Abstract
Many reports have indicated that the abnormal expression of microRNAs (miRNAs) is associated with the progression of disease and have identified miRNAs as attractive targets for therapeutic intervention. However, the bifunctional mechanisms of miRNA guide and passenger strands in RNA interference (RNAi) therapy have not yet been clarified. Here, we show that miRNA (miR)-582-5p and -3p, which are strongly decreased in high-grade bladder cancer clinical samples, regulate tumor progression in vitro and in vivo. Significantly, the overexpression of miR-582-5p or -3p reduced the proliferation and invasion of UM-UC-3 human bladder cancer cells. Furthermore, transurethral injections of synthetic miR-582 molecule suppressed tumor growth and metastasis in an animal model of bladder cancer. Most interestingly, our study revealed that both strands of miR-582-5p and -3p suppressed the expression of the same set of target genes such as protein geranylgeranyltransferase type I beta subunit (PGGT1B), leucine-rich repeat kinase 2 (LRRK2) and DIX domain containing 1 (DIXDC1). Knockdown of these genes using small interfering RNA (siRNA) resulted in the inhibition of cell growth and invasiveness of UM-UC-3. These findings uncover the unique regulatory pathway involving tumor suppression by both strands of a single miRNA that is a potential therapeutic target in the treatment of invasive bladder cancer.
Introduction
RNA interference (RNAi), a natural cellular process that regulates gene expression, is the most significant recent contribution to the field of cell biology.1 Recently, RNAi-based therapies, which are harnessed to control the expression of pathogenic proteins, have been demonstrated in humans and have provided alternative powerful approaches to the traditional small molecule therapies. There has been an increase in the development of RNAi therapies for accessible tissues, such as the skin, retina, liver, and airways, due to their ability to be efficiently and safely delivered without unwanted side effects.2,3,4
Urinary bladder cancer remains one of the most costly cancers with regard to treatment and the monitoring of cytological changes, such as surveillance cystoscopy and periodic imaging.5 However, despite the existence of appropriate therapies, patients are continually under the threat of ongoing recurrence and progression to muscle.5,6 Therefore, the development of new treatment strategies to reduce the risk of recurrence and progression based on novel molecular networks is strongly desired. The facts described above provide an insight into an innovative approach that harnesses the power of the RNAi pathway; i.e., the bladder maximizes the effect of RNAi therapy because of its accessibility and closed environment. Although some previous studies showed that the intravesical injection of small interfering RNA (siRNA) has potential as a treatment,7,8 whether an intravesical strategy can overcome the progression to muscle and metastasis in invasive bladder cancer remains unclear.
In cancer research, the copy number variation of DNA was a focus during the 1990s and the beginning of the 2000s as a result of the development of technologies such as array-based comparative genomic hybridization and microsatellite analysis,9,10,11 because copy number gains or losses were believed to be specific markers for functional protein-coding genes. However, if protein-coding genes were not located in these aberrant regions, disease candidate regions were excluded from the functional analysis. On the other hand, recent studies found that microRNAs (miRNAs), which are key post-transcriptional regulators, are the main candidates for cancer-predisposing genes and that approximately half of the miRNAs are located at chromosomal regions that are genetically altered in cancers.12,13,14 Indeed, since their discovery, a widespread dysregulation of miRNAs caused by copy number variation is commonly observed in human cancers and has been shown to be involved in diverse physiological/pathological processes.15,16,17,18,19 In this study, we hypothesized that a novel tumor-suppressive miRNA or onco-miR can be identified more efficiently and more expeditiously by utilizing a vast amount of previously generated information about aberrant chromosomal regions related to bladder cancer.
Cytogenetic studies of bladder cancer have revealed a number of genetic aberrations.10 For discrimination of miRNAs which denote the abnormal expression in poorly prognostic patients on bladder cancer, we focused on the copy number variation in UM-UC-3 cells which is one of the most invasive bladder cancer cell lines. It was reported that a number of genomic regions were deficient or amplified in that cell line.20 Furthermore, in those genomic regions, 3p11-12, 4q33-34, and 5q12-13 were selected as analysis sets because the correlations between the copy number loss and bladder cancer progression in clinical samples had been reported by at least two groups.11,21,22,23,24 Although these regions include some miRNA genes, such as miR-1305, miR-1324, miR-578, miR-582-5p, and miR-582-3p, the modulation of the miRNA expression profiles and the exact functional mechanism are not thoroughly understood. In the present study, we report that miR-582-5p and miR-582-3p are concurrently downregulated in invasive bladder cancer, which is correlated with a copy number loss of chromosome 5q12. These significant reductions of miR-582-5p and -3p were also observed in clinical samples and strongly correlated with tumor grade. Furthermore, the restorations of miR-582-5p or -3p strongly inhibit cell proliferation and invasion. In the lung metastasis mouse model of human bladder cancer, involving orthotopic transplantation, the injection of the miR-582/cationic liposome complex prevented tumor growth and lung metastasis. This demonstration is the first, to our knowledge, to show inhibition of tumor metastasis in an orthotopic model by intravesical miRNA injection.
Two mature miRNAs can be generated from the same stem-loop pre-miRNA.25 These 5p and 3p miRNAs, although excised from a single primary transcript, have different sequences and therefore target different mRNAs. Despite nearly a decade of studies on miRNA, the effect of strand-specific mature miRNAs has not yet been fully understood. In the present study, we also provide evidence that the expression of protein geranylgeranyltransferase type I beta subunit (PGGT1B), leucine-rich repeat kinase 2 (LRRK2), and DIX domain containing 1 (DIXDC1), which are involved in the cell growth and the invasiveness, is bifunctionally regulated by the effects of both strands of miR-582-5p and -3p. These findings suggest that a novel pathway involving miR-582 and cancer-related genes has the potential to be a critical target for the therapeutic treatment of invasive bladder cancer.
Results
Downregulation of miR-582-5p and -3p in bladder cancer cell lines
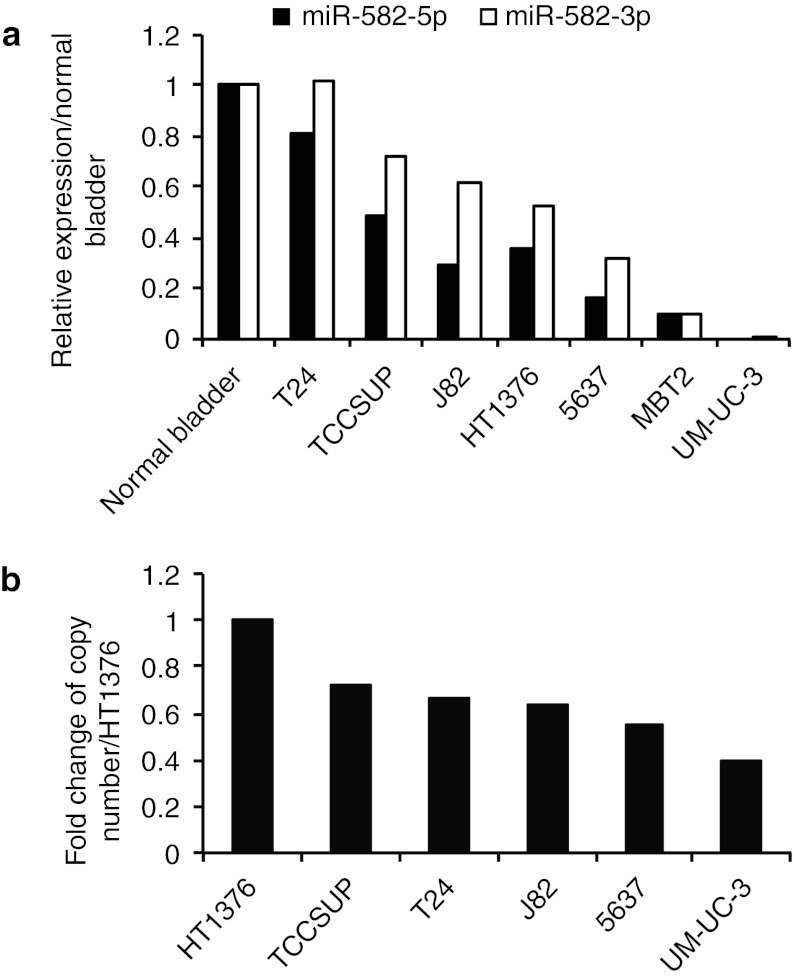
To identify miRNAs downregulated in bladder cancer cell lines, we performed quantitative reverse transcription-PCR (qRT-PCR) for miR-1305, miR-1324, miR-578, and miR-582-5p and -3p, which are located in aberrant genomic regions correlated with tumor progression (Supplementary Tables S1 and S2). The results showed no downregulation of miR-1305 in bladder cancer cells (Supplementary Figure S1a), and the expression of miR-1324 and miR-578 was not detected in either bladder cancer cell lines or normal bladder tissue RNA, but we found that miR-582-5p and -3p are strongly downregulated in invasive bladder cancer cell lines (UM-UC-3, 5637, MBT2, J82, TCCSUP) (Figure 1a). Furthermore, the copy number of chromosome 5q12, in which the miR-582 gene is located, was lower in UM-UC-3 cells than in HT1376 cells (Figure 1b). HT1376 cells were reported to not have a loss in 5q12.24 However, the degree of downregulation of miR-582 expression in UM-UC-3 cells seems too strong compared with the reduction of the copy number. This means that a remarkable reduction of miR-582-5p and -3p expression might be invoked by a combination of copy number loss and other factors. To identify whether the expression of these mature miRNAs are reduced in a transcriptional or biogenic process, we assessed the expression levels of pri-miR-582 and phosphodiesterase 4D (PDE4D) because miR-582 is located in the intronic region of PDE4D. The expression levels of pri-miR-582 and PDE4D are strongly reduced in UM-UC-3 and 5637 cells (Supplementary Figure S1b,c). These results indicate that the expression of miR-582-5p and -3p is reduced by the combination of copy number loss and transcriptional attenuation.
Figure 1.
Expression level of miR-582-5p and -3p in human bladder cancer cell lines. (a) Expression level of miR-582-5p and -3p in human bladder cancer cell lines and a mouse bladder cancer cell line (MBT2). The relative expression of miR-582 for each of the cell lines was calculated compared with the level in normal bladder tissue RNA. (b) Copy number change of the miR-582 loci on chromosome 5q12. The HT1376 cell line was used as the control for this experiment.
Downregulation of miR-582-5p and -3p in clinical samples
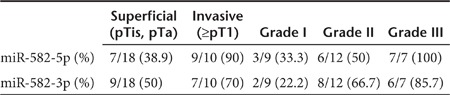
We examined the expression levels of miR-582-5p and -3p in laser capture-microdissected bladder cancer tissue regions (n = 53) and matched adjacent normal regions (n = 31) derived from 28 patients (Supplementary Table S3). Relative to normal regions, the tumor regions showed markedly lower miR-582-5p and -3p expression levels (Figure 2a). In particular, we found that these levels tend to decrease more remarkably in invasive tumors (tumor stage ≥pT1) than in superficial tumors (tumor stage pTis and pTa) (Figure 2b) (miR-582-5p: superficial tumor P = 0.006, invasive tumor P = 0.003; miR-582-3p: superficial tumor P = 0.035, invasive tumor P < 0.001). Furthermore, a strong correlation was also observed between tumor grade and the downregulation of miR-582-5p and -3p (Figure 2c) (miR-582-5p: Grade I P = 0.042, Grade II P = 0.028, Grade III P = 0.006; miR-582-3p: Grade I P = 0.206, Grade II P = 0.270, Grade III P = 0.046). In addition to this, the ratio of samples showing strong downregulation greater than a 2.5-fold change increased as the tumor stage and grade advanced (Table 1). These data suggested that miR-582-5p and -3p levels vary with the malignancy potential and are potential therapeutic markers in the treatment of invasive bladder cancer.
Figure 2.
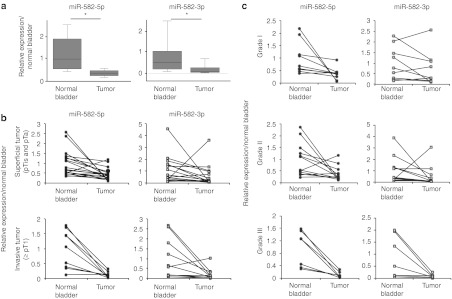
Expression level of miR-582-5p and -3p in bladder cancer tissue samples. (a) Expression of miR-582-5p and -3p in clinical samples, which show the downregulation in the tumor region compared with the adjacent normal tissues (tumor: n = 53, normal bladder: n = 31). The boxes and whiskers range from 25% to 75% and from 10% to 90%, respectively. *P < 0.01 versus normal bladder. (b) Comparison with the tumor stages in carcinoma cells paired with normal bladder tissue (superficial tumor: n = 18, invasive tumor: n = 10). (c) Comparison with the tumor grades in carcinoma cells paired with normal bladder tissue. All values for the miRNA expression levels were normalized to hsa-miR-103 as an internal control (Grade I: n = 9, Grade II: n = 12, Grade III: n = 7).
Table 1. Downregulation in clinical samples (fold change >2.5).
Effect of miR-582-5p and -3p on cell proliferation and invasive ability
To investigate the functional role of miR-582-5p and -3p in bladder cancer, we performed a cell proliferation assay and a cell invasion assay. UM-UC-3, J82, and TCCSUP cells were transfected transiently with miR-582, miR-582-5p, miR-582-3p, and negative control (NC) siRNA. miR-582 is synthesized to mimic endogenous mature miR-582, whereas miR-582-5p and -3p are designed to match perfectly and complementarily to each strand, similar to siRNA. Three days after transfection, the cell viability was decreased from 30% to 80% in the miR-582-, miR-582-5p-, or miR-582-3p-transfected cells compared with the NC cells (Figure 3a). The matrigel invasion assay showed that the cell invasion ability was significantly (>70%) decreased in miR-582-, miR-582-5p-, or miR-582-3p-transfected UM-UC-3, J82, and TCCSUP cells (Figure 3b,c). These results suggest that the cell proliferation and invasive abilities of bladder cancer cells were severely affected only by either strand of miR-582-5p or miR-582-3p. In addition, a reduction of invasiveness was observed in 5637 cells, although T24 cells did not show a significant response to miR-582 in the cell proliferation and cell invasion assays (Supplementary Figure S2a,b). The expression and functional data suggest that the tumor-suppressive role of miR-582 is correlated with the reduction of its expression level in bladder cancer cells.
Figure 3.
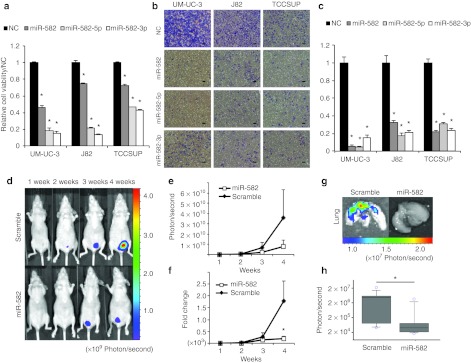
Function of miR-582 in human bladder cancer in vitro and in vivo. (a) Effect of miR-582 on cell proliferation of UM-UC-3, J82 and TCCSUP. The proliferation values were normalized to the values from cells treated with NC (n = 6). *P < 0.01 versus NC. (b,c) Effect on cell invasion. The invasive values were normalized to the values from cells treated with NC (n = 4). *P < 0.01 versus NC (scale bar: 100 µm). (d) Inhibition of tumor growth by miR-582 treatment in a murine orthotopic xenograft model of human bladder cancer. The mice were injected with 5 × 106 UM-UC-3-luc cells into the bladder on day 0. miR-582 and miR-582-scramble (10 µg) with cationic liposomes in a 70 µl volume were injected into the bladder on days 4, 6, 8, 11, 13, and 15 after tumor injection. (e,f) The growth curves of the orthotopically transplanted UM-UC-3-luc cells were measured by IVIS. Filled diamonds: treatment with miR-582-scramble; Open squares: treatment with miR-582 (scramble: n = 8, miR-582: n = 10). (g) Inhibition of lung metastasis by miR-582 treatment in an orthotopic bladder cancer mouse model. The images were obtained by IVIS on day 26 after transplantation. (h) Quantitation of bioluminescence emitted from the whole lungs of mice on day 26. The boxes and whiskers range from 25% to 75% and from 10% to 90%, respectively. *P < 0.05 versus scramble. NC, negative control.
To ensure that the functions of miR-582-5p and -3p are not at a supraphysiological level, we repeated these experiments using a stable miRNA vector that synthesizes mature miRNAs by biological processing. UM-UC-3 cells were transduced with a lentiviral construct stably expressing precursor miR-582, and the expression level was examined (Supplementary Figure S3a). Furthermore, the functional analysis revealed that UM-UC-3 cells stably expressing miR-582 (UM-UC-3-miR-582) show a significant repression of cell proliferation and invasion (Supplementary Figure S3b,c). By contrast, as shown in a loss-of-function assay using the locked nucleic acid–modified anti-miRNA oligonuleotides against each strand of miR-582-5p and -3p, cell proliferation and invasion are promoted in UM-UC-3-miR-582 cells (Supplementary Figure S3d,e). These results indicate that the presence of either miR-582-5p or -3p was sufficient to suppress tumor growth and invasiveness.
Inhibition of tumor growth and metastasis in vivo with miR-582 treatment
To assess the therapeutic potential of miR-582, we used an orthotopic bladder cancer mouse model featuring UM-UC-3-luc cells, which have the capacity to form tumors in the bladder and spread to the lungs of mice. The cationic liposome, LIC-101 (Nippon Shinyaku, Tsukuba, Japan), was used to deliver miR-582 into the tumor cells in the mouse bladder. The miR-582/LIC-101 or miR-582-scramble/LIC-101 complexes were injected transurethrally into the bladder at 5, 7, 9, 11, 13, and 15 days after tumor transplantation (Figure 3d). These results demonstrated that miR-582 successfully suppressed tumor growth in vivo; in particular, we found that miR-582 provided a greater than fourfold reduction of the tumor growth in the fourth week (Figure 3e). Furthermore, there was a significant difference in fold changes relative to the photon counts in the first week (miR-582 = 2,253.5 ± 851.7, scramble = 187.1 ± 108.9, P = 0.042) (Figure 3f). At the end of the experiment, 4 weeks after transplantation, the mice treated with miR-582-scramble showed a more frequent presence of tumors in the lung (5 of 8; 63%) than in the miR-582 group (1 of 10; 10%) (Figure 3g). Moreover, there was a significant difference between the two groups regarding the luminescence of the lung (Figure 3h). These results indicated that the intravesical administration of the miR-582/LIC101 complex could be a novel therapeutic strategy for the inhibition of tumor progression and metastasis in bladder cancer.
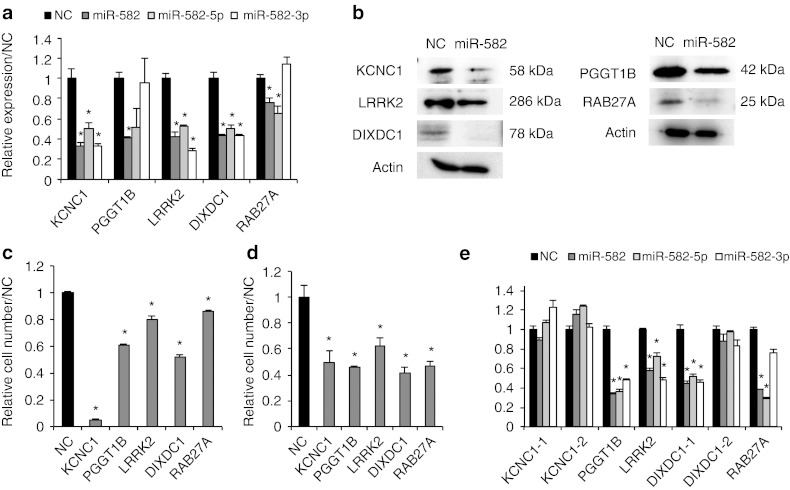
Figure 4.
Identification of PGGT1B, LRRK2, DIXDC1, RAB27A, and KCNC1 as miR-582-5p and -3p target genes. Target validations of KCNC1, PGGT1B, LRRK2, RAB27A, and DIXDC1 were confirmed by qRT-PCR, western blot analysis, and cell proliferation, cell invasion, and luciferase reporter assays. (a) qRT-PCR was performed in UM-UC-3 cells at 24 hours after transfection with miR-582. The relative expressions of the target mRNAs were calculated compared with the level in NC (n = 3). (b) Western blot analysis was performed in UM-UC-3 cells at 48 hours after transfection. Actin was used as the loading control. (c) Effect of target siRNAs on the proliferation of UM-UC-3. The proliferation values were normalized to values from cells treated with NC (n = 6). (d) Effect of target siRNAs on the invasion of UM-UC-3. The invasive values were normalized to the values from cells treated with NC (n = 3). (e) Each luciferase reporter construct containing a target 3′UTR was cotransfected with miR-582, miR-582-5p or -3p, or NC into UM-UC-3 cells (n = 6). *P < 0.05 versus NC. NC, negative control; qRT-PCR, quantitative reverse transcription-PCR.
Identification of miR-582-5p and -3p target genes
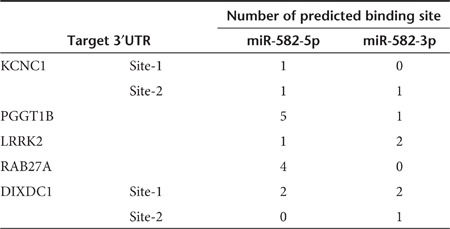
To identify miR-582-5p and -3p target genes, hemagglutinin (HA)-tagged Ago2-immunoprecipitation (Ago2-IP) and mRNA array analysis were performed. The UM-UC-3-Ago2 cells stably expressing HA-tagged-Ago2 were transfected with miR-582, and the Ago2-RNA complexes were precipitated with the anti-HA antibody, which was followed by the microarray analysis. The total RNA from UM-UC-3-Ago2 cells transiently transfected with miR-582 and from UM-UC-3-miR-582 cells was also analyzed (Supplementary Figure S4a). As a result, 259 genes were identified as candidates for directly targeted genes, and 1,559 genes were identified as candidates for indirectly targeted genes that are downstream of the direct targets (Supplementary Tables S4 and S5). In addition to these experimental approaches, we used two target prediction algorithms (TargetScan: http://www.targetscan.org, miRanda: http://www.microrna.org) as in silico approaches. After calculating the fold change and conducting an overlapping prediction analysis, we validated the identified 42 genes by qRT-PCR (Supplementary Figure S4b), western blotting analysis, and cell proliferation, cell invasion, and 3′UTR reporter assays. The results of the qRT-PCR and western blotting analyses demonstrated that the expression levels of the mRNA and protein of five genes—potassium voltage-gated channel subfamily C member 1 (KCNC1), PGGT1B, LRRK2, DIXDC1, and ras-related GTP-binding protein (RAB27A)—were more downregulated by transfection with miR-582, or miR-582-5p, or -3p than the NC siRNA-treated cells (Figure 4a,b). Furthermore, silencing of these target genes by siRNAs showed a remarkable inhibition of cell proliferation and cell invasion in UM-UC-3 cells (Figure 4c,d). These five genes have several putative binding sites for miR-582-5p or -3p in their 3′UTRs (Table 2 and Supplementary Table S6). Indeed, the expression of each luciferase reporter gene fused with the 3′UTR of PGGT1B, LRRK2 and DIXDC1 was suppressed by transfection with miR-582, miR-582-5p, or -3p (Figure 4e). Conversely, the expression of that of RAB27A was suppressed by miR-582 and miR-582-5p. There was no significant difference in the expression of that of KCNC1. These results indicated that miR-582-5p and -3p regulate the expression of PGGT1B, LRRK2, and DIXDC1 by binding their 3′UTRs. One strand, miR-582-5p, can directly bind 3′UTR of RAB27A and regulate the expression. In contrast, the downregulation of KCNC1 by transfection with miR-582-5p and -3p is not a direct consequence of miR-582.
Table 2. Summary of the number of target sites for miR-582-5p and -3p.
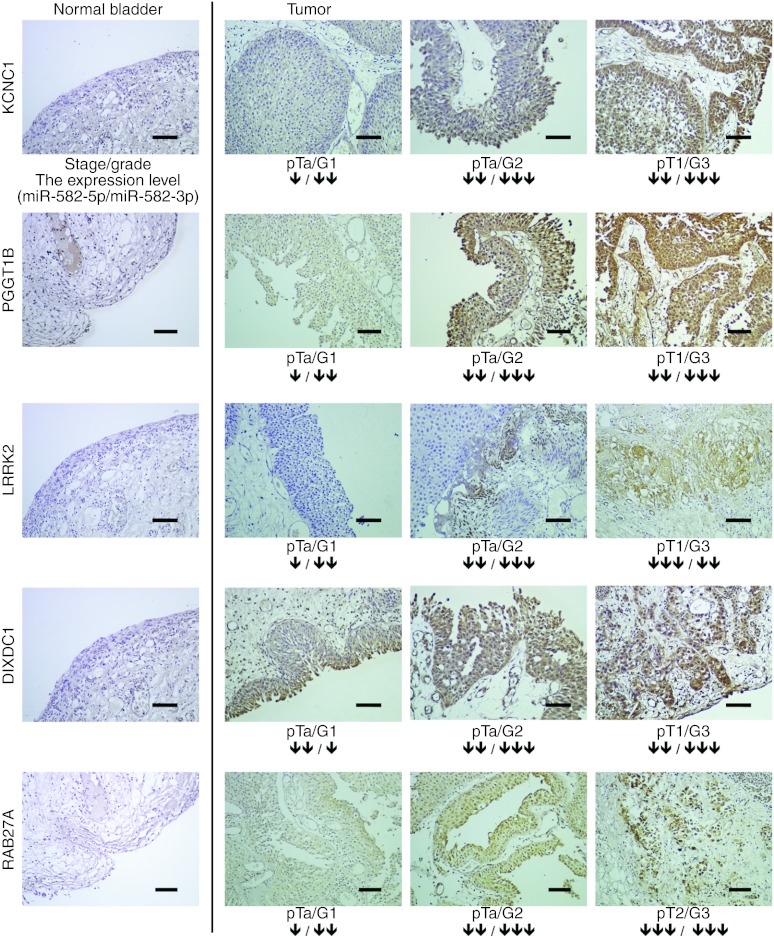
In addition, we performed an immunohistochemical analysis to observe the expression of these target genes in clinical samples of human bladder cancer. Strong nuclear or cytoplasmic staining was observed in the tumors but not in the adjacent normal bladder tissues (Figure 5). Furthermore, there was a positive correlation between the expression levels of target genes and tumor stage or grade of bladder cancer, and miR-582 was downregulated, whereas, in the low-stage or -grade bladder cancer, a high expression of target genes was not notably observed. These results suggest that the expressions of these cancer-related genes are a clinically relevant biological process, and the levels of the expressions are correlated with malignancy potential in bladder cancer.
Figure 5.
Representative examples of immunohistochemical expression of different gene products in human bladder cancer. Target gene localization in clinical samples by IHC. Left panel: representative normal bladder biopsy (scale bar: 100 µm); Right panel: representative bladder cancer with strong staining observed. A black arrow shows the fold change of the downregulation of miR-582-5p or -3p in tumor tissue compared with the adjacent normal tissues (single black arrow: fold change <2.0, double black arrows: fold change = 2.0 ~ 5.0, triple black arrows: fold change >5.0). Magnification: ×20. IHC, immunohistochemistry.
Discussion
Genetic changes potentially affect all medical conditions and are associated with thousands of diseases.26 The amplification of oncogenes or deletion of tumor-suppressor genes can broadly influence tumor initiation and progression.21,24 Although many previous reports have indicated the correlation between region-specific gain or loss of DNA and tumor progression in bladder cancer, it has not yet been fully elucidated as to which genes, including noncoding RNAs, are responsible for these different observations. In particular, the losses of chromosomes 3p11-12, 4q32-34, and 5q12-13 loci were reported to be associated with clinicopathological factors in bladder cancer.11,21,22,23,24 Here, we provided evidence on the relationship between the copy number loss of 5q12 and tumor progression via post-transcriptional regulation by miRNA. We demonstrated a novel pathway of paired tumor-suppressive miRNAs, miR-582-5p and -3p located on 5q12, which are specifically attenuated in invasive bladder cancer cells by copy number loss. Furthermore, the expression levels of miR-582-5p and -3p were remarkably lower in bladder cancer tissues and correlated with the tumor grade. These clear correlations suggest an important role for miR-582 on bladder cancer progression.
Despite these considerable dysregulations, there are no reports concerning the expression profiling and functional role of miR-582 in bladder cancer. Thus, focusing on miR-582 as a new candidate tumor-suppressive miRNA in bladder cancer is natural. As expected, the restoration of miR-582-5p or -3p expression inhibited the cell proliferation and invasion in UM-UC-3 cells. Generally, most miRNA duplexes can be functionally asymmetric due to unequal election for assembly into RISC and distinct sequence features. Although previous reports showed that miR199a-5p and -3p or miR-297b-5p and -3p target same mRNAs,27,28 for this reason, it is extremely rare that each strand of mature miRNA, 5p and 3p, has an identical property in the same physiological/pathological processes. The present experiment indicated that both strands of miR-582-5p and -3p have tumor-suppressive activities based on their sequences.
The number of RNAi-based preclinical and clinical trials has grown over the past several years, and these trials provide opportunities for success.2,4 These studies were conducted mainly in specific organs, such as eyes, dermis, and lung, which were relatively accessible by topical or local administration. Thus, accessibility is a key requirement for successful RNAi in vivo to be delivered tissue specifically or cell specifically. Here, in a closed environment of the bladder, the intravesical injection of miRNA is expected to offer high specificity to a tissue and noninvasive administration. Indeed, a previous study showed that the low-toxic cationic liposome, LIC-101, and the dsRNA complex could be successfully delivered into the bladder tissue and were able to affect tumor growth.7 However, whether transurethral injection is effective against a severe problem, such as invasion into the muscle and metastasis, remains unknown. In this study, we were successful at providing evidence for this procedure by showing that the instillation of the synthetic miR-582/LIC-101 complex inhibited both lung metastasis and tumor growth. For the first time, our findings have provided a new insight into the availability of a bladder cancer lung metastasis model mouse in RNAi therapeutic trials.
miRNAs are known to have the potential to target thousands of different mRNAs, and each mRNA is also targeted by multiple miRNAs.29,30 Although target prediction algorithms (TargetScan, miRanda) such as in silico approaches based on prior experimental studies can help to predict miRNA-binding targets, computational algorithms are imperfect and insufficient to identify some essential or novel target genes. Hence, in addition to in silico analysis, as an optimized and high-throughput biochemical assay, we evaluated the use of Ago2-IP.31,32,33 The validation studies revealed that miR-582-5p and -3p have a tumor-suppressive function by both the inhibition of translation and the degradation of KCNC1, PGGT1B, LRRK2, DIXDC1, and RAB27A. Although miR-582 did not regulate KCNC1 expression directly, it is very interesting that KCNC1 has a novel remarkable function in cell proliferation (Figure 4c), because it usually works as a potassium ion transporter. PGGT1B is involved in the geranylgeranylation of RhoA and the activation of MMP-9, resulting in the disruption of F-actin organization and the progression of cell motility, invasion, and metastasis.34,35 The kinase domain of LRRK2 is homologous to B-RAF kinase, which is well known to drive malignant melanoma. Furthermore, LRRK2 is related to the direct phosphorylation of Akt1, resulting in cell survival and the prevention of apoptosis.36,37 DIXDC1, which works as a receptor of the Wnt signaling pathway, is related to the upregulation of CyclinD1 and the downregulation of p21. The siRNA knockdown of DIXDC1 was reported to cause G1/S phase arrest in colon cancer.38,39 RAB27A, a member of the RAS oncogene family, is well known to be a driver of melanoma and breast cancer, promoting cell proliferation, invasion, and metastasis potential.40,41 It is also involved in the regulation of membrane trafficking and exosome formation.42 This study carries considerable potential to provide novel insights into the pathogenic mechanisms of exosomes regulated by miR-582-5p in invasive bladder cancer.
In conclusion, this study provides two novel findings in bladder cancer. First, we presented evidence of a unique regulatory pathway involving a couple of tumor-suppressive miRNAs, miR-582-5p and -3p, that are downregulated in invasive bladder cancer. The significant dysregulation of the miR-582 network may contribute to the progression of other invasive cancers, and we are actively investigating this hypothesis. Second, an in vivo experiment showed the possibility of the application to a therapeutic target against the continued threat of progression to muscle-invasion and metastasis. Importantly, our findings provide an insight into the unique regulatory pathway, which has a coordinated role between miRNA-5p and -3p. These novel findings also provide a potential value of an extreme contribution to the development of the cancer therapy as a prominent class of RNAi therapy.
Materials and Methods
Cell culture. The human bladder cancer cell lines UM-UC-3, 5637, J82, TCCSUP, T24, HT1376, and RT4 and the mouse bladder cancer cell line MBT2 were obtained from the American Type Culture Collection (Manassas, VA). The UM-UC-3, J82, TCCSUP, T24, and MBT2 cells were cultured in Modified Eagle's Medium (Sigma-Aldrich, St Louis, MO) containing 10% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA) and an antibiotic-antimycotic (Invitrogen) at 37 °C in 5% CO2. The 5637 cells were cultured in RPMI (Invitrogen) containing 10% heat-inactivated fetal bovine serum and an antibiotic-antimycotic at 37 °C in 5% CO2. The T24 and RT4 cells were cultured in McCoy'5A (Sigma-Aldrich) containing 10% heat-inactivated fetal bovine serum and an antibiotic-antimycotic at 37 °C in 5% CO2.
Patients and human samples. All human bladder tissue samples were derived from the resected bladders of 29 patients who underwent transurethral resection or radical cystectomy at the Department of Urology, St. Marianna University between 2009 and 2011. The samples of tumor and normal epithelium were fixed in formalin, embedded in paraffin, and sectioned for use in microscopic analysis and laser capture microdissection. All materials were obtained with written informed consent, and the study protocols were approved by the Institutional Review Board at the National Cancer Center Research Institute and St. Marianna University.
RNA extraction. Total RNA was extracted from cultured cells using the QIAzol and miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol.
Real-time PCR (qRT-PCR). The expression of miRNA was quantified by TaqMan miRNA assays (Applied Biosystems, Foster City, CA). The PCR was performed in 96-well plates using the 7300 Real-Time PCR System (Applied Biosystems). All reactions were performed in duplicate. Human-RNU6B or hsa-miR-103 was used as an invariant control.43
For the mRNA expression analysis, total RNAs were reverse-transcribed with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) using a random hexamer primer. The synthesized cDNAs were quantified by TaqMan Gene expression analysis or SYBR Green I qRT-PCR. The β-actin housekeeping gene was used to normalize the variation in the cDNA levels. The primer pairs used for gene amplification are listed in Supplementary Table S7.
Quantitative PCR of miR-582 loci on chromosome 5q12. Genomic DNA was extracted from cultured cells using a GenElute Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich). The qPCR for the miR-582 locus on chromosome 5q12 was performed using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen), and the primer sequences were 5′-ccacaacaagtcaatctgtgc-3′ and 5′-tattgaagggggttctggtg-3′. The housekeeping gene RNase P was also quantified as a control reference gene using Platinum Quantitative PCR SuperMix-UDG (Invitrogen) and the TaqMan RNase P Detection Reagents Kit (Applied Biosystems).
Transient miRNA/siRNA transfection. Synthetic hsa-miR-582, hsa-miR-582-5p, hsa-miR-582-3p, and hsa-miR-582-scramble duplexes were obtained from the Bonac (Kurume, Japan). The Allstars Negative Control siRNA was purchased from Qiagen. The siRNAs targeting KCNC1, PGGT1B, DIXDC1, LRRK2, and RAB27A were purchased from the Bonac. The cells were transfected with 25 nmol/l of either the miRNA or siRNA using DharmaFECT 1 (Thermo Fisher Scientific, San Jose, CA) according to the manufacturer's protocol. The miRNA and siRNA sequences are given in Supplementary Table S8.
Cell proliferation assay (MTS assay). For the cell proliferation assay, 24 hours after transfection, 3 × 103 cells were seeded in a 96-well plate. After 3 days of culture, the cell viability was measured using the Tetra Color One assay kit (Seikagaku Kohgyo, Tokyo, Japan) according to the manufacturer's instructions. The absorbance at 450 nm was measured using Envision (PerkinElmer, Norwalk, CT).
Cell invasion assay. The invasive ability of the bladder cancer cells was assayed in 24-well Biocoat Matrigel invasion chambers (8 µm pore size; Becton Dickinson, Franklin Lakes, NJ) according to the manufacturer's protocol. Briefly, the cells were transfected with miRNA or siRNA and, on the following day, 1 × 105 cells were plated in the upper chamber. The lower chamber was supplemented with a medium containing 10% fetal bovine serum. After 24 hours incubation, the cells on the upper surface were scraped off, and the invasive cells attached to the lower surface of the membrane inserts were fixed and stained with Diff-Quik (Sysmex, Kobe, Japan). The invading cells were observed and counted under a microscope in four random fields. All assays were performed in triplicate.
Generation of stable cell lines expressing miR-582 or HA-tagged Ago2 or luciferase. To construct a lentiviral vector for miR-582, pre-miRNA encompassing the stem-loop was amplified from genomic DNA isolated from HT1376 cells by PCR. The PCR product was digested and cloned into the pCDH cDNA cloning lentivector (Cat#CD513B-1; SBI, Mountain View, CA). To construct a lentiviral vector for luciferase, the luc construct was amplified from the pGL4 luciferase reporter vector by PCR. The PCR product was digested and cloned into the pLenti6/V5 Directional TOPO vector (Promega, Madison, WI). The lentiviral vector Lenti-miR-582, Lenti-scramble shRNA (Cat#MZIP000PA-1, SBI), Lenti-HA-Ago2 (RA703B-1, SBI), or Lenti-Luc and the lentiviral packaging plasmids (Invitrogen) were cotransfected into L293T cells. After 48 hours, the lentiviruses in the supernatant were collected and used to infect the UM-UC-3 cells. After antibiotic selection for 2 weeks, stable clones were obtained.
Analysis of miR-582 treatment in a mouse model of bladder cancer. The animal experiments in this study were performed in compliance with the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute. For the generation of the lung metastasis mouse model, we referred to some previous studies.7,44,45,46 Six- to seven-week-old female Balb/c athymic nude mice (CLEA Japan, Shizuoka, Japan) were anesthetized by exposure to 3% isoflurane on day zero and subsequent days. The murine bladder was injected with UM-UC-3-luc cells intravesically at 5 × 106 cells/50 µl/bladder after 15 minutes of trypsin treatment. The development of subsequent tumor growth and metastasis was monitored once a week by in vivo imaging. In brief, the mice were injected with 150 mg/kg D-luciferin (Promega) intraperitoneally and imaged 10 minutes later to count the photons from the whole bladder or lung using the IVIS imaging system (Xenogen, Alameda, CA) according to the manufacturer's instructions. The data were analyzed using the LivingImage software (version 2.5; Xenogen). On day 4, the bioluminescence from the implanted cancer cells was measured, and the mice were divided into two treatment groups with equivalent levels of bioluminescence. The transurethral treatment with the miR-582 and LIC101 (Nippon Shinyaku, Kyoto, Japan) complexes at a ratio of 1:16 (w/w) in a volume of 70 µl (10 µg/site) was performed on days 5, 7, 9 11, 13, and 15.
Ago2-IP. The RNA-binding protein immunoprecipitation was performed using an immunoprecipitation kit (RNA-binding protein immunoprecipitation–assay kit for microRNA; MBL, Nagoya, Japan) following the manufacturer's instructions. In brief, UM-UC-3 cells stably expressing HA-Ago2 were transfected with either miR-582 or NC for 48 hours and immunoprecipitated using anti-HA agarose beads (Wako, Osaka, Japan). The Ago2-bound RNA was eluted from beads with the HA peptide (Wako), and the QIAzol reagent was added to extract the total RNA. Ago2-bound total RNA was cleaned further using miRNeasy columns and then subjected to microarray analysis.
Microarray analysis. Total RNA was harvested from UM-UC-3-miR-582, UM-UC-3-shNC, and UM-UC-3-HA-Ago2 cells that were transfected transiently with miR-582 or NC for 48 hours. The Ago2-bound RNA was prepared from the Ago2-IP experiments. The total RNA was labeled with Cy3 using a Low Input Quick Amp Labeling Kit (Agilent, Palo Alto, CA) and hybridized to a SurePrint G3 Human GE 8×60 K array (Agilent) according to the manufacturer's instructions. The data analysis was performed using GeneSpring GX11.5.
Luciferase reporter assay. The 3′-UTRs of human KCNC1, DIXDC1, LRRK2, PGGT1B, and RAB27A were amplified by PCR from genomic DNA and cloned at the NotI and XhoI sites into pGMT Easy vector (Promega). The PCR primers and oligonucleotide sequences for the constructs are listed in Supplementary Table S9. All the constructs were further confirmed by sequencing.
For the luciferase activity analysis, each construct was cotransfected with the miRNA duplex in a 96-well plate using the DharmFECT Duo transfection reagent (Thermo Fisher Scientific) for 24 hours, and the luciferase assays were performed with the Dual-Glo Luciferase assay system (Promega) according to the manufacturer's instructions.
Western blotting. Forty-eight hours after transfection, the cells were homogenized in an M-PER mammalian protein extraction reagent (Pierce, Rockford, IL). The proteins in the total cell lysate or bound to the anti-HA agarose beads were separated by SDS-PAGE gels, which were calibrated using Precision Plus protein standards (Bio-Rad, Richmond, CA). The primary antibodies against KCNC1 (1: 250, ab84823), DIXDC1 (1: 250, ab67763), LRRK2 (1: 250, ab57329), PGGT1B (1: 250, ab55615), and RAB27A (1: 200, ab55667) were purchased from Abcam (Cambridge, MA), and the primary antibody against ACTIN (1:10,000, MAB1501) was purchased from Millipore (Billerica, MA). The dilution ratio of each antibody is indicated in parentheses. The HRP-linked anti-mouse secondary antibody (GE Healthcare, Buckinghamshire, UK) was used at a dilution of 1:5,000. The bound antibodies were visualized by chemiluminescence using the ECL Plus Western blotting detection system (GE HealthCare), and luminescent images were analyzed using a LuminoImager (LAS-3000; Fuji Film, Inc., Tokyo, Japan).
Immunohistochemistry. All tumors resected from human bladders were fixed with 10% buffered formalin and embedded in paraffin. Sections with a 5-µm thickness were examined using immunohistochemistry. The sections were deparaffinized, the antigens were retrieved by autoclaving in a 10 mmol/l citrate buffer (pH 6.0), and the endogenous peroxidase activity was blocked with the Immuno Pure Peroxidase Suppressor (Pierce, Chester, UK). The primary antibodies used in this study were the same as those used in the western blotting analysis (1:50), followed by incubation with peroxidase-coupled anti-mouse IgG (ImmPRESS Reagent; Vector labs, Burlingame, CA). The immunoreactions were visualized with diaminobenzidine, and the sections were counterstained with hematoxylin.
Statistical analysis. Results are expressed as the mean ± SE. The statistical analyses were conducted using the Bonferroni multiple-comparison test, and the analyses of the luminescence of the lung were conducted using the nonparametric Mann–Whitney–Wilcoxon test. These analyses were performed with the Expert StatView analysis software (version 4; SAS Institute, Cary, NC). P < 0.05 was considered to be statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. Expression of miR-1305, pri-miR-582 and PDE4D in human bladder cancer cell lines. Figure S2. Functions of miR-582 in T24 and 5637 cells. Figure S3. Functional analysis of UM-UC-3-miR-582 cells. Figure S4. Identification of miR-582-5p and -3p target genes using HA-Ago2 IP and microarray analysis. Table S1. Copy number losses detected in UM-UC-3 cell line. Table S2. Copy number losses correlated with pathological stage in human bladder cancer. Table S3. Stage and Grade distribution of the patients. Table S4. A total of 259 genes as candidates for miR-582 direct targets. Table S5. A total of 1,559 genes as candidates for miR-582 indirect targets. Table S6. Summary of miR-582-5p and -3p target site. Table S7. PCR primer sequences and oligonucleotide sequences. Table S8. miRNA and siRNA oligonucleotide sequences. Table S9. PCR primer sequences for Luciferase assay.
Acknowledgments
This work was supported in part by a Grant-in-Aid for the Third-Term Comprehensive 10-Year Strategy for Cancer Control, a Grant-in-Aid for Scientific Research on Priority Areas Cancer, and a Grant-in-Aid for Scientific Research on Innovative Areas (“functional machinery for non-coding RNAs”) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology, the National Cancer Center Research and Development Fund, the Program for the Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NiBio), the Project for Development of Innovative Research on Cancer Therapeutics, and the Japan Society for the Promotion of Science (JSPS) through the “Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program)” initiated by the Council for Science and Technology Policy (CSTP). We thank Ayako Inoue and Maki Abe for their excellent technical assistance. The authors declared no conflict of interest.
Supplementary Material
Expression of miR-1305, pri-miR-582 and PDE4D in human bladder cancer cell lines.
Functions of miR-582 in T24 and 5637 cells.
Functional analysis of UM-UC-3-miR-582 cells.
Identification of miR-582-5p and -3p target genes using HA-Ago2 IP and microarray analysis.
Copy number losses detected in UM-UC-3 cell line.
Copy number losses correlated with pathological stage in human bladder cancer.
Stage and Grade distribution of the patients.
A total of 259 genes as candidates for miR-582 direct targets.
A total of 1,559 genes as candidates for miR-582 indirect targets.
Summary of miR-582-5p and -3p target site.
PCR primer sequences and oligonucleotide sequences.
miRNA and siRNA oligonucleotide sequences.
PCR primer sequences for Luciferase assay.
REFERENCES
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE., and, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Davidson BL., and, McCray PB., Jr Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner A. Applications of RNA interference: current state and prospects for siRNA-based strategies in vivo. Appl Microbiol Biotechnol. 2007;76:9–21. doi: 10.1007/s00253-007-0984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita F., and, Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006;97:689–696. doi: 10.1111/j.1349-7006.2006.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingrich JR. Bladder cancer: chemohyperthermia for bladder cancer–clinically effective. Nat Rev Urol. 2011;8:414–416. doi: 10.1038/nrurol.2011.104. [DOI] [PubMed] [Google Scholar]
- Pasin E, Josephson DY, Mitra AP, Cote RJ., and, Stein JP. Superficial bladder cancer: an update on etiology, molecular development, classification, and natural history. Rev Urol. 2008;10:31–43. [PMC free article] [PubMed] [Google Scholar]
- Nogawa M, Yuasa T, Kimura S, Tanaka M, Kuroda J, Sato K.et al. (2005Intravesical administration of small interfering RNA targeting PLK-1 successfully prevents the growth of bladder cancer J Clin Invest 115978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth S, Matsui Y, Fosnaugh K, Liu Y, Vaish N, Adami R.et al. (2011RNAi-based therapeutics targeting survivin and PLK1 for treatment of bladder cancer Mol Ther 19928–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J, Beffa L, Wagner U, Schraml P, Gasser TC, Moch H.et al. (1998Patterns of chromosomal imbalances in advanced urinary bladder cancer detected by comparative genomic hybridization Am J Pathol 1531615–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaveri E, Brewer JL, Roydasgupta R, Fridlyand J, DeVries S, Koppie T.et al. (2005Bladder cancer stage and outcome by array-based comparative genomic hybridization Clin Cancer Res 1119 Pt 17012–7022. [DOI] [PubMed] [Google Scholar]
- Caballi MR, Egozcue J, Gelabert A, Prat E., and, Bernue M. Detection of chromosomal imbalances in papillary bladder tumors by comparative genomic hybridization. Urology. 2001;4295:5–11. doi: 10.1016/s0090-4295(01)00909-8. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S.et al. (2004Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers Proc Natl Acad Sci USA 1012999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S, Sevignani C, Nnadi SC, Siracusa LD., and, Calin GA. Cancer-associated genomic regions (CAGRs) and noncoding RNAs: bioinformatics and therapeutic implications. Mamm Genome. 2008;19:526–540. doi: 10.1007/s00335-008-9119-8. [DOI] [PubMed] [Google Scholar]
- Sevignani C, Calin GA, Nnadi SC, Shimizu M, Davuluri RV, Hyslop T.et al. (2007MicroRNA genes are frequently located near mouse cancer susceptibility loci Proc Natl Acad Sci USA 1048017–8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E.et al. (2002Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia Proc Natl Acad Sci USA 9915524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Yanagisawa K, Tokumaru S, Taguchi A, Nimura Y, Osada H.et al. (2008Detailed characterization of a homozygously deleted region corresponding to a candidate tumor suppressor locus at 21q11-21 in human lung cancer Genes Chromosomes Cancer 47810–818. [DOI] [PubMed] [Google Scholar]
- Agueli C, Cammarata G, Salemi D, Dagnino L, Nicoletti R, La Rosa M.et al. (201014q32/miRNA clusters loss of heterozygosity in acute lymphoblastic leukemia is associated with up-regulation of BCL11a Am J Hematol 85575–578. [DOI] [PubMed] [Google Scholar]
- Weiss GJ, Bemis LT, Nakajima E, Sugita M, Birks DK, Robinson WA.et al. (2008EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines Ann Oncol 191053–1059. [DOI] [PubMed] [Google Scholar]
- Takeshita F, Patrawala L, Osaki M, Takahashi RU, Yamamoto Y, Kosaka N.et al. (2010Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes Mol Ther 18181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst CD, Fiegler H, Carr P, Williams S, Carter NP., and, Knowles MA. High-resolution analysis of genomic copy number alterations in bladder cancer by microarray-based comparative genomic hybridization. Oncogene. 2004;23:2250–2263. doi: 10.1038/sj.onc.1207260. [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo C. Molecular alterations in bladder cancer. Cancer Surv. 1998;32:115–131. [PubMed] [Google Scholar]
- Wada T, Louhelainen J, Hemminki K, Adolfsson J, Wijkström H, Norming U.et al. (2001The prevalence of loss of heterozygosity in chromosome 3, including FHIT, in bladder cancer, using the fluorescent multiplex polymerase chain reaction BJU Int 87876–881. [DOI] [PubMed] [Google Scholar]
- Polascik TJ, Cairns P, Chang WY, Schoenberg MP., and, Sidransky D. Distinct regions of allelic loss on chromosome 4 in human primary bladder carcinoma. Cancer Res. 1995;55:5396–5399. [PubMed] [Google Scholar]
- Richter J, Jiang F, Görög JP, Sartorius G, Egenter C, Gasser TC.et al. (1997Marked genetic differences between stage pTa and stage pT1 papillary bladder cancer detected by comparative genomic hybridization Cancer Res 572860–2864. [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A., and, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34 Database issue:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almal SH., and, Padh H. Implications of gene copy-number variation in health and diseases. J Hum Genet. 2012;57:6–13. doi: 10.1038/jhg.2011.108. [DOI] [PubMed] [Google Scholar]
- Sakurai K, Furukawa C, Haraguchi T, Inada K, Shiogama K, Tagawa T.et al. (2011MicroRNAs miR-199a-5p and -3p target the Brm subunit of SWI/SNF to generate a double-negative feedback loop in a variety of human cancers Cancer Res 711680–1689. [DOI] [PubMed] [Google Scholar]
- Zhang T, Luo Y, Wang T., and, Yang JY. MicroRNA-297b-5p/3p target Mllt3/Af9 to suppress lymphoma cell proliferation, migration and invasion in vitro and tumor growth in nude mice. Leuk Lymphoma. 2012;53:2033–2040. doi: 10.3109/10428194.2012.678005. [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP.et al. (2005The widespread impact of mammalian MicroRNAs on mRNA repression and evolution Science 3101817–1821. [DOI] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP., and, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Davila J, Swerdel MR, Moore JC, Cohen RI, Wu H.et al. (2009Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors PLoS ONE 4e7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Wilfred BR, Hu Y, Stromberg AJ., and, Nelson PT. Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA. 2010;16:394–404. doi: 10.1261/rna.1905910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya N, Izumiya M, Ogata-Kawata H, Okamoto K, Fujiwara Y, Nakai M.et al. (2011Tumor suppressor miR-22 determines p53-dependent cellular fate through post-transcriptional regulation of p21 Cancer Res 714628–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusama T, Mukai M, Tatsuta M, Matsumoto Y, Nakamura H., and, Inoue M. Selective inhibition of cancer cell invasion by a geranylgeranyltransferase-I inhibitor. Clin Exp Metastasis. 2003;20:561–567. doi: 10.1023/a:1025898316728. [DOI] [PubMed] [Google Scholar]
- Virtanen SS, Sandholm J, Yegutkin G, Kalervo Väänänen H., and, Härkönen PL. Inhibition of GGTase-I and FTase disrupts cytoskeletal organization of human PC-3 prostate cancer cells. Cell Biol Int. 2011;34:815–826. doi: 10.1042/CBI20090288. [DOI] [PubMed] [Google Scholar]
- Pan T, Li X., and, Jankovic J. The association between Parkinson's disease and melanoma. Int J Cancer. 2011;128:2251–2260. doi: 10.1002/ijc.25912. [DOI] [PubMed] [Google Scholar]
- Ohta E, Kawakami F, Kubo M., and, Obata F. LRRK2 directly phosphorylates Akt1 as a possible physiological substrate: impairment of the kinase activity by Parkinson's disease-associated mutations. FEBS Lett. 2011;585:2165–2170. doi: 10.1016/j.febslet.2011.05.044. [DOI] [PubMed] [Google Scholar]
- Wang L, Cao XX, Chen Q, Zhu TF, Zhu HG., and, Zheng L. DIXDC1 targets p21 and cyclin D1 via PI3K pathway activation to promote colon cancer cell proliferation. Cancer Sci. 2009;100:1801–1808. doi: 10.1111/j.1349-7006.2009.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zheng L, Zeng Z, Zhou G, Chien J, Qian C.et al. (2006DIXDC1 isoform, l-DIXDC1, is a novel filamentous actin-binding protein Biochem Biophys Res Commun 34722–30. [DOI] [PubMed] [Google Scholar]
- Wang JS, Wang FB, Zhang QG, Shen ZZ., and, Shao ZM. Enhanced expression of Rab27A gene by breast cancer cells promoting invasiveness and the metastasis potential by secretion of insulin-like growth factor-II. Mol Cancer Res. 2008;6:372–382. doi: 10.1158/1541-7786.MCR-07-0162. [DOI] [PubMed] [Google Scholar]
- Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC.et al. (2010An integrated approach to uncover drivers of cancer Cell 1431005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A.et al. (2010Rab27a and Rab27b control different steps of the exosome secretion pathway Nat Cell Biol 1219–30; sup pp 1. [DOI] [PubMed] [Google Scholar]
- Peltier HJ., and, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Shinohara N, Sazawa A, Harabayashi T, Ogiso Y, Koyanagi T.et al. (2000An improved intravesical model using human bladder cancer cell lines to optimize gene and other therapies Cancer Gene Ther 71575–1580. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Gee JR, De La Cerda J, Rosser CJ, Zhou JH, Benedict WF.et al. (2003Noninvasive detection of bladder cancer in an orthotopic murine model with green fluorescence protein cytology J Urol 170975–978. [DOI] [PubMed] [Google Scholar]
- Horiguchi Y, Kikuchi E, Ozu C, Nishiyama T, Oyama M, Horinaga M.et al. (2008Establishment of orthotopic mouse superficial bladder tumor model for studies on intravesical treatments Hum Cell 2157–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of miR-1305, pri-miR-582 and PDE4D in human bladder cancer cell lines.
Functions of miR-582 in T24 and 5637 cells.
Functional analysis of UM-UC-3-miR-582 cells.
Identification of miR-582-5p and -3p target genes using HA-Ago2 IP and microarray analysis.
Copy number losses detected in UM-UC-3 cell line.
Copy number losses correlated with pathological stage in human bladder cancer.
Stage and Grade distribution of the patients.
A total of 259 genes as candidates for miR-582 direct targets.
A total of 1,559 genes as candidates for miR-582 indirect targets.
Summary of miR-582-5p and -3p target site.
PCR primer sequences and oligonucleotide sequences.
miRNA and siRNA oligonucleotide sequences.
PCR primer sequences for Luciferase assay.