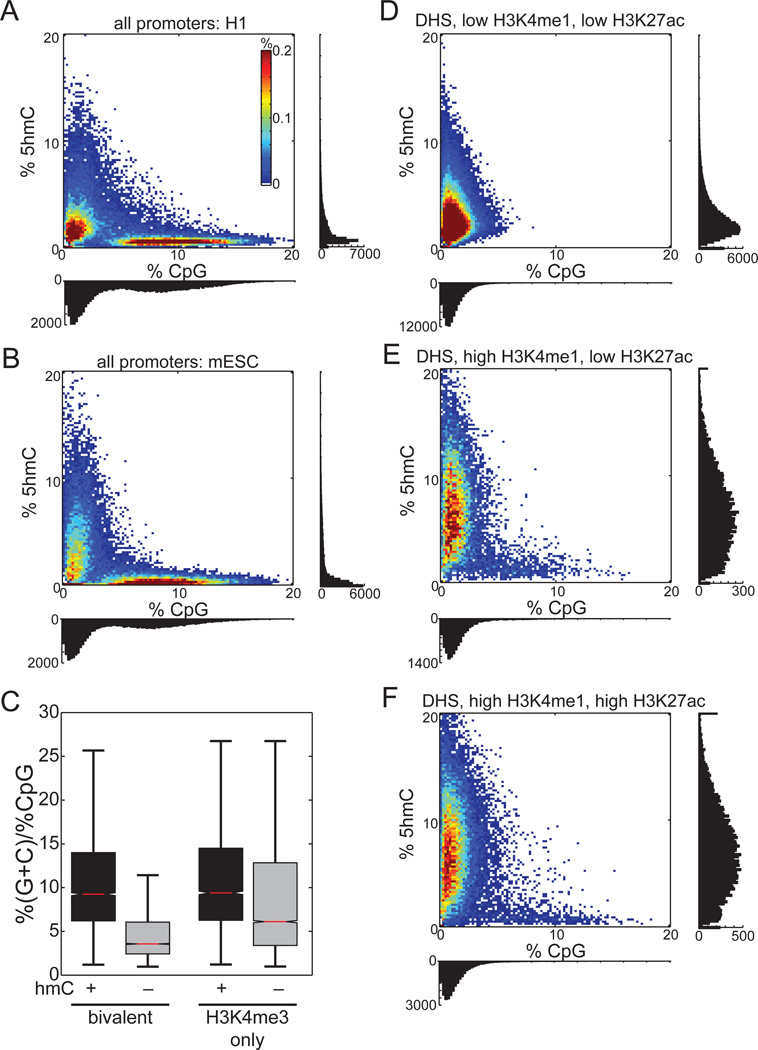
5hmC is Most Enriched near Low CpG Regions
Recent affinity-based studies in mouse ESCs have observed 5hmC to be frequently enriched at CpG island-containing promoters (Ficz et al., 2011; Pastor et al., 2011; Williams et al., 2011), and that the highest levels of 5hmC correspond to the highest density of CpGs (Ficz et al., 2011). In contrast, an affinity-based map of 5hmC produced in H1 found 5hmC-rich regions to be depleted of CpG dinucleotides (Szulwach et al., 2011a). These confounding results prompted us to examine the relationship between absolute steady-state 5hmC level and CpG content at promoters. We found that promoters with the highest levels of 5hmCG are almost exclusively of low CpG content (Figure 7A), which are also the promoters most likely to have the highest 5mCG (Figure S7A). In agreement with this observation, when we divide promoters by CpG content, we observe that the density of 5hmC is lowest at HCPs, while at LCPs and ICPs 5hmC is at least 3.3 times more abundant (Figure S7H). Analyses of mouse ESCs give similar results (Figure 7B). In both human and mouse ESCs, CpG-rich promoters are almost devoid of steady-state 5hmC. Moreover, these results apply to promoters containing H3K4me3 or bivalent chromatin modifications (Figure S7G).
Figure 7. 5hmCG is Biased towards Low CpG Regions.
Shown are heatmaps of percent 5hmCG (±250bp from TSS or DHS) as a function of CpG density for
A) Promoters in H1 ES cells, B) promoters in mouse ES cells, D) DHS sites lacking H3K4me1 and H3K27ac, E) DHS sites with a poised enhancer chromatin signature, and F) DHS sites with an active enhancer chromatin signature.
(C) The GC content relative to the CpG content for the 5hmC-enriched versus the 5hmC not enriched promoters.
See also Figure S7.
Together with our observation of an increased local density of guanine on the strand of hydroxymethylation, we postulated that promoters with high GC content but low CpG density are more likely to be hydroxymethylated. Indeed, such bivalent (p < 1E-300) and H3K4me3-only promoters (p = 7.8E-286) are more enriched with 5hmC (Figure 7C).
To determine if hydroxymethylation at distal regulatory elements is also biased towards low CpG density, we examined three classes of DNase I hypersensitive sites (DHSs): 1) those lacking the enhancer histone modifications H3K4me1 and H3K27ac; 2) putative poised enhancers bearing only H3K4me1; and 3) putative active enhancers with both modifications (Hawkins et al., 2010; Myers et al., 2011). Poised and active enhancers exhibit the strongest enrichment of 5hmC (Figure 7D–F), which almost exclusively corresponds to low CpG density regions. Like promoters, the few distal DHSs with high CpG density are generally composed of low 5hmCG content. We also observed similar results at distal p300 binding sites (Figure S7B–C). Together, these results suggest that the highest levels of 5hmC occur at regions of the genome with low CpG density.
Comparing DNase I hypersensitive sites lacking the H3K4me1 and H3K27ac enhancer marks to poised enhancers having only H3K4me1, 5mCG drops by 12.6% and 5hmC increases by 2.7-fold (Figure S7D–F). In contrast, active enhancers having both H3K4me1 and H3K27ac have 8.3% less 5mCG than poised enhancers, but with only a 1.08-fold increase in 5hmC. These results suggest that while 5mCG is inversely related to both H3K4me1 and H3K27ac, 5hmC is primarily proportional to H3K4me1.
DISCUSSION
Bisulfite sequencing has been broadly used to analyze the genomic distribution and abundance of 5mC (Bernstein et al., 2007; Clark et al., 1994; Lister et al., 2008; Meissner, 2010; Pelizzola and Ecker, 2011). However, because traditional bisulfite sequencing cannot distinguish 5mC from 5hmC, results from such approaches cannot yet accurately reveal 5mC abundance (Huang et al., 2010; Jin et al., 2010). Recent experiments show that 5hmC is widespread in the mammalian genome, and at least two functions have been proposed for this cytosine modification: 1) 5hmC serves as an intermediate in the process of DNA demethylation, either passively (Inoue and Zhang, 2011), or actively through further oxidation (He et al., 2011; Ito et al., 2011; Maiti and Drohat, 2011; Zhang et al., 2012); 2) 5hmC may be recognized by chromatin factors (Frauer et al., 2011; Yildirim et al., 2011), and that its presence could reduce binding of certain methyl-CpG-binding proteins (Hashimoto et al., 2012; Kriaucionis and Heintz, 2009; Valinluck et al., 2004). These functions implicate two opposing notions about the relative stability of 5hmC at distinct genomic loci. As the first step toward understanding these molecular mechanisms associated with 5hmC function, it is important to not only precisely locate 5hmC in the genome, but also to determine the relative abundance at each modified site. Here we describe a modified bisulfite sequencing method that when combined with traditional bisulfite sequencing can determine the location of 5hmC at single-base resolution and quantitatively assess the abundance of 5mC and 5hmC at each modified cytosine.
Using synthetic model DNA we demonstrated that coupling βGT-mediated protection of 5hmC with mTet1-based oxidation of 5mC allows for the distinction of 5hmC from unmodified cytosine and 5mC by sequencing. 5fC and 5caC presented in the original genomic DNA do not interfere with TAB-Seq since they behave like unmodified cytosine under bisulfite treatment (He et al., 2011). We also utilized this method to examine previously reported 5hmC enriched loci and successfully identified genuine 5hmC sites. These results show the general utility of TAB-Seq to assess 5hmC in a loci-specific manner, much the same as traditional bisulfite sequencing is currently used.
We applied this technique to mammalian genomes by generating single-base resolution maps of 5hmC in human and mouse ESCs. We show that these maps agree well with previous maps generated using affinity-based 5hmC profiling. Importantly, these single-base maps also revealed a significant number of new 5hmC sites. Analyses of two 5hmC maps in ESCs identified several novel sequence-based characteristics of 5hmC that were previously unknown. We observed that, much like 5mC, 5hmC tends to occur primarily at CpG-dinucleotides yet, unlike 5mC, exhibits an asymmetric strand bias. We also observed a relatively strong local sequence preference surrounding 5hmC, with 5hmC occurring within a G-rich context. This observation is consistent with previous report that 5hmC regions are GC-skewed (Stroud et al., 2011). These sequence-based features associated with 5hmC may provide a basis for future mechanistic insight into the means by which 5hmC is deposited, recognized, and dynamically regulated.
The ability to quantify 5hmC abundance with base resolution offered the unique opportunity to assess its relative abundance at various regulatory elements and genomic annotations without bias. In contrast to the nearly uniform distribution of 5mC outside of promoter regions, we found that the abundance of 5hmC varies among different classes of functional sequences. It is most enriched at distal regulatory regions where levels of 5mC are correspondingly lower than the genome average. This observation agrees with recent findings from others (Stadler et al., 2011), and suggests that active demethylation occurs at active regulatory elements through 5hmC. This active demethylation is distributed around, but not within, transcription factor consensus motifs. Supporting the notion of active demethylation, total DNA methylation exhibits a strong negative correlation with 5hmC at distal regulatory elements (Spearman correlation = −0.30). One interesting observation of these distal cis-regulatory elements is that 5hmC and 5mC often occur together at the same cytosine. Currently, the exact mechanisms that determine the dynamics of 5hmC and 5mC at these cis-regulatory sequences are unclear.
Previous affinity-based studies have suggested enrichment of 5hmC at CpG-rich transcription start sites. However, these observations relied heavily on antibody-base detection, which has been shown to exhibit bias toward 5hmC dense regions. Here we find that, in general, 5hmC is most abundant at regions of low CpG content. Furthermore, even promoters with relatively high 5hmC content tend to have low CpG content in both mouse and human ESCs. These findings highlight the utility of a base-resolution method for measuring 5hmC abundance, and provide new insight into its dynamic regulation at promoter sites with distinct CpG content.
Tahiliani and colleagues (Tahiliani et al., 2009) recently estimated the genome-wide abundance of 5hmC to be about 14 times less than that of 5mC, which would correspond to ~4.4 million 5hmCs in human. However, as our results indicate that the base-level abundance of 5hmC is several times lower than 5mC, this is likely an under-estimate. The comparatively low number of 5hmCs confidently detected in our study (691,414) is likely explained by the frequent hydroxymethylation of gene bodies previously observed in affinity-based studies (Ficz et al., 2011; Pastor et al., 2011; Stroud et al., 2011; Szulwach et al., 2011a; Williams et al., 2011; Wu et al., 2011; Xu et al., 2011). Since genic cytosines likely exist at a relatively low abundance of 5hmC (between 3–4%), they would have escaped detection at our current sequencing depth. In order to resolve low abundance 5hmCs at single-base precision, significantly more sequencing would be required. This observation highlights the biases inherent in affinity-based 5hmC mapping, which can amplify frequent weak signals found in gene bodies to overshadow rare but stronger ones at distal regulatory elements.
In summary, we have developed a genome-wide approach to determine 5hmC distribution at base resolution, and generated the first base-resolution maps of 5hmC in both human and mouse ESCs. These maps provide a template for further understanding the biological roles of 5hmC in stem cells as well as gene regulation in general. In conjunction with methylC-Seq, the TAB-Seq method described here represents a general approach to measure the absolute abundance of 5mC and 5hmC at specific sites or genome-wide, which could be widely applied to various cell types and tissues.
EXPERIMENTAL PROCEDURES
Glucosylation and Oxidation of Genomic DNA
Glucosylation reaction was performed in a 50 µl solution with 50 mM HEPES buffer (pH 8.0), 25 mM MgCl2, 100 ng/µl sonicated genomic DNA with spike-in control, 200 µM UDP-Glc, and 1 µM wild-type βGT. The reaction was incubated at 37 °C for 1 h. After the reaction, the DNA was purified by QIAquick Nucleotide Removal Kit (Qiagen). The oxidation reaction was performed in a 50 µl solution with 50 mM HEPES buffer (pH 8.0), 100 µM ammonium iron (II) sulfate, 1 mM α-ketoglutarate, 2 mM ascorbic acid, 2.5 mM DTT, 100 mM NaCl, 1.2 mM ATP, 10 ng/µl glucosylated DNA and 3 µM recombinant mTet1. The reaction was incubated at 37 °C for 1.5 h. After proteinase K treatment, the DNA was purified with Micro Bio-Spin 30 Columns (Bio-Rad) and then by QIAquick PCR Purification Kit (Qiagen).
Quantifying %5hmCG and %5mCG
For a given genomic interval, the abundance of hydroxymethylation (%hmCG) is estimated as the number of cytosine base calls in the interval divided by the number of cytosine plus thymine base calls in the interval from TAB-Seq reads, where the reference is in CG context. To estimate %5mC level, we subtracted the total methylation level from methylC-Seq by the %5hmC level from TAB-Seq. In all instances, only base calls with Phred score ≥ 20 were considered.
Supplementary Material
HIGHLIGHTS.
Genome-wide Tet-assisted bisulfite sequencing for single-base detection of 5hmC
Single-base resolution maps of 5hmC vs 5mC in human and mouse ESCs
5hmC is enriched at distal regulatory elements and promoters with low-CpG content
5hmC deposition is asymmetric and strand-biased towards G-rich sequences
Acknowledgements
This study was supported by National Institutes of Health (GM071440 to C.H., NS051630 and P50AG025688 to P.J., U01 ES017166 to B.R.), a Catalyst Award (C.H. and J.-H.M.) from the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust, the Emory Genetics Discovery Fund (P.J.), the Simons Foundation Autism Research Initiative (P.J.), the Autism Speaks grant (#7660 to X.L.), and the Ludwig Institute for Cancer Research (B.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Sequencing data have been deposited to GEO (accession GSE36173).
REFERENCES
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bird A. The dinucleotide CG as a genomic signalling module. J Mol Biol. 2011;409:47–53. doi: 10.1016/j.jmb.2011.01.056. [DOI] [PubMed] [Google Scholar]
- Chen ZX, Riggs AD. DNA methylation and demethylation in mammals. J Biol Chem. 2011;286:18347–18353. doi: 10.1074/jbc.R110.205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, Leonhardt H. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, Wold B, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelizzola M, Ecker JR. The DNA methylome. FEBS Lett. 2011;585:1994–2000. doi: 10.1016/j.febslet.2010.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AB, Dahl JA, Ougland R, Klungland A. Pull-down of 5-hydroxymethylcytosine DNA using JBP1-coated magnetic beads. Nat Protoc. 2012;7:340–350. doi: 10.1038/nprot.2011.443. [DOI] [PubMed] [Google Scholar]
- Robertson AB, Dahl JA, Vagbo CB, Tripathi P, Krokan HE, Klungland A. A novel method for the efficient and selective identification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2011;39:e55. doi: 10.1093/nar/gkr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011a;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011b;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell. 1981;24:33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nature Chem Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.