Abstract
We developed a strategy to treat hepatitis C virus (HCV) infection by replacing five endogenous microRNA (miRNA) sequences of a natural miRNA cluster (miR-17–92) with sequences that are complementary to the HCV genome. This miRNA cluster (HCV-miR-Cluster 5) is delivered to cells using adeno-associated virus (AAV) vectors and the miRNAs are expressed in the liver, the site of HCV replication and assembly. AAV-HCV-miR-Cluster 5 inhibited bona fide HCV replication in vitro by up to 95% within 2 days, and the spread of HCV to uninfected cells was prevented by continuous expression of the anti-HCV miRNAs. Furthermore, the number of cells harboring HCV RNA replicons decreased dramatically by sustained expression of the anti-HCV miRNAs, suggesting that the vector is capable of curing cells of HCV. Delivery of AAV-HCV-miR-Cluster 5 to mice resulted in efficient transfer of the miRNA gene cluster and expression of all five miRNAs in liver tissue, at levels up to 1,300 copies/cell. These levels achieved up to 98% gene silencing of cognate HCV sequences, and no liver toxicity was observed, supporting the safety of this approach. Therefore, AAV-HCV-miR-Cluster 5 represents a different paradigm for the treatment of HCV infection.
Introduction
It is estimated that nearly 3% of the world population are chronically infected with hepatitis C virus (HCV).1 Successfully treating this virus has been challenging because HCV mutates rapidly and generates a heterogeneous population of viral variants. Until recently, the standard therapy for HCV infection was a year-long treatment with a combination of pegylated interferon-α (IFN-α) and ribavirin. This treatment is <50% effective against the major HCV genotype (genotype 1), and is poorly tolerated. Recently, the US Food and Drug Administration approved two HCV NS3/4A protease inhibitors (Boceprevir; Merck, Whitehouse Station, NJ and Telaprevir; Vertex, Cambridge, MA) for use with IFN-α and ribavirin. However, although these new direct-acting antiviral (DAA) agents have good antiviral properties they were not approved as monotherapies, because drug resistance develops rapidly when they are used alone.2 Despite the improved response rates of the triple drug regimens, the new standard-of-care treatment is not broadly applicable to all HCV genotypes and serious side effects persist. Second generation protease inhibitors and other DAAs that inhibit other viral proteins (e.g., NS5A and NS5B) or host factors required for HCV replication (e.g., cyclophilins and miR-122) are in clinical development.3,4 However, most of these are unlikely to be effective as monotherapies, and combinations of these DAAs are currently being evaluated, with the ultimate goal of developing an IFN-free drug regimen.
We are developing an alternative strategy to treat HCV that exploits the mechanism of RNA interference (RNAi), and uses a combination of exogenous or “artificial” anti-HCV microRNAs (miRNAs) to target five different regions of the HCV genome. Recombinant adeno-associated virus (AAV) vectors are used for delivery of this cluster of anti-HCV miRNAs. Unlike other DAAs, this strategy has the potential to prevent the selection of drug-resistant escape mutants, since five regions of the HCV genome are targeted simultaneously. This is an important consideration because, based on the high error rate of the HCV polymerase (10−4–10−5 errors/copied base) and the high daily productive capacity of HCV (1012 virions/day), it has been calculated that HCV genomes with all possible single and double nucleotide substitutions are generated multiple times each day. The viable variants pre-exist in HCV-infected individuals before drug treatment is initiated, and one additional nucleotide change is expected to arise during treatment.5 Therefore, to avoid resistance emergence, the number of substitutions that a combination of DAAs would need to overcome is ≥4 (ref. 5), and the miRNA cluster created in this work has the potential to do this.
Previously, we described the utilization of an endogenous miRNA cluster (miR-17–92) as a scaffold for RNAi effectors targeting the HCV genome.6 We replaced the first five mature miRNA sequences of the miR-17–92 cluster with sequences complementary to the HCV genome (positive strand). Three of the five miRNAs target conserved sequences in the 5′ untranslated region (UTR) of HCV genotype 1b (UTR1, UTR2, and UTR3), and the two others target sequences in one structural (Core) and one nonstructural (NS5B) gene . All five anti-HCV miRNAs were shown to have gene silencing activity when endogenous miR-17, miR-19A, miR-20, or miR-19B was used as a scaffold. However, if miR-18 was used as a scaffold, mature miRNAs were not observed and no gene silencing activity was detected.6 We now report the construction of a new polycistronic anti-HCV miRNA (HCV-miR-Cluster 5) that uses the last miRNA in the miR17-92 cluster, i.e., miR-92, rather than miR-18, as a scaffold for an anti-HCV miRNA. This optimized cluster was capable of inhibiting HCV replication in the HCV cell culture system, and sustained expression of miRNAs completely eliminated HCV from the culture, indicating inhibition of viral spread. In vivo, the new cluster showed potent gene silencing activity following AAV gene transfer, and all five miRNAs were expressed in mouse liver at levels that are predicted to inhibit HCV replication. Therefore, this vector represents a viable alternative treatment for HCV infection.
Results
In vitro gene silencing activity of HCV-miR-Cluster 5
The endogenous miR 17-92 cluster encodes six mature miRNAs (Figure 1a). We used five of the miRNAs (miR-17, miR-19a, miR-20, miR-19b, and miR-92) as scaffolds for miRNAs that target the HCV (+) genome. Our goal was to develop the most efficient and minimal set of miRNAs required to inhibit HCV replication and prevent the emergence of escape mutants. The endogenous sequences of the mature regions of the miRNAs (22–23 nucleotides) were replaced with HCV sequences (19–21 nucleotides) that have previously been shown to be effective small interfering RNAs and short hairpin RNAs, whereas the miR17–92 sequences that encode the lower stems, loops, and intervening sequences of the primary miRNA transcript were left intact. The guide strands were designed to have low internal stability at their 5′ ends because this feature promotes efficient entry of the guide strand of mature small interfering RNAs into RNA-induced silencing complex (RISC) .7,8 In some cases, this involved creating wobble or mismatches by changing the sequence at the 3′ end of the passenger strand to manipulate the internal stability of the hairpin. We also mimicked the secondary structure of the endogenous miRNAs by introducing mismatches and bulges into the stem of the anti-HCV miRNAs, as this has been shown to increase the probability that the guide strand will be incorporated into the RISC.9 Since the exogenous mature miRNAs are 2–4 nucleotides shorter than the endogenous mature miRNAs they replaced, it was possible to insert them at the 5′ or 3′ end of the original miRNA sequence. We chose to insert them at the 5′ end because it has been demonstrated that this orientation results in the generation of more efficient miRNAs10 and because it is important that the miRNA seed region (2–8 nucleotides from the 5′ end) contains HCV antisense sequences rather than endogenous miRNA sequences. This has implications for how the mature miRNAs will be cleaved by Dicer, as will be discussed later. Unlike the endogenous miRNAs, the anti-HCV miRNA guide strands were designed to be entirely complementary to their targets, and thus are predicted to mediate site-specific cleavage of their cognate targets.
Figure 1.
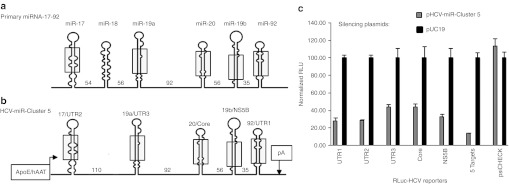
In vitro gene silencing activity of pHCV-miR-Cluster 5. (a) Structure of endogenous miR-17–92 cluster. (b) Structure of HCV-miR-Cluster 5. Numbers in between pre-miRNAs represent nucleotides. The shaded regions schematically represent sequences that differ between endogenous miR-17–92 and HCV-miR-Cluster 5. (c) Huh-7 cells were co-transfected with 125 µg of a reporter plasmid (RLuc-UTR1, RLuc-UTR2, RLuc-UTR3, RLuc-Core, RLuc-NS5B, RLuc-5 Targets, or psiCHECK2), and 125 µg of a plasmid expressing 5 anti-HCV miRNAs (HCV-miR-Cluster 5) or a nonsilencing control plasmid, pUC19. Twenty-four hours post-transfection, dual luciferase (FFLuc and RLuc) assays were performed on cell lysates. ApoE, human apolipoprotein E hepatic control region; hAAT, human alpha-one antitrypsin promoter; HCV, hepatitis C virus; pA, bovine growth hormone polyadenylation signal; RLuc, Renilla luciferase; UTR, untranslated region.
The polycistronic anti-HCV miRNA plasmid constructed (pHCV-miR-Cluster 5) encodes five anti-HCV miRNAs (miR-UTR1, miR-UTR2, miR-UTR3, miR-Core, and miR-NS5B) (Figure 1b). The HCV (+) strand sequences targeted by miR-UTR1, miR-UTR2, miR-UTR3, and miR-Core are highly conserved among all HCV genotypes, and miR-NS5B targets the NS5B gene of HCV genotype 1b.6 pHCV-miR-Cluster 5 uses two regulatory elements that promote liver-specific expression (i.e., human apolipoprotein E hepatic control region fused to the human α1 antitrypsin promoter), with the goal of limiting production of the anti-HCV miRNAs to the major site of HCV replication.
The activities of the five miRNAs were evaluated in Huh-7 cells using dual luciferase reporter plasmids, which encode a Renilla luciferase (RLuc) gene fused to an HCV DNA sequence, to determine gene silencing activity, and a firefly luciferase gene, to normalize transfection efficiencies.6 Five different reporter plasmids were constructed to evaluate the activity of each miRNA individually (RLuc-UTR1, RLuc-UTR2, RLuc-UTR3, RLuc-Core, and RLuc-NS5B). In addition, a reporter plasmid that contains the target sites of all five miRNAs was constructed (RLuc-5 Targets). To evaluate the miRNAs for nonspecific silencing, a reporter plasmid that did not contain HCV sequences (psiCHECK2) was used. As shown in Figure 1c, the miRNAs expressed from pHCV-miR-Cluster 5 induced 56–72% gene silencing of their cognate targets, relative to the pUC19 negative control plasmid, and 86% silencing of the reporter plasmid containing all five targets. No gene knock-down was observed using a plasmid that lacked HCV sequences (psiCHECK2).
Inhibition of HCV genotype 2a cell culture replication in vitro by AAV-HCV-miR-Cluster 5
In order to evaluate HCV-miR-Cluster 5 for its ability to inhibit bona fide HCV replication, we used the JFH-1 HCV genotype 2a cell culture (HCVcc) system.11 It should be noted that the JFH-1 HCVcc used in these experiments contains multiple cell culture-adaptive mutations, and replicates to high levels in Huh-7.5 cells.12 At the dose of HCVcc used in these experiments ~3,000–5,000 copies of HCVcc/cell were observed, which is ~50-fold higher than the levels observed in HCV-infected human hepatocytes13; thus this in vitro assay represents a stringent system for evaluating the anti-HCV activity of the miRNAs. Sequencing of the JFH-1 HCVcc genotype 2a RNA confirmed the presence of the three conserved 5′UTR sequences, and a near identical target sequence for miR-Core (20/21 nucleotides; identical seed sequence). Therefore, we predicted that miR-UTR1, miR-UTR2, miR-UTR3, and miR-Core would have activity against HCVcc. However, miR-NS5B would not be active against JFH-1 HCVcc RNA, since it is directed against HCV genotype 1b. Self-complementary adeno-associated viral vectors of serotype 2 (scAAV2) were used to deliver the exogenous cluster to Huh-7.5 cells. Cells were co-infected with HCVcc and scAAV2-HCV-miR-Cluster 5 at one of four doses (1 × 103, 1 × 104, 1 × 105, or 1 × 106 vector genomes (vg)/cell). Two hours later, the cells were washed and incubated for an additional 48 hours, at which time RNA from cell supernatants and cell pellets was purified and analyzed for HCVcc RNA by a quantitative real time reverse transcription PCR assay (QRT-PCR). A dose-dependent reduction in HCVcc RNA levels was observed in cell supernatants as the AAV dose increased, with a ~95% decrease observed using the two highest vector doses (Figure 2a). Similar results were observed when the cellular RNA was assayed for HCVcc RNA levels (Figure 2b).
Figure 2.
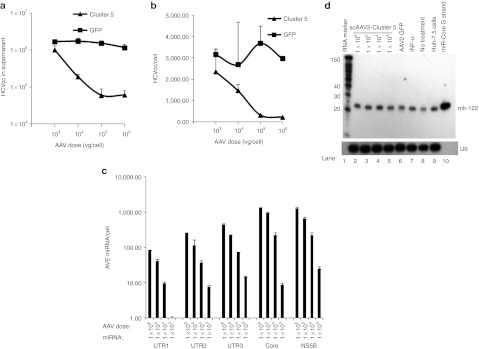
Adeno-associated virus (AAV) vectors expressing HCV-miRNA-Cluster 5 inhibit hepatitis C virus genotype 2a cell culture (HCVcc) replication. Huh-7.5 cells were co-infected with HCVcc (~0.5 FFU/cell) and either scAAV2-HCV-miR-Cluster 5 or the control vector, scAAV2-eGFP, at one of four doses (1 × 103, 1 × 104, 1 × 105, and 1 × 106 vg/cell). Two hours following infection, the cells were washed, the media was replaced, and the cells were incubated for 48 hours. (a) HCV RNA levels in supernatants quantified by QRT-PCR. Data represent triplicate HCV RNA measurements of duplicate infections. (b) HCV RNA levels in cellular RNA quantified by QRT-PCR. (c) Anti-HCV miRNA levels in cellular RNA quantified by custom QRT-PCR assays. (d) Northern blot analysis of miR-122 in Huh-7.5 cells co-infected with HCVcc and decreasing doses of scAAV2-HCV-miR-Cluster 5 (lanes 2–5), or scAAV2-eGFP (lane 6), or treated with 200 U/ml IFN-α (lane 7), or not treated with drug (lane 8). RNA from naive Huh-7.5 cells was also included (lane 9). RNA markers included γ-P32-labeled Decade RNA molecular weight markers (lane 1) and a miR-122 oligonucleotide (lane 10). eGFP, enhanced green fluorescent protein; miRNA, microRNA; scAAV2, self-complementary adeno-associated viral vectors of serotype 2.
Custom QRT-PCR assays were used to quantify the intracellular levels of the anti-HCV miRNAs. All five miRNAs were expressed in a dose-dependent manner (Figure 2c). Similar to what has been observed for the endogenous miR-17-92 cluster, the miRNAs were not expressed at identical levels.14 For example, in cells treated with a scAAV2-HCV-miR-Cluster 5 dose of 1 × 106 vg/cell, miR-Core and miR-NS5B levels reached ~1,300 copies per cell (Figure 2c), whereas 82 copies/cell of miR-UTR1 were observed in the same cells. Therefore, in this two day HCVcc assay, where HCV levels of over 3,000 copies/cell can be produced, miR-Core levels of ~1,300 copies/cell resulted in ~95% inhibition of HCV replication. Therefore, our goal was to achieve similar levels of anti-HCV miRNAs in mouse liver using an AAV serotype optimized for liver delivery (see below).
The above data indicate that scAAV2-HCV-miR-Cluster 5 induces the expression of high levels of miRNAs, and inhibits HCV replication by ~95%. However, the expression of exogenous miRNAs has the potential to disrupt the endogenous miRNA biogenesis pathway.15 Because HCV replication requires the highly abundant liver-specific miR-122 for RNA accumulation,16 it was important to rule out the possibility that an inadvertent decrease in miR-122, led to a decrease in HCVcc RNA. To evaluate this, miR-122 levels were analyzed by northern blot. As shown in Figure 2d, no differences in miR-122 levels were seen between cells transduced with increasing doses of scAAV2-HCV-miR-Cluster 5 and untransduced cells. In addition, using a QRT-PCR assay, the level of miR-122 observed in cells transduced with the high dose (1 × 106 vg/cell) of scAAV2-HCV-miR-Cluster 5 was 4.95 × 105 ± 1.18 × 104 copies/ng RNA, whereas the level in cells transduced with the control vector (scAAV2-eGFP) was 4.1 × 105 ± 1.7 × 104 copies/ng RNA, indicating that the inhibition of HCV replication was due to the expression of anti-HCV miRNAs, and not an indirect effect caused by altered miR-122 biogenesis.
Prevention of HCV spread by AAV-HCV-miR-Cluster 5
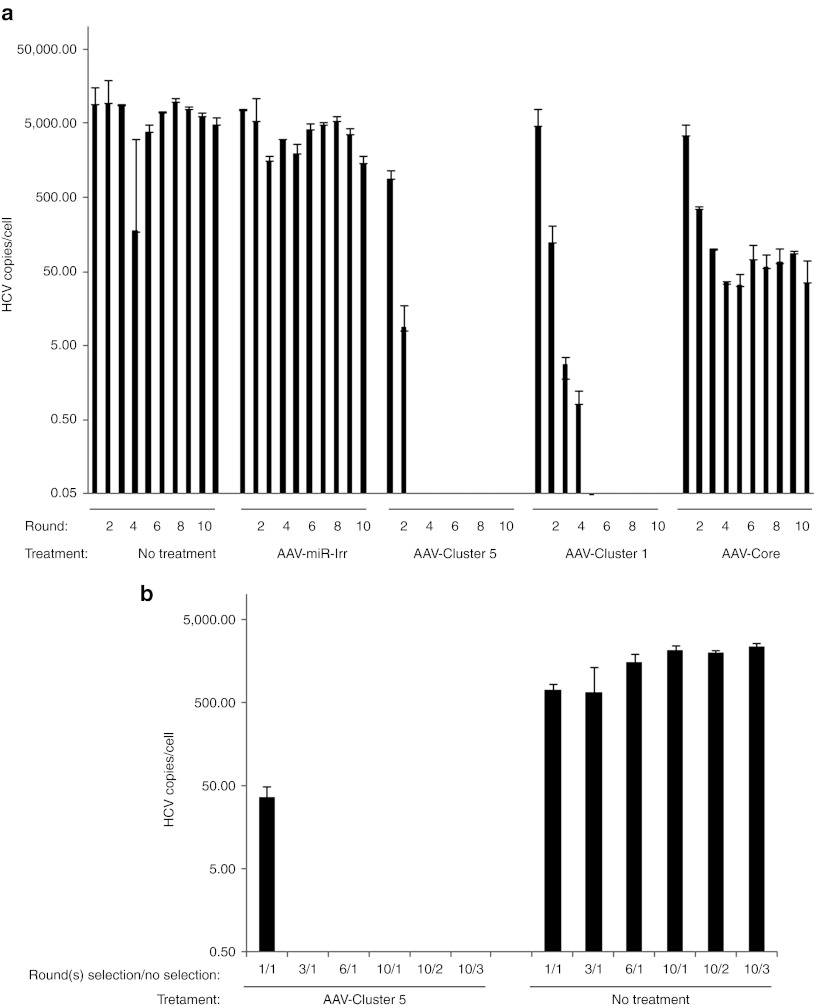
We determined the fate of HCV in cells continuously expressing the miRNAs, the situation that would be expected in vivo using AAV vectors for transgene delivery.17 Modeling this condition in the Huh-7.5/HCVcc system poses several challenges. First, HCVcc replicates to super-physiological levels in Huh-7.5 cells. Second, since AAV vectors are maintained as nuclear episomes,18 AAV genomes are lost over time in actively dividing cells in vitro, such as Huh-7.5 cells. In contrast, in adult liver, hepatocytes divide slowly and AAV episomes are more stably maintained, providing sustained expression of transgenes.17 Therefore, to mimic the in vivo situation, Huh-7.5 cells were co-infected with HCVcc and scAAV2-HCV-miR-Cluster 5, scAAV2-miR-Core, or a scAAV2-eGFP control vector. Additional controls included cells that were treated with IFN-α or were left untreated. Forty-eight hours later, an aliquot of the media, containing HCVcc, was used to infect separate wells of Huh-7.5 cells that were treated one day earlier with the respective scAAV vector, IFN-α, or were not treated. This was repeated for a total of six cycles. Cells were recovered after each round of infection, RNA was isolated, and HCVcc levels were quantified using QRT-PCR. The data in Supplementary Figure S1 demonstrate that in this system, a greater than five log decrease in HCVcc levels was observed within four rounds of propagation when cells were transduced with scAAV2-HCV-miR-Cluster 5, or were serially treated with IFN-α. This is in contrast to what was observed using scAAV vectors that expressed just a single miRNA (scAAV2-miR-Core), where HCVcc replication was inhibited by two logs, but the vector was incapable of eradicating the infection. This could be due to the effectiveness of miR-Core being below a critical treatment value and/or the emergence of miR-Core-resistant escape mutants. To evaluate the latter, we performed 454/Roche pyrosequencing on PCR fragments amplified from RNA isolated from HCV-infected cells that had been transduced with either scAAV2-HCV-miR-Cluster 5 (Rounds 1 and 3) or scAAV2-miR-Core (Rounds 1, 3, and 6). RNA from HCV-infected cells that were untreated (Rounds 1, 3, and 6) or treated with IFN-α (Rounds 1 and 3) was also amplified and sequenced. The results of deep sequencing are tabulated in Supplementary Table S1, with the sequence variation observed within the miR-Core target sequence only, shown for simplicity. The nucleotide changes are highlighted, and the possibility that a change may lead to an escape mutant is also indicated. The frequency of true miR-Core resistant escape mutants is expected to increase over six rounds, whereas they are expected to be absent in cells treated with IFN-α or untreated cells. A comparison of the sequence changes occurring between cells treated with AAV-miR-Core and cells treated with IFN-α or left untreated, suggests that miR-Core escape mutants emerged between rounds 1, 3, and 6. For example, at nucleotide 284 an increase in the percentage of variants containing a C→A transversion increased from 4 to 9% between rounds 3 and 6, and the reciprocal change (A→C transversion) at nucleotide 285 also increased during this time. In addition, variants with a deletion of a G at nucleotide 288, an A→T transversion at nucleotide 289, and a deletion of an A at nucleotide 292 also increased over time. At most positions the percentage of HCV genomes containing the correct nucleotide was greater than those with a mutated base. This suggests that the inability of this single miRNA to control an HCV infection is due to at least two factors: the emergence of escape mutants and the activity of the miRNA falling below a critical threshold value.
The data for AAV-Cluster 5 also suggests that HCV variants with a mutation (C→T transition) at nucleotide 295 emerge following round 3. However, since we were unable to amplify HCV RNA beyond this point, this suggests that this variant was either nonviable or was sensitive to another anti-HCV miRNA, and underscores the notion that the use of multiple miRNAs targeting conserved HCV sequences is important to eliminate HCV from the culture.
One caveat to the sequencing data is that many of the observed changes occurred within homopolymeric runs. Although the data was processed with an algorithm (PyroNoise) to improve the accuracy of the sequences, it will be important to isolate mutant clones and sequence them multiple times, as well as insert the mutated sequences into the HCV genome and confirm their resistance to AAV-miR-Core, to definitively state that the observed changes are true miR-Core resistant escape mutants.
To be sure that Cluster 5-resistant variants could not be generated at even later time points, in either the presence or absence of selective pressure from miRNAs, a follow-up study was conducted in which Huh-7.5 cells were co-infected with HCVcc and scAAV2-HCV-miR-Cluster 5, scAAV2-miR-Core, or a vector expressing an irrelevant miRNA (scAAV2-miR-Irr) for a total of 10 cycles. For comparison, a previously constructed AAV cluster (scAAV2-HCV-miR-Cluster 1), which expresses four active miRNAs (three against HCV genotype 2a) and one inactive miRNA (miR-UTR2), was also included.6 Additional controls included cells that were left untreated. RNA was isolated from cells after each round of infection and HCVcc levels were quantified using QRT-PCR. Similar to what was seen in the previous experiment, a greater than five log decrease in HCVcc levels was observed, this time, within three rounds of propagation when cells were transduced with scAAV2-HCV-miR-Cluster 5, and within five rounds using scAAV2-HCV-miR-Cluster 1 (Figure 3a). In contrast using scAAV2-miR-Core, HCVcc replication was once again inhibited by two logs, but the infection was not eradicated. These data confirm that expression of one miRNA is insufficient to inhibit HCV spread, and indicate that expression of four miRNAs is more potent than expression of just three, with miR-UTR2, specifically, enhancing the activity of scAAV2-HCV-miR-Cluster 5.
Figure 3.
Adeno-associated virus (AAV) vectors expressing HCV-miRNA-Cluster 5 prevent hepatitis C virus (HCV) spread. (a) In presence of selective pressure: Huh-7.5 cells were co-infected with HCV genotype 2a cell culture (HCVcc) (~0.2 IU/cell) and either scAAV2-HCV-miR-Cluster 5, scAAV2-HCV-miR-Cluster 1, scAAV2-miR-Core, or scAAV2-miR-Irr at an AAV dose of 1 × 105 vg/cell. Negative control wells consisted of Huh-7.5 cells that were infected with HCVcc but were not treated with a drug. Two hours later, the media was replaced and the cells were incubated for 48 hours. Supernatants were serially passaged (total of 10 rounds) onto Huh-7.5 cells that had been transduced 24 hours previously with the corresponding AAV vector. After each round, total cellular RNA was isolated and HCVcc RNA levels were measured by QRT-PCR (limit of sensitivity = 0.05 HCV copies/cell). Data represent triplicate HCV RNA measurements of duplicate infections. (b) In absence of selective pressure: supernatants collected from rounds 1, 3, 6, and 10 of cells treated with scAAV2-HCV-miR-Cluster 5 or not treated (a) were applied to naive Huh7.5 cells (in total volume of 2 ml). Forty-eight hours later, total cellular RNA was isolated and HCVcc RNA levels were measured by QRT-PCR. In addition, the supernatants from the cells treated with the round 10 supernatants were serially passaged (two additional rounds) onto naive Huh7.5 cells and 48 hours later intracellular RNAs levels were measured. Data represent triplicate HCV RNA measurements of duplicate infections. miRNA, microRNA; scAAV2, self-complementary adeno-associated viral vectors of serotype 2.
Since the goal of anti-HCV therapy is to completely eradicate the virus from infected cells and prevent a resurgence of replication once treatment is terminated, we also evaluated HCVcc replication in these cultures in the absence of selective pressure from the miRNAs. Media was collected from cells that had been repeatedly treated with scAAV2-HCV-miR-Cluster 5 or were left untreated following rounds 1, 3, 6 and 10 (from Figure 3a), and it was used to infect naïve Huh7.5 cells. Forty-eight hours later, cellular RNA was isolated and evaluated for HCV sequences by QRT-PCR (Figure 3b). In addition, supernatants from some of these infections (from the original round 10 infections) were serially passaged for two additional rounds on naive Huh7.5 cells and analyzed in the same manner. The data in Figure 3b demonstrate that no replication competent HCV was present in the supernatants (collected following rounds 3, 6, and 10) from cells serially transduced with AAV-Cluster 5. The lack of HCV in the absence of selective pressure indicates that scAAV2-HCV-miR-Cluster 5 can prevent the propagation of HCV in vitro.
Clearance of HCV by AAV-HCV-miR-Cluster 5
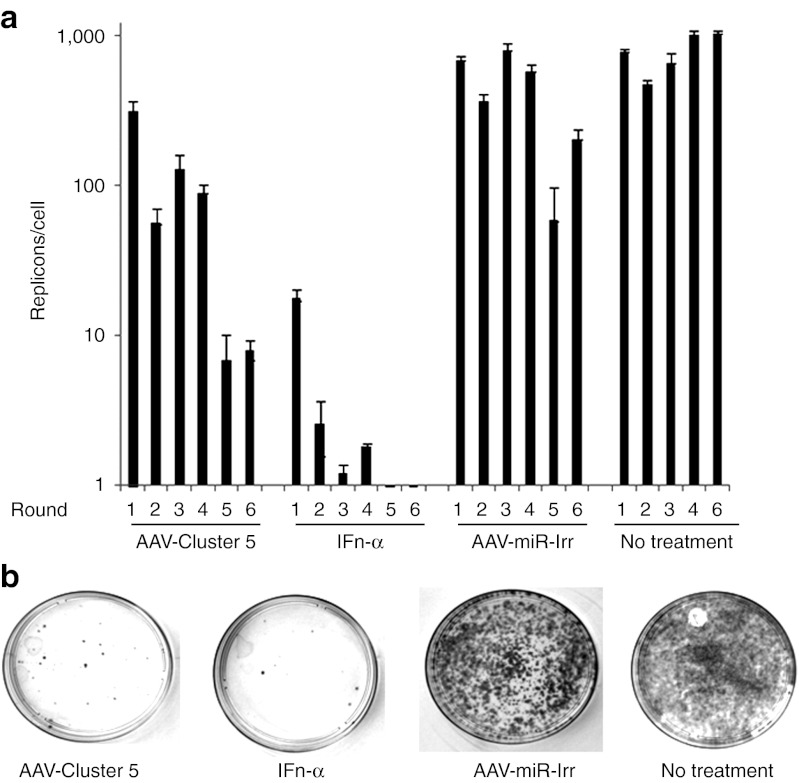
To investigate the ability of scAAV2-HCV-miR-Cluster 5 to cure HCV-infected cells, we adopted an HCV replicon clearance and rebound assay that was developed to evaluate the NS3/4A protease inhibitor VX-950 (Telaprevir),19 The assay uses Huh-7 cells harboring an autonomously replicating subgenomic HCV genotype 1b replicon, and unlike the HCVcc system, does not produce infectious HCV. This replicon also encodes a neomycin phosphotransferase gene, which allows for selective growth of the HCV replicon-containing cells in the presence of G418. Replicon cells were treated with scAAV2-HCV-miR-Cluster 5, scAAV2-miR-Irr, IFN-α, or were left untreated. Three days later, the replicon cells were re-plated in fresh media and retreated with the corresponding AAV vector, IFN-α, or were left untreated. This was repeated for a total of six rounds. Cellular RNA was isolated after each round, and was analyzed for HCV RNA by QRT-PCR. At the end of the sixth round, cells were plated in the absence of drug (AAV or IFN-α), but in the presence of G418 to allow for selective growth of those cells still harboring replicons. As shown in Figure 4a, a time-dependent decline in HCV replicon RNA levels was observed when scAAV2-HCV-miR-Cluster 5 or IFN-α was used. A slight decrease was also observed in cells treated with scAAV2-miR-Irr relative to untreated cells. This may be due to the passenger strand of miR-Irr, which has partial complementary to the HCV sequence. Although we have not detected passenger strands for the anti-HCV miRNAs, the stability of the guide and passenger strands of miR-Irr has not been analyzed. Upon growth in G418, a dramatic reduction in the number of replicon-containing colonies was observed when cells were treated with scAAV2-HCV-miR-Cluster 5 or IFN-α, as compared with cells treated with an irrelevant miRNA or left untreated (Figure 4b). Although, complete eradication of HCV from all replicon-containing cells was not observed, these data demonstrate that scAAV2-HCV-miR-Cluster 5 significantly reduced the intracellular HCV RNA copy number and the absolute number of HCV-containing cells.
Figure 4.
Adeno-associated virus (AAV) vectors expressing HCV-miRNA-Cluster 5 decrease hepatitis C virus (HCV) from infected cells. (a) Huh-7.5 cells containing HCV genotype 1b replicons were transduced with scAAV2-HCV-miR-Cluster 5 or scAAV2-miR-Irr at an AAV dose of 1 × 105 vg/cell, in the absence of G418. Positive controls were treated with interferon-α(IFN-α) (200 U/ml) and negative controls were left untreated. Cells were split every 3 days, and retreated with drug (AAV or IFN-α) for a total of six rounds. Total cellular RNA was isolated and HCV RNA levels were measured by QRT-PCR at each round. (b) After six passages, cells (1 × 105) were plated in 10-cm dishes in the presence of G418 and were incubated for 2 weeks. HCV replicon-containing colonies were stained with 0.05% crystal violet. miRNA, microRNA; scAAV2, self-complementary adeno-associated viral vectors of serotype 2.
In vivo gene silencing activity by scAAV vectors expressing HCV-miRNA-Clusters
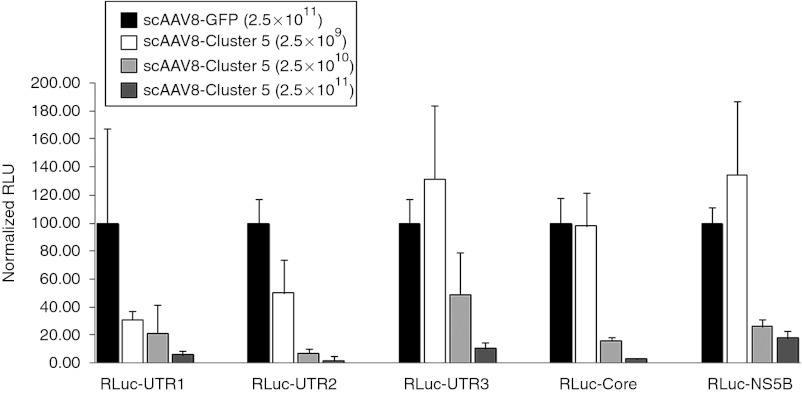
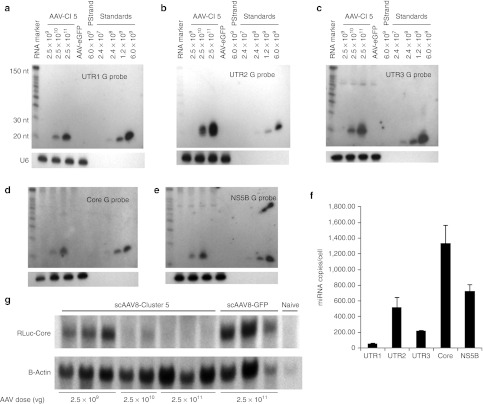
The in vivo activity of AAV-HCV-miR-Cluster 5 was evaluated by monitoring gene silencing of the RLuc-HCV reporter plasmids. scAAV vectors20 of serotype 8 were used for delivery, as this serotype efficiently transduces mouse liver.21 Mice were injected via the tail vein with three different doses of scAAV8-HCV-miR-Cluster 5 (2.5 × 109, 2.5 × 1010, and 2.5 × 1011 vg/mouse) or a control vector (scAAV8-eGFP; 2.5 × 1011 vg/mouse). Two weeks later, one of the five RLuc-HCV reporter plasmids was delivered by hydrodynamic tail vein injection, which results in transduction of ~40% hepatocytes.22 Mice were killed 2 days later, livers were harvested, and DNA, protein, and RNA were isolated for evaluation of gene transfer, dual luciferase activity, and miRNA and mRNA levels, respectively. At the low dose of AAV (2.5 × 109 vg/mouse), the AAV copy number was estimated to be ~0.4 copies/diploid genome by quantitative Southern blot (Supplementary Figure S2), and only two of the five reporter plasmids were inhibited (RLuc-UTR1 by 70% and RLuc-UTR2 by 51%). No gene silencing of the three other reporters was seen (Figure 5). However, using a 10-fold higher dose of AAV (2.5 × 1010 vg/mouse), the AAV copy number increased to ~6.0 AAV copies/diploid genome and moderate to high levels of gene silencing of all five reporters was observed (RLuc-UTR1: 80%; RLuc-UTR2: 93%; RLuc-UTR3: 54%; RLuc-Core: 81%; and RLuc-NS5B: 73%) (Figure 5). Furthermore, using the highest dose of vector (2.5 × 1011 vg/mouse), the AAV copy number was estimated to be ~81 copies/diploid genome and 83–98% gene silencing of all five reporters was observed (Figure 5). The higher levels of reporter inhibition that were observed at the two highest AAV doses are consistent with more efficient gene transfer, and correlate with the higher intracellular miRNA levels, as shown by both northern blot analyses and QRT-PCR assays. As shown in Figure 6a–e, northern blot analyses demonstrated a dose-dependent increase in the expression of all five mature (~20 nt) miRNAs (miR-UTR1, miR-UTR2, miR-UTR3, miR-Core, and miR-NS5B), but no precursor miRNAs (~70 nt) were detected. Furthermore, the use of strand-specific probes showed that only the guide (or antisense) strands of the five miRNAs were stably produced (negative data using passenger (or sense) strand probes are not shown). Of note, the mature miR-UTR3 guide strand appeared longer than the 19 nucleotide small interfering RNA it was designed to mimic. We believe this is due to Dicer cleavage occurring 23 nucleotides from the Drosha cleavage site, as would occur for endogenous miR-19a, the miRNA that miR-UTR3 replaced. Since the additional nts are likely to be added to the 3′ end, they would not affect the seed sequence, and therefore the activity of the miRNA is not expected to be adversely affected. Custom QRT-PCR assays revealed miRNA levels ranging from 60/cell for miR-UTR1 to 1,300/cell for miR-Core in mice injected with the highest dose of AAV (Figure 6f). It should be noted that the miRNA levels estimated from the northern blots, using synthetic RNA standards, were higher than those determined by custom QRT-PCR assays. The reason for this discrepancy is unclear, but it is possible that the miRNAs contain heterogeneous 3′ ends that would not all be amplified in the PCR assay.
Figure 5.
Adeno-associated virus (AAV) vectors expressing HCV-miRNA-Cluster 5 mediate hepatitis C virus (HCV) gene silencing in vivo. Female C57/Bl6 mice (6–8 weeks) were injected with 2.5 × 109, 2.5 × 1010, or 2.5 × 1011 vg/mouse of scAAV8-HCV-miR-Cluster 5 or scAAV-GFP (2.5 × 1011 vg/mouse) via the tail vein. Two weeks later, separate cohorts of mice (n = 5) were injected with one of five RLuc-HCV reporter plasmids (RLuc-UTR1, RLuc-UTR2, RLuc-UTR3, RLuc-Core, and RLuc-NS5B) using the HDTV procedure. Mice were killed 2 days later and liver lysates were analyzed for dual luciferase activity. Two independent liver lysates were prepared and each lysate was analyzed in triplicate; results are reported as the mean and SD of a representative lysate. Normalized RLuc expression in mice injected with AAV-GFP was set as 100% activity. GFP, green fluorescent protein; HDTV, hydrodynamic tail vein.
Figure 6.
MicroRNA (miRNA) and RLuc mRNA analysis of scAAV2-HCV-miR-Cluster 5-injected mice. (a–e) Northern blot analyses of miRNA guide strands. Twenty-five µg of total RNA from mice injected with increasing doses of scAAV8-HCV-miR-Cluster 5 (2.5 × 109, 2.5 × 1010, and 2.5 × 1011 vg/mouse) or scAAV8-eGFP (2.5 × 1011 vg/mouse) was resolved on 15% denaturing polyacrylamide TBE-urea gels. Increasing amounts (2.4 × 107, 2.4 × 108, 1.2 × 109, and 6 × 109 copies) of the synthetic specific miRNA guide strands were loaded as standards. The synthetic specific miRNA passenger strand was loaded as a negative control. A γ-P32-labeled Decade RNA molecular weight marker was also loaded. The miRNA transcripts were detected using γ-P32-labeled DNA oligonucleotide probes specific for the antisense strand (miRNA guide strand): (a) miR-UTR1 guide probe, (b) miR-UTR2 guide probe, (c) miR-UTR3 guide probe, (d) miR-Core guide probe, (e) miR-NS5B guide probe. Blots were stripped and reprobed with a U6 snRNA probe to confirm equal sample loading. (f) Custom QRT-PCR analysis of miRNAs in animals injected with 2.5 × 1011 vg/mouse of scAAV8-HCV-miR-Cluster 5 (g) Analysis of RLuc-Core mRNA. Twelve micrograms of total cellular RNA was separated on a 1% agarose gel. Millennium RNA markers were used as a molecular weight standard. The RLuc-Core mRNA was detected using an α-P32-labeled RLuc DNA probe. The membrane was stripped and reprobed with an α-P32-dCTP labeled β-actin probe to evaluate equal loading of samples. eGFP, enhanced green fluorescent protein; HCV, hepatitis C virus; miRNA, microRNA; RLuc, Renilla luciferase; scAAV, self-complementary adeno-associated viral vector; snRNA, small nuclear RNA; UTR, untranslated region.
To investigate the mechanism by which in vivo gene silencing occurs, the fate of one of the reporter plasmid transcripts (RLuc-HCV-Core mRNA) was evaluated by northern blot. Because the miRNAs were designed to be completely complementary to their HCV target sequences, it is predicted that the RLuc-HCV-Core mRNA expressed from the reporter plasmid will be cleaved, rather than translationally repressed.23 To evaluate this, total cellular liver RNA from mice injected with increasing doses of scAAV8-HCV-miR-Cluster 5 and the RLuc-HCV-Core reporter plasmid was analyzed by northern blot for the presence of the RLuc-HCV-Core mRNA transcript. As seen in Figure 6g, as the dose of AAV increased, the level of this transcript decreased and became undetectable, consistent with an RNAi mechanism (mRNA cleavage or destabilization),24 rather than a nonspecific gene silencing mechanism, such as innate immune response induction.25
Self-complementary AAV-miR-Cluster 5 and single-stranded AAV-miR-Cluster 5 vectors induce similar levels of gene silencing
The initial in vivo studies were performed using scAAV vectors because they have been reported to transduce mouse liver ~10 times more efficiently than traditional single-stranded (ss) AAV vectors.20 In our hands, scAAV vectors are more challenging to produce, as they result in lower yields. For example, using an unbiased methodology (quantitative silver stained PAGE), as opposed to QPCR,26 for quantification, the average number of vector particles produced per roller bottle for scAAV vectors expressing miRNAs was 5.14 × 1011 ± 3.15 × 1011 (n = 53 preps), which was significantly different (P = 0.001) than the yield of ssAAV vectors expressing miRNAs (3.34 × 1012 ± 5.3e12; n = 7 preps). Therefore, our preference is to use ssAAV vectors for clinical development, if they are equally effective as scAAV vectors. To directly compare these two types of vectors, the identical HCV-miR-Cluster 5 expression cassette was inserted into either scAAV or ssAAV backbone plasmids and vectors were produced. Since the physical titer of scAAV vectors is often underestimated using QPCR,26 three additional methods were employed: quantitative PAGE/silver stain,27 optical density,28 and quantitative alkaline agarose gel electrophoresis followed by Southern blot hybridization (Figure 7a). Based on the results of the four assays, the 95% confidence intervals for the doses of scAAV8-HCV-miR-Cluster 5 and ssAAV8-HCV-miR-Cluster 5 used in this study were 4.4 ± 2.4 × 1010 vg/mouse and 2.7 × 1010 ± 9.1 × 109 vg/mouse, respectively. Control mice were injected with scAAV-eGFP (2.5 × 1010). Two weeks following vector administration, either the RLuc-Core or RLuc-NS5B reporter plasmid was injected into the mice via hydrodynamic tail vein. Livers were harvested 2 days later for analysis of dual luciferase activity. As shown in Figure 7b, the scAAV8-HCV-miR-Cluster 5 and ssAAV8-HCV-miR-Cluster 5 vectors inhibited the expression of the RLuc-Core reporter plasmid by 82 and 80% respectively, relative to the control mice, and inhibited the RLuc-NS5B reporter plasmid by 72 and 63%, respectively. No statistically significant differences in gene silencing activity were observed between the two types of vectors. In addition, the levels of miR-Core in mouse liver, as determined by QRT-PCR, were similar (Figure 7c). An additional study was performed to determine the level of gene silencing of the other three RLuc-HCV reporter plasmids (RLuc-UTR1, RLuc-UTR2, and RLuc-UTR3) by the ssAAV-HCV-miR-Cluster 5 vector (Figure 7d). The data in Figure 7b–d, as well as a comparison between Figure 5a and Figure 7d, indicate that the ssAAV8-HCV-miR-Cluster 5 vector and the scAAV8-HCV-miR-Cluster 5 vector have similar potencies. The cumulative data demonstrate that, at least for this expression cassette, the self-complementary configuration of AAV does not provide any advantage over the traditional single-stranded vector.
Figure 7.
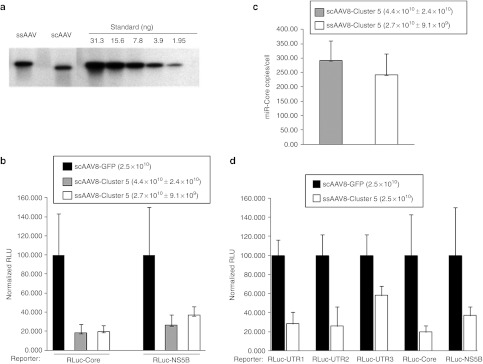
Comparison of self-complementary and single-stranded AAV-HCV-miR-Cluster 5 vectors in vivo. (a) Quantitative alkaline agarose Southern blot of scAAV8-HCV-miR-Cluster 5 (5 µl of 1:20 dil stock), ssAAV8-HCV-miR-Cluster 5 (5 µl of 1:40 dil stock), and decreasing amounts (31.3–1.95 ng) of a 4,175 bp DNA fragment containing the HCV-miR-Cluster 5 expression cassette, which were used as copy number standards. (b) Female C57/Bl6 mice (6–8 weeks) were injected with AAV-HCV-miR-Cluster 5 vectors whose titers were determined using four independent methods and dosed according to the 95% confidence intervals of the results: scAAV8-HCV-miR-Cluster 5 (95% confidence intervals (CI) = 4.4 × 1010 ± 2.4 × 1010 vg/mouse), ssAAV8-HCV-miR-Cluster 5 (95% CI = 2.7 × 1010 ± 9.1 × 109 vg/mouse), or scAAV-GFP (2.5 × 1010 vg/mouse) via the tail vein. Two weeks later, separate cohorts of mice (n = 5) were injected with either the RLuc-Core or RLuc-NS5B reporter plasmid using the hydrodynamic tail vein (HDTV) procedure. Mice were sacrificed 2 days later and liver lysates were analyzed for dual luciferase activity. Two independent liver lysates were prepared and each lysate was analyzed in triplicate; results are reported as the mean and SD. Normalized RLuc expression in mice injected with AAV-GFP was set as 100% activity. (c) Levels of miR-Core in mouse liver quantified using a custom QRT-PCR assay. (d) Female C57/Bl6 mice (6–8 weeks) were injected with of ssAAV8-HCV-miR-Cluster 5 (2.5 × 1010 vg/mouse) or scAAV-GFP (2.5 × 1010 vg/mouse) via the tail vein. Two weeks later, separate cohorts of mice (n = 5) were injected with either the RLuc-UTR1, RLuc-UTR2, RLuc-UTR3, RLuc-Core, or RLuc-NS5B reporter plasmid using the HDTV procedure. Mice were killed 2 days later and liver lysates were analyzed for dual luciferase activity. Two independent liver lysates were prepared and each lysate was analyzed in triplicate; results are reported as the mean and SD. Normalized RLuc expression in mice injected with AAV-GFP was set as 100% activity. AAV, adeno-associated virus; GFP, green fluorescent protein; HCV, hepatitis C virus; RLuc, Renilla luciferase; scAAV, self-complementary adeno-associated viral vector; UTR, untranslated region.
Evaluation of AAV8-Cluster 5 for potential liver toxicity
Previous studies using AAV vectors to deliver short hairpin RNAs to mouse liver and brain resulted in toxicity;15,29 however, expression of artificial miRNAs mitigated these toxic responses.15 Liver toxicity was assessed in mice injected with either scAAV8-HCV-miR-Cluster 5 or ssAAV8-HCV-miR-Cluster 5 vectors. Animals were injected with three different vector doses (2.5 × 109, 2.5 × 1010, and 2.5 × 1011 vg/mouse), and blood was collected at six different time points (pretreatment, 1, 2, 4, 8, and 27 weeks postinjection), for the analysis of eight different serum proteins. The data in Figure 8a–d demonstrate that no elevations or decreases in alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, and albumin and were observed at any time point following administration of scAAV8-HCV-miR-Cluster 5. The same was true for the four other serum proteins monitored (γ-glutamyl transferase, bilirubin, total protein, creatine phosphokinase; data not shown). In addition, no elevations of any serum proteins were observed following administration of similar doses of the ssAAV8-HCV-miR-Cluster 5 vector (data not shown). These data support the safety of AAV-mediated delivery and expression of anti-HCV miRNAs.
Figure 8.
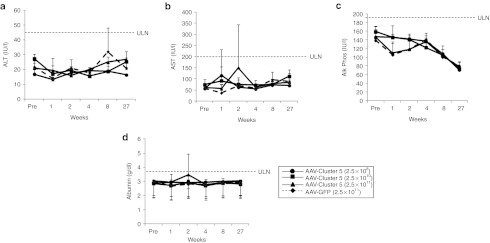
Toxicity Evaluation of scAAV8-HCV-miR-Cluster 5. Female C57/Bl6 mice (6–8 weeks) were injected with one of three doses of scAAV8-HCV-miR-Cluster 5: 2.5 × 109 vg/mouse (circles), 2.5 × 1010 vg/mouse (squares), 2.5 × 1011 vg/mouse (triangles), or 2.5 × 1011 vg/mouse of scAAV8-eGFP (dashed line, diamonds) via the tail vein. Blood collected before vector injection and at five different time points postinjection (1, 2, 4, 8, and 27 weeks) was analyzed for (a) alanine amino transferase (ALT), (b) aspartate amino transferase (AST), (c) alkaline phosphatase (Alk Phos), and (d) albumin. eGFP, enhanced green fluorescent protein; scAAV, self-complementary adeno-associated viral vector; ULN, upper limit of normal.
Discussion
The new HCV protease and polymerase DAA agents represent a new strategy in HCV therapy over traditional IFN-based treatments, because they inhibit specific HCV enzymes important for viral replication. Unfortunately, many of the DAAs quickly select for drug resistance, and have not eliminated the need for IFN-α in a treatment regimen. Because HCV has both high replication and mutation rates, all possible single and double mutants are generated multiple times each day in an infected individual.5 Therefore, resistant variants of single DAA agents naturally exist in infected individuals before drug treatment has initiated, and more will be generated during treatment. To avoid resistance emergence, it has been suggested that DAA drug combinations will need to overcome a genetic barrier of at least four mutations.5 RNAi therapeutics against HCV can also be considered DAA agents, and thus are susceptible to viral escape.30 Viral evolution around RNAi might be prevented through the simultaneous use of a mixture of antiviral RNAi molecules targeting different regions of the viral and/or cellular sequences,31,32 and in particular by targeting essential viral elements.33 By exploiting the endogenous miR-17–92 cluster, we have created an RNAi-based therapeutic for the treatment of HCV infection, which relies on viral vector-mediated delivery. The anti-HCV miRNA cluster described here expresses five biologically active anti-HCV miRNAs, and is more potent than previously constructed clusters.6 In addition, it is effective against both HCV genotypes 1b and 2a, is capable of preventing HCV spread, reduces HCV RNA in replicon-containing cells, and safely induces HCV gene silencing in vivo.
Using AAV2 vectors for in vitro delivery, scAAV2-HCV-miR-Cluster 5 inhibited bona fide HCV replication by up to 95% when HCV-infected cells were co-infected with scAAV2-HCV-miR-Cluster 5 at doses of 1 × 105–1 × 106 vg/cell. These doses of AAV led to the expression of all five anti-HCV miRNAs at levels as high as 1,300 copies/cell (miR-Core). Based on the data in Figure 2a–c and using four parameter curve fitting, the EC50 of scAAV2-HCV-miR-Cluster 5 is 1.5 × 104 vg/cell and the levels of each miRNA expressed at this dose of vector are estimated to be ~12, 47, 90, 278, and 277 miRNAs/cell for miR-UTR1, miR-UTR2, miR-UTR3, miR-Core, and miR-NS5B, respectively. However, the contribution of each miRNA toward HCV inhibition is not presently known.
Serial passage of HCVcc (genotype 2a) onto scAAV2-HCV-miR-Cluster 5-transduced cells prevented viral spread in vitro, and at a faster rate than the scAAV2-HCV-miR-Cluster 1 vector. This suggests that in HCV-infected patients, sustained expression of the miRNAs from scAAV vectors may prevent viral spread from HCV-infected hepatocytes to neighboring uninfected cells. In contrast to what was observed using scAAV2-HCV-miR-Cluster 5, a vector expressing just a single miRNA (scAAV2-HCV-miR-Core) was incapable of preventing virus spread. In that case, the HCV levels were reduced by ~100-fold, but the virus was not eliminated from the culture. Evidence for the emergence of HCV variants with sequence changes in the miR-Core target site was observed when cells were treated with either AAV-miR-Cluster 5 or AAV-miR-Core. However, in the former case, the variants disappeared from the culture, suggesting that they were either nonviable or eliminated by a different anti-HCV miRNA. In cells serially treated with AAV-miR-Core, some of the variants increased over time, suggesting that they may be true miR-Core resistant escape mutants. However follow-up studies are necessary to confirm that these sequence changes confer drug resistance, when they are in the context of an HCV genome. The data clearly demonstrate that the effectiveness of a single miRNA in inhibiting HCV replication is below a critical treatment value, and that AAV vectors expressing clusters of miRNAs prevent the spread of an HCV infection. The kinetics of AAV-Cluster 5, which expresses four miRNAs (miR-UTR1, miR-UTR2, miR-UTR3, and miR-Core) against HCV 2a was faster than AAV-Cluster 1 which expresses three anti-HCV 2a miRNAs (miR-UTR1, miR-UTR3, and miR-Core), suggesting that one additional miRNA improves efficacy. scAAV2-HCV-miR-Cluster 5 was also capable of reducing the level of HCV genotype 1b RNA in replicon-containing cells, as well as the number of replicon-containing cells, suggesting that sustained expression of the miRNAs may eventually cure cells of HCV.
HCV is a liver disease, with the majority of viral infection and replication occurring in hepatocytes. Therefore, as with small molecule drugs, it is crucial that RNAi-based therapeutics achieve adequate liver exposure. Therefore, we used AAV serotype 8 vectors to deliver HCV-miR-Cluster 5. This serotype has been shown to be more effective in transducing mouse,21 dog35 and nonhuman primate 36 liver than AAV2, and recently has been used successfully to deliver a coagulation factor IX transgene to the livers of patients with hemophilia B.37 Efficient gene transfer to mouse liver was observed using the scAAV8-HCV-miR-Cluster 5 vector at doses at or >2.5 × 1010 vg/mouse (1 × 1012 vg/kg). Mature miRNAs of ~20 bases were observed and no precursor miRNAs (~70 nt) were detected, indicating that the miRNAs were processed efficiently, in contrast to what has been observed using short hairpin RNAs.15,29 Although, in theory the HCV (−) strand can also be targeted by RNAi, the primary HCV-miR-Cluster 5 was designed to produce miRNAs whose guide strands would be selected by RISC and target the positive strand of HCV. Indeed, no passenger (or sense) strands, which are not fully complementary to HCV, were observed, suggesting that the predicted (guide) strand was loaded into the RISC, and that the passenger strand was degraded. This feature, as well as the efficient miRNA processing and liver-specific expression, should minimize the potential for off-target effects by these anti-HCV miRNAs.
Although all five miRNAs were expressed from the same primary transcript, the levels of the mature miRNAs varied by at least a log (based on QRT-PCR assays). For example, miR-UTR1 was expressed at the lowest levels both in vitro and in vivo (60–80 copies/cell) and miR-Core showed the highest levels in both situations (1,300 copies/cell). This has also been observed with the endogenous miR-17–92 transcripts, and it has been suggested that the primary RNA assumes a compact globular tertiary structure and the internalized miRNAs are processed less efficiently than the surface-exposed miRNAs.14 Of note, the miRNA levels observed in vivo are similar to or higher than those observed in HCVcc-infected Huh-7.5 cells under conditions where >95% inhibition of HCVcc replication and elimination of virus spread was observed. Because the HCV level in human hepatocytes is estimated to be ~50-fold lower than the levels observed in HCVcc-infected Huh-7.5 cells,13,38 the miRNA levels achieved in vivo at the mid and high doses of AAV may provide therapeutic benefit to HCV-infected individuals. Despite the varied miRNA levels, similar levels of gene silencing of RLuc reporter plasmids were observed in vivo by all five miRNAs, implying that the miRNAs may differ in their potency. More detailed biochemical studies are required to determine how much of the mature miRNAs are loaded into RISC and how efficiently they cleave their cognate mRNA targets.
Unlike mice, complete liver transduction in HCV-infected patients by AAV vectors is unlikely, and probably undesirable. However, this may not be necessary if the anti-HCV miRNAs can be transported via the intercellular exosomal pathway. Exosomes are 40–100 nm vesicles of endocytic origin that are formed through fusion of multivesicular bodies with the plasma membrane.39 Exosomes can mediate the transfer of functional miRNAs between many cell types, including hepatocytes.40 If used, this mechanism of miRNA transport would have significant implications for the treatment of HCV using AAV-miRNA vectors, as it would be unnecessary to transduce every HCV-infected hepatocyte.
One surprising finding was the observation that ssAAV vectors produce similar levels of miRNAs and induce gene silencing to the same levels as scAAV vectors. This contradicts a number studies that have demonstrated higher transduction levels using scAAV vectors.20,41,42,43 One complication of using the latter vectors is that assays based on QPCR often underestimate of the titers of scAAV, but not ssAAV vectors.26,44 Thus, relying on QPCR-derived titers to dose animals would result in the administration of higher doses of scAAV vectors relative to their ssAAV vector counterparts. For this reason, the titers of scAAV and ssAAV vectors containing identical expression cassettes were determined using four independent methods. When equal doses of each vector were evaluated for miRNA expression and gene silencing activity, similar levels of miR-Core and similar gene silencing of all five reporter plasmids were observed. It is too early to conclude that all scAAV vectors are affected in this way, but our data indicate that for the HCV-miR-Cluster 5 expression cassette, scAAV vectors do not have enhanced activity relative to ssAAV vectors.
AAV vectors are gaining credibility as therapeutic products.45 A variety of different AAV serotypes with different tissue tropisms are being assessed in both gene replacement and gene inhibition applications, and preclinical and clinical data are encouraging.37,46,47 Using AAV-mediated gene delivery, we have developed a treatment for HCV infection which exploits the endogenous miRNA pathway to produce five functional anti-HCV miRNAs capable of cleaving independent HCV sequences. Therefore, AAV-HCV-miR-Cluster 5 has the potential to cleave HCV genomes that have up to four nucleotide substitutions giving it a high genetic barrier to resistance, which is one of the requirements for effective treatment of HCV infection.5 The overall efficacy of the therapy will be based on efficient delivery of AAV-HCV-miR-Cluster 5 to HCV-infected cells and on the probability of each individual miRNA for cleaving its target. The model of Leonard and Schaffer33 predicts that as the cleavage probability increases, the number of RNAi effectors required for therapeutic success decreases, and that increasing the number of RNAi targets dramatically improves RNAi efficacy. Since four of the five miRNAs expressed from AAV-HCV-miR-Cluster 5 target conserved regions of the HCV genome, mutations in these regions may generate nonviable variants, preventing drug selection. The use of conserved target sites also provides the vector broad specificity toward multiple HCV genotypes. Furthermore, the use of miRNAs and a liver-specific promoter, add elements of safety to AAV-HCV-miR-Cluster 5. Assessments of hepatic toxicity for both scAAV8 and ssAAV8-HCV-miR-Cluster 5 vectors revealed no safety issues at doses up to 2.5 × 1011 vg/mouse. Significantly, the safety of peripheral vein administration of AAV8 vectors was recently demonstrated in a human clinical trial.37 Finally, the demonstration that ssAAV vectors are as effective as scAAV vectors for delivering the miRNA cluster will not only simplify the manufacture of clinical grade vector, but may avoid innate immune responses induced by the latter.48 Therefore, AAV8-HCV-miR-Cluster 5 represents a different paradigm for the treatment of HCV infection and evaluation of this approach in the clinic is warranted.
Materials and Methods
DNA constructs. The plasmid pUC19MCSD-ApoE/hAAT-HCV-miR-Cluster 26 was used to create the Cluster 5 vector plasmids that were used to generate scAAV and ssAAV vectors. First, a DNA fragment that encodes miR-UTR1 embedded in endogenous miR-92 sequences (miR-92/UTR1) was synthesized with PmeI sites at the 5′ and 3′ ends (GenScript, Piscataway, NJ). This fragment was cloned into the single PmeI site of pUC19MCSD-ApoE/hAAT-HCV-miR-Cluster 2, creating pUC19MCSD-ApoE/hAAT-HCV-miR-Cluster 4D. A DNA fragment encoding miR-17/UTR2 was synthesized with StuI and ClaI ends and was used to replace the same fragment in pUC19MCSD-ApoE/hAAT-HCV-miR-Cluster 4D, creating pUC19MCSD-ApoE/hAAT-HCV-miR-Cluster 5. This step resulted in the removal of the inactive miR-18/UTR1 from the Cluster 4D plasmid.
For construction of the AAV vector plasmid, pscAAV-HCV-miR-Cluster 5, the plasmid pUC19MCSD-ApoE/hAAT-HCV-miR-Cluster 5 was digested with AgeI and PmeI, releasing three fragments. The AgeI–PmeI fragment, which encodes miR-17/UTR2, miR-19a/UTR3, miR-20/Core, and miR-19b/NS5B, and the PmeI-PmeI fragment, which encodes miR-92/UTR1, were cloned in a three-part ligation with the AgeI–PmeI backbone fragment of pscAAV-HCV-miR-Cluster 1.6 The salient features of pscAAV-HCV-miR-Cluster 5 are in order from 5′ to 3′: (i) a WT AAV2 ITR at the 5′ end, (ii) a transcriptional regulatory element consisting of a 248 bp ApoE HCR and a 403 bp hAAT promoter, (iii) a cluster of anti-HCV miRNAs in the order of miR-17/UTR2, miR-19a/UTR3, miR-20/Core, miR-19b/NS5B, and miR-92/UTR1, (iv) a bovine growth hormone poly(A) sequence, (v) a deleted AAV4 ITR, and (vi) an ampicillin resistance marker in the plasmid backbone.
Construction of pssAAV-HCV-miR-Cluster 5 required a long series of steps, to ensure that the expression cassette was identical to that in pscAAV-HCV-miR-Cluster 5. It was also necessary to introduce a stuffer fragment between the two AAV ITRs to increase the insert size from 2.06 to 4.47 kb. A 2.4 kb fragment from intron 1 of the hypoxanthine-guanine phosphoribosyltransferase (HGPRT) gene was PCR amplified from human genomic DNA and was inserted downstream of the poly(A) sequence. It was also necessary to replace the deleted AAV4 ITR at the 3′ end of scAAV-HCV-miR-Cluster 5 with a WT AAV2 ITR. These two steps ensured that a single-stranded DNA molecule was packaged into AAV.
AAV vector production, purification and quantitation. Recombinant AAV vectors were produced by triple transfection of HEK293 cells, followed by cesium chloride-based purification, as previously described.49 Vectors were pseudotyped with either AAV2 or AAV8 capsids and physical titers were determined by QPCR using primers and probes specific to the ApoE HCR sequence (Forward primer: 5′ccactcgaccccttggaatt3′ Reverse primer: 5′aaccacgccaggacaacct3′ Probe: 5′cggtggagagg agca3′) and using linearized plasmid DNA as copy number standards.27 Vector titers were also determined by quantitative sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)/silver staining using reference vector standards.27 In addition, some vectors were also quantified by optical density28 and quantitative alkaline agarose gel, followed by Southern blotting, using different amounts (1.95–31.25 ng) of a 4,175 bp DNA fragment containing the Cluster 5 expression cassette as copy number standards. Gels were transferred to nylon membranes and hybridized to a α-P32-CTP-labeled miR-Core DNA fragment in UltraHyb buffer (Ambion, Austin, TX) according to the manufacturer's instructions. The membranes were then exposed to a phosphor screen and imaged using a Typhoon 9400 imaging system and Image Quant software (GE Healthcare Bio-Sciences, Piscataway, NJ).
In vitro gene silencing and HCVcc inhibition assays. All in vitro gene silencing assays were performed as previously described.6 The in vitro HCVcc inhibition assays were also performed as previously described,6 with a few modifications. Briefly, Huh-7.5 cells were plated in 6 well plates at 2 × 105 cells/well. Twenty-four hours later, cells were infected with either scAAV2-HCV-miR-Cluster 5 or scAAV2-eGFP, at one of four doses (1 × 103, 1 × 104, 1 × 105, and 1 × 106 vector genomes (vg)/cell), and with JFH-1 HCVcc (~0.5 FFU/cell) (kindly provided by Dr. George Luo, University of Kentucky, Lexington, Kentucky). Two hours later, the media was replaced, and the cells were incubated for an additional 48 hours. Supernatants were collected and viral RNA was isolated using the QIAamp viral RNA mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Total cellular RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. HCV RNA levels were quantified by QRT-PCR as described below. All infections were performed in duplicate.
HCV spread assay. Huh-7.5 cells were plated in 6 well plates at 2 × 105 cells/well. Twenty-four hours later, cells were infected with either scAAV2-HCV-miR-Cluster 5, scAAV2-HCV-miR-Cluster 1, scAAV2-miR-Core, or scAAV2-miR-Irr, at a dose of 1 × 105 vg/cell, and with JFH-1 HCVcc (~0.5 FFU/cell) (kindly provided by Dr. George Luo, University of Kentucky, Lexington, Kentucky). Negative control cells were left untreated. Two hours later, the media was replaced, and the cells were incubated for an additional 48 hours. At this time, 1 ml of supernatant from each treatment group was used to infect HCVcc-naïve Huh-7.5 cells that had been transduced 24 hours earlier with either scAAV2-HCV-miR-Cluster 5, scAAV2-HCV-miR-Cluster 1, scAAV2-miR-Core, or scAAV2-miR-Irr, at a dose of 1 × 105 vg/cell, or were untreated (total volume media = 2 ml). Total cellular RNA from the initially infected cells was isolated using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, and HCV RNA levels were quantified in triplicate by QRT-PCR. This procedure was continued for a total of 10 rounds. All infections were performed in duplicate. A preliminary HCV spread assay was performed for a total of six rounds, and evaluated the generation of escape mutants (Supplementary Materials and Methods).
In a separate experiment, to evaluate HCV replication in the absence of miRNA selective pressure, 0.5 ml of supernatant from the scAAV2-HCV-miR-Cluster 5 treatment group (rounds 1, 3, 6, and 10) and from the same rounds of the nontreatment group was used to infect naïve Huh-7.5 cells (total volume media = 2 ml). Forty-eight hours later, cellular RNA was isolated and HCV RNA levels were quantified by QRT-PCR. In addition, the supernatants from the round 10 samples were serially passaged for two additional rounds and intracellular HCV RNA levels were measured 48 hours later.
HCV replicon clearance and rebound assay. Huh-7 cells harboring an autonomously replicating subgenomic HCV genotype 1b replicon (kindly provided by Dr. George Luo, University of Kentucky, Lexington, Kentucky) were maintained in Dulbecco's modified Eagle medium, 10% heat-inactivated fetal bovine serum, 2mmol/l L-glutamine, nonessential amino acids, and 0.5 mg/ml G418. For the HCV replicon clearance and rebound assay, the replicon cells were plated in the absence of G418 in 6 well plates at a density of 2 × 105 cells/well. Twenty-four hours after plating, the cells were transduced with scAAV2-HCV-miR-Cluster 5 or AAV-miR-Irr at a dose of 1 × 105 vg/cell (in replicates of six). Positive control cells were treated with IFN-α (300 U/ml) and negative controls were left untreated. Three days later, cells from three of the six wells were harvested for RNA isolation using Trizol, and HCV levels were measured using QRT-PCR. The cells from the other three wells were trypsinized, pooled and re-plated in all six wells of six well plates at a density of 2 × 105 cells/well. Each entire six well plate was then re-transduced with either scAAV2-HCV-miR-Cluster 5 or scAAV-miR-Irr at 1 × 105 vg/cell, retreated with IFN-α or not treated. This procedure was repeated for a total of six rounds of propagation. At the end of the sixth round, 1 × 105 cells from each treatment group were plated into one 10-cm dish in the presence of G418 (0.5 mg/ml) and were cultured for 2 weeks. The G418-resistant colonies were fixed and stained with 0.05% crystal violet.
Animal procedures. All animal studies were conducted at the Children's Hospital of Philadelphia with approval from the Institutional Animal Care and Use Committee. The in vivo gene silencing studies, using female C57Bl/6 mice (Charles River Labs; Wilmington, MA), relied on hydrodynamic tail injection22 of RLuc-HCV reporter plasmids, as previously described.6 Briefly, scAAV8-HCV-miR-Cluster 5 and ssAAV8-HCV-miR-Cluster 5 vectors, at doses ranging from 2.5 × 109 vg/mouse 2.5 × 1011 vg/mouse, were injected into the tail vein of mice using low pressure (n = 5). Control animals received scAAV8-eGFP vectors (2.5 × 1011 vg/mouse) (n = 5). Two weeks later, an hydrodynamic tail vein injection of one of five RLuc-HCV reporter plasmids was performed. Two days later, the animals were sacrificed, livers were harvested and stored at −80 °C until processing. For assessment of liver toxicity, eight other cohorts of mice were injected with scAAV8-HCV-miR-Cluster 5 or ssAAV8-HCV-miR-Cluster 5 at one of three doses (2.5 × 109, 2.5 × 1010, and 2.5 × 1011 vg/mouse), and serum was analyzed for eight different serum proteins at six different time points (pretreatment, 1, 2, 4, 8, and 27 weeks postinjection) by IDEXX (North Grafton, MA). Control animals were injected with scAAV8-eGFP (2.5 × 1011 vg/mouse).
Biochemical analysis of mouse liver. Frozen (−80 °C) whole mouse livers were ground using a freezer/mill (Spex CertiPrep, Metuchen, NJ) in liquid nitrogen using three, 1-minute grinding cycles spaced by three, 1-minute cooling cycles. Ground livers were stored at −80 °C until used for protein, DNA, or RNA isolation. Protein lysates of frozen ground liver were prepared by adding 200 µl of Passive Lysis Buffer (Promega, Madison, WI) to ~100 mg frozen liver. The luciferase activity in 10 µl lysate was determined using the Dual Luciferase Assay (Promega) on a Veritas luminometer (Turner Biosystems, Sunnyvale, CA). Relative RLuc activity (RLuc/FFLuc) in each sample was normalized to control animal lysates, which were set as 100% activity. Two to three independent lysates were prepared for each animal and each lysate was analyzed in triplicate for dual luciferase activity. Results are reported as the mean and SD of one representative lysate. Liver DNA was analyzed for AAV copy number by Southern blotting (Supplementary Materials and Methods).
Northern blot analysis of mouse liver and Huh-7.5 cellular RNA. For analysis of anti-HCV miRNAs from mouse liver, 2 ml of TRizol reagent (Invitrogen, Carlsbad, CA) was added to ~200 mg frozen ground liver tissue, and total RNA was extracted according to the manufacturer's protocol and quantified on a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE). Twenty-five micrograms of total RNA was resolved on 15% denaturing polyacrylamide TBE -urea gels (Invitrogen, Carlsbad, CA). Decade RNA molecular weight markers (Ambion) were labeled with γ-P32-ATP as according to the manufacturer's instructions, and were run adjacent to liver RNA samples. RNA was transferred to nylon membranes, which were prehybridized using UltraHyb-Oligo buffer (Ambion) for 1 hour at 65 °C and subsequently probed with γ-P32-ATP labeled DNA oligonucleotide probes at room temperature overnight. The blots were washed three times at room temperature and once at 42 °C for 30 minutes with 6× SSC/0.2% SDS, exposed to film, and developed using a Kodak processor. The DNA oligonucleotide sequences used as probes were as follows: miR-UTR1 guide probe: 5′-CCATAGTGGTCTGCGGAAC-3′, miR- UTR2 guide probe: 5′-AAAGGCCTTGTGGTACTGCCT-3′, miR-UTR3 guide probe: 5′-AGGTCTCGTAGACCGTGCA-3′, miR-Core guide probe: 5′-AACCTCAAAGAAAAACCAAAC-3′, miR-NS5B guide probe: 5′-GAC ACTGAGACACCAATTGAC-3′, U6 small nuclear RNA probe: 5′-TATGG AACGCTTCACGAATTTGC-3′. The RNA oligonucleotides used as positive controls were as follows: miR-UTR1 (guide strand): 5′-GUUCC GCAGACCACUAUGG-3′, miR-UTR2 (guide strand) 5′-AGGCAG UACCACAAGGCCUUU-3′, miR-UTR3 (guide strand) 5′-UGCACGG UCUACGAGACCU-3′, miR-Core (guide strand) 5′-GUUUGGUUUUU CUUUGAGGUU-3′, miR-NS5B (guide strand) 5′-GUCAAUUGGUGU CUCAGUGUC-3′.
For analysis of RLuc-Core mRNA, 12 µg of total cellular RNA was separated on a 1% agarose gel prepared in Northern Max denaturing gel buffer and run in Northern Max gel running buffer (Ambion). Millennium RNA marker (Ambion) was used as a molecular weight standard. The RNA fragments were transferred to Bright Star Plus positively charged nylon membranes (Ambion) and hybridized to a α-P32-dCTP labeled RLuc DNA probe, isolated from psiCHECK2 (Promega), in UltraHyb buffer (Ambion), according to the manufacturer's instructions. The blots were washed two times at 42 °C with 2× SSC/0.1% SDS for 5 minutes and twice with 0.1× SSC/0.1% SDS for 5 minutes. The membrane was exposed to film, and developed using a Kodak processor. The membrane was stripped and reprobed with a α-P32-dCTP labeled β-actin probe to evaluate equal loading of samples.
Northern blot analysis of miR-122 from Huh-7.5 cells was performed by purifying cellular RNA from cells using Trizol according to the manufacturer's instructions, resolving 1 µg RNA on a 15% denaturing polyacrylamide TBE-urea gel, and performing a Northern blot using a γ-P32- ATP labeled oligonucleotide probe specific to miR-122 guide strand (5′ACAAA CACCATTGTCACACTCCA3′) under conditions identical to those used for the anti-HCV miRNAs. The membrane was stripped and reprobed with a U6 small nuclear RNA probe to evaluate equal loading of samples.
QRT-PCR analyses of HCV RNA and miRNAs. HCV RNA was measured using the TaqMan One Step RT-PCR kit (Applied Biosystems, Foster City, CA) and the primers and probe detailed in Takeuchi et al.50 In vitro transcribed JFH-1 RNA was generated from plasmid pJFH-1 (Apath, Brooklyn, NY) according to Kato et al.,11 and was used to generate a standard curve. The assay sensitivity was 1 × 103 HCV RNA copies/200 ng cellular RNA (~0.05 copies/cell).
Quantitation of the anti-HCV miRNAs in Huh-7.5 cell extracts and mouse liver RNA was performed using custom TaqMan small RNA QRT-PCR assays (Applied Biosystems) according to the manufacturer's instructions, using synthetic oligonucleotides to generate standard curves. The RNA oligonucleotides that were used as positive controls in the miRNA Northern blots were used to generate the standard curves. The assay sensitivity was 67 miRNA copies/13.3 ng cellular RNA (~0.05 copies/cell) for miR-UTR, miR-UTR2, miR-Core, and miR-NS5B, and was 10-fold less sensitive for miR-UTR3.
Quantitation of miR-122 in Huh-7.5 cells was performed using a custom TaqMan small RNA QRT-PCR assay (Applied Biosystems) according to the manufacturer's instructions, using a synthetic miR-122 oligonucleotide (5′UGGAGUGUGACAAUGGUGUUUGU3′) to generate a standard curve. The assay sensitivity was 1 × 103 miRNA copies/13.3 ng cellular RNA (~0.75 copies/cell).
Statistical analysis. Two tailed Student's t-tests were performed. P values of 0.05 or 0.01 were used to assess statistical significance.
SUPPLEMENTARY MATERIAL Figure S1. AAV vectors expressing HCV-miRNA-Cluster 5 prevent HCV spread. Figure S2. Southern blot analyses of mouse liver DNA. Table S1. Sequence variation in miR-Core target site of HCV2a genome after various treatments (454/Roche PyroSequencing Analysis). Materials and Methods.
Acknowledgments
We thank the research vector core at CHOP for manufacture of AAV vectors. We also thank Dr. George Luo (University of Kentucky, Lexington, KY) for generously providing the Huh-7.5 and HCV genotype 1b replicon cell lines, and for HCVcc. In addition, we are grateful to Dr. Laura Steel (Drexel University College of Medicine, Philadelphia, PA) for providing Huh-7 cells. We also acknowledge Drs. Tapan Ganguly, Scott Sherrill-Mix, and Rohini Sinha (University of Pennsylvania School of Medicine, Philadelphia, PA) for pyrosequencing and bioinformatics analysis. We also thank Drs. Joseph Couto, Federico Mingozzi, and Greg Podsakoff for helpful discussions and for critical reading of the manuscript. This work was funded by the Center for Cellular and Molecular Therapeutics, Children's Hospital of Philadelphia, Philadelphia, PA. and a grant (QED A1001) from the Philadelphia Science Center (L.B.C.).
Supplementary Material
References
- Wasley A., and, Alter MJ. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin Liver Dis. 2000;20:1–16. doi: 10.1055/s-2000-9506. [DOI] [PubMed] [Google Scholar]
- De Francesco R., and, Migliaccio G. Challenges and successes in developing new therapies for hepatitis C. Nature. 2005;436:953–960. doi: 10.1038/nature04080. [DOI] [PubMed] [Google Scholar]
- Nunnari G., and, Schnell MJ. MicroRNA-122: a therapeutic target for hepatitis C virus (HCV) infection. Front Biosci (Schol Ed) 2011;3:1032–1037. doi: 10.2741/207. [DOI] [PubMed] [Google Scholar]
- Bourlière M, Khaloun A, Wartelle-Bladou C, Oules V, Portal I, Benali S.et al. (2011Chronic hepatitis C: treatments of the future Clin Res Hepatol Gastroenterol 35 Suppl 2S84–S95. [DOI] [PubMed] [Google Scholar]
- Rong L, Dahari H, Ribeiro RM., and, Perelson AS. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci Transl Med. 2010;2:30ra32. doi: 10.1126/scitranslmed.3000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Haurigot V, Zhou S, Luo G., and, Couto LB. Inhibition of hepatitis C virus replication using adeno-associated virus vector delivery of an exogenous anti-hepatitis C virus microRNA cluster. Hepatology. 2010;52:1877–1887. doi: 10.1002/hep.23908. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A., and, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N., and, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Aagaard LA, Zhang J, von Eije KJ, Li H, Saetrom P, Amarzguioui M.et al. (2008Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs Gene Ther 151536–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP, Haasnoot J, ter Brake O, Berkhout B., and, Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Date T, Murayama A, Morikawa K, Akazawa D., and, Wakita T. Cell culture and infection system for hepatitis C virus. Nat Protoc. 2006;1:2334–2339. doi: 10.1038/nprot.2006.395. [DOI] [PubMed] [Google Scholar]
- Jiang J., and, Luo G. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J Virol. 2009;83:12680–12691. doi: 10.1128/JVI.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Williams O, Mittler J, Quintanilla A, Carithers RL, Jr, Perkins J.et al. (2003Dynamics of hepatitis C virus replication in human liver Am J Pathol 163433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaulk SG, Thede GL, Kent OA, Xu Z, Gesner EM, Veldhoen RA.et al. (2011Role of pri-miRNA tertiary structure in miR-17~92 miRNA biogenesis RNA Biol 81105–1114. [DOI] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I.et al. (2008Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi Proc Natl Acad Sci USA 1055868–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM., and, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Niemeyer GP, Herzog RW, Mount J, Arruda VR, Tillson DM, Hathcock J.et al. (2009Long-term correction of inhibitor-prone hemophilia B dogs treated with liver-directed AAV2-mediated factor IX gene therapy Blood 113797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai H, Yant SR, Storm TA, Fuess S, Meuse L., and, Kay MA. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J Virol. 2001;75:6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perni RB, Almquist SJ, Byrn RA, Chandorkar G, Chaturvedi PR, Courtney LF.et al. (2006Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease Antimicrob Agents Chemother 50899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM, Monahan PE., and, Samulski RJ. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8:1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J., and, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Song Y., and, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- Meister G., and, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N., and, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, Ambegia E, Choi C, Yaworski E, Palmer L.et al. (2008Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation Hum Gene Ther 19991–999. [DOI] [PubMed] [Google Scholar]
- Fagone P, Wright JF, Nathwani AC, Nienhuis AW, Davidoff AM., and, Gray JT. Systemic errors in quantitative polymerase chain reaction titration of self-complementary adeno-associated viral vectors and improved alternative methods. Hum Gene Ther Methods. 2012;23:1–7. doi: 10.1089/hgtb.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JF., and, Zelenaia O. Vector characterization methods for quality control testing of recombinant adeno-associated viruses. Methods Mol Biol. 2011;737:247–278. doi: 10.1007/978-1-61779-095-9_11. [DOI] [PubMed] [Google Scholar]
- Sommer JM, Smith PH, Parthasarathy S, Isaacs J, Vijay S, Kieran J.et al. (2003Quantification of adeno-associated virus particles and empty capsids by optical density measurement Mol Ther 7122–128. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR.et al. (2006Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways Nature 441537–541. [DOI] [PubMed] [Google Scholar]
- Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S.et al. (2003RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells Proc Natl Acad Sci USA 1002783–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B. RNA interference as an antiviral approach: targeting HIV-1. Curr Opin Mol Ther. 2004;6:141–145. [PubMed] [Google Scholar]
- Lee NS., and, Rossi JJ. Control of HIV-1 replication by RNA interference. Virus Res. 2004;102:53–58. doi: 10.1016/j.virusres.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Leonard JN., and, Schaffer DV. Computational design of antiviral RNA interference strategies that resist human immunodeficiency virus escape. J Virol. 2005;79:1645–1654. doi: 10.1128/JVI.79.3.1645-1654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedj J, Rong L, Dahari H., and, Perelson AS. A perspective on modelling hepatitis C virus infection. J Viral Hepat. 2010;17:825–833. doi: 10.1111/j.1365-2893.2010.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Calcedo R, Nichols TC, Bellinger DA, Dillow A, Verma IM.et al. (2005Sustained correction of disease in naive and AAV2-pretreated hemophilia B dogs: AAV2/8-mediated, liver-directed gene therapy Blood 1053079–3086. [DOI] [PubMed] [Google Scholar]
- Gao G, Lu Y, Calcedo R, Grant RL, Bell P, Wang L.et al. (2006Biology of AAV serotype vectors in liver-directed gene transfer to nonhuman primates Mol Ther 1377–87. [DOI] [PubMed] [Google Scholar]
- Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC.et al. (2011Adenovirus-associated virus vector-mediated gene transfer in hemophilia B N Engl J Med 3652357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiffler JD, Nguyen M, Sohn JA, Liu C, Kaplan D., and, Seeger C. Focal distribution of hepatitis C virus RNA in infected livers. PLoS ONE. 2009;4:e6661. doi: 10.1371/journal.pone.0006661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorey JS., and, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde-Vancells J, Rodriguez-Suarez E, Embade N, Gil D, Matthiesen R, Valle M.et al. (2008Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes J Proteome Res 75157–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ma HI, Li J, Sun L, Zhang J., and, Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- Nathwani AC, Gray JT, Ng CY, Zhou J, Spence Y, Waddington SN.et al. (2006Self-complementary adeno-associated virus vectors containing a novel liver-specific human factor IX expression cassette enable highly efficient transduction of murine and nonhuman primate liver Blood 1072653–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM, Fu H, Monahan PE, Toulson CE, Naik P., and, Samulski RJ. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- Allay JA, Sleep S, Long S, Tillman DM, Clark R, Carney G.et al. (2011Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for a hemophilia B clinical trial Hum Gene Ther 22595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F., and, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- McBride JL, Pitzer MR, Boudreau RL, Dufour B, Hobbs T, Ojeda SR.et al. (2011Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington's disease Mol Ther 192152–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonelli F, Maguire AM, Testa F, Pierce EA, Mingozzi F, Bennicelli JL.et al. (2010Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration Mol Ther 18643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino AT, Suzuki M, Markusic DM, Zolotukhin I, Ryals RC, Moghimi B.et al. (2011The genome of self-complementary adeno-associated viral vectors increases Toll-like receptor 9-dependent innate immune responses in the liver Blood 1176459–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Zhou S, Nakai H, Thomas CE, Storm TA, Fuess S.et al. (2003Preclinical in vivo evaluation of pseudotyped adeno-associated virus vectors for liver gene therapy Blood 1022412–2419. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Katsume A, Tanaka T, Abe A, Inoue K, Tsukiyama-Kohara K.et al. (1999Real-time detection system for quantification of hepatitis C virus genome Gastroenterology 116636–642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.