Abstract
Squamous cell carcinoma of the head and neck (HNSCC) is a relatively common human cancer characterized by high morbidity, high mortality, and few therapeutic options outside of surgery, standard cytotoxic chemotherapy, and radiation. Although the most important risk factors are tobacco use and alcohol consumption, the disease is also linked to infection with high-risk types of human papilloma viruses (HPVs). Recent genetic analyses have yielded new insights into the molecular pathogenesis of this disease. Overall, while somatic activating mutations within classical oncogenes including PIK3CA and RAS occur in HNSCC, they are relatively uncommon. Instead genetic data point to a contribution of multiple tumor suppressor pathways, including p53, Rb/INK4/ARF, and Notch, in tumor initiation, progression, and maintenance. The increasingly refined knowledge of HNSCC genetics, combined with ever-more-sophisticated animal models and newer drug targeting strategies, should promote novel therapeutic approaches and improved disease outcomes.
Clinical and epidemiological features
Squamous cell carcinoma of the head and neck (HNSCC) is the sixth most common cancer worldwide, affecting 600,000 new patients each year. In the United States, 50,000 new cases are diagnosed, and nearly 10,000 deaths are attributable to this disease annually. Despite their common origin in the squamous mucosa of the upper aerodigestive tract, there are several sources for the heterogeneity of HNSCC that complicate their consideration as a single disease entity. Anatomically, the head and neck region consists of several distinct structures (e.g., lip, tongue, nasopharynx, oropharynx, larynx, hypopharynx) with distinct microscopic features and lymphatic and venous drainage. As a result, treatment approaches and outcomes vary significantly among the different subsites. In addition, etiologic risk factors vary among the different subsites, and HNSCC can occur in young patients without any known risk factors. Biologic heterogeneity independent of subsite has been suggested by molecular analyses that have uncovered distinct classes of HNSCC with unique mRNA expression profiles or patterns of DNA copy number alterations that correlate with clinical behavior (1–4).
Although the most important risk factors are tobacco use and alcohol consumption, infection with high-risk types of human papilloma viruses (HPVs) has also been recognized as an increasingly important risk factor, particularly for oropharyngeal squamous cell carcinoma (OPSCC). Importantly, survival for patients with HPV-positive OPSCC is significantly better than for HPV-negative cancers, irrespective of treatment modality (5–8). Possible explanations include both host-intrinsic factors (healthier patients with fewer comorbidities) and tumor-intrinsic factors (distinct genetic pathogenesis yielding enhanced treatment sensitivity). The relatively good prognosis of HPV-related cancers highlights an unfortunate paradox — despite improvements in outcome for patients with oropharynx cancers, little or no improvement has been observed for patients with HPV-negative tumors over the past 30 years (9). This fact is attributable at least in part to our limited understanding of molecular pathways that both promote HSNCC pathogenesis and can be targeted therapeutically.
Linking pathways to genetics
Recent advances in our understanding of the molecular pathogenesis of HNSCC were provided by whole-exome sequencing (i.e., sequencing exons of all known protein-coding genes) conducted on a total of approximately 100 HNSCC specimens independently by two groups (10, 11). While the two studies analyzed etiologically similar tumors with related sequencing platforms, there was a five-fold difference in the average number of mutations reported per tumor. This difference likely reflects distinct bioinformatic and validation approaches used in the studies, and therefore a subset of identified changes may represent “passenger” mutations (as a result of increased mutation rates in cancer cells, or even mutation “miscalls”) rather than true “driver” mutations with an etiologic role in HNSCC. Nevertheless, several key findings were shared by these studies. This work, together with a large body of previous genomic and functional analyses of HNSCC, highlights the relatively small number of oncogenes targeted by activating mutations and supports the fundamental roles of tumor suppressor pathways including p53, Rb/INK4/ARF, and Notch in disease pathogenesis. These and other bona fide HNSCC cancer genes play major roles in at least four key functional pathways: cellular proliferation, squamous epithelial differentiation, cell survival, and invasion/metastasis, with many of the genes impacting more than a single pathway (Figure 1). These pathways are critical to the pathogenesis of HNSCC and, not surprisingly, reflect normal developmental programs within the stratified squamous epithelium. Given the paucity of driver oncogenes in HNSCC, targeting these pathways therapeutically represents a substantial and critical challenge for improving outcomes of this disease.
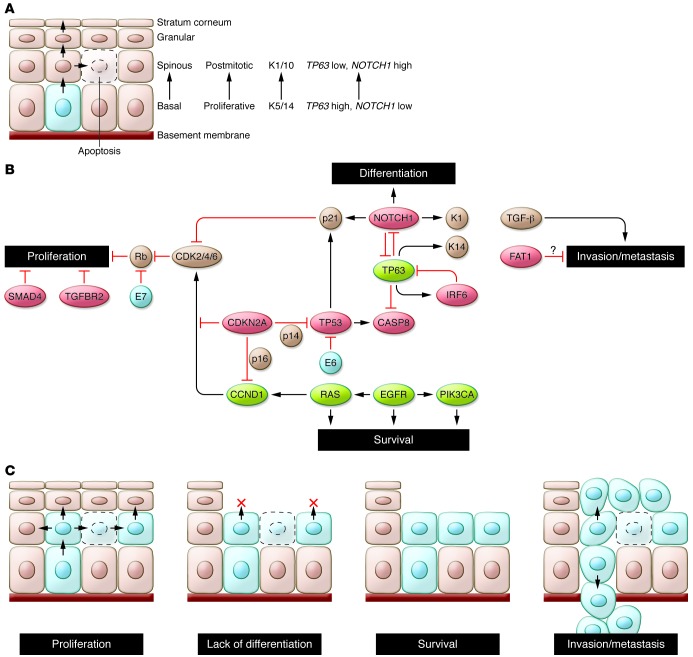
Figure 1. Hallmarks of head and neck squamous tumorigenesis.
(A) The normal process of squamous morphogenesis in the adult mucosa is controlled in part by TP63 and NOTCH1. The former is expressed in keratinocytes of the basal layer, where it maintains their proliferative potential and controls expression of basal markers (e.g., keratins 5/14 [K5/14]); expression of the latter results in terminal differentiation into cells of the spinous (K1/10) and granular layers. Rare stem cells in the basal layer (light blue) undergo terminal differentiation through asymmetric cell division. Abnormal proliferation is prevented primarily by differentiation-associated cell cycle exit and by apoptosis. (B) Pathways altered in HNSCC pathogenesis identified in whole-exome sequencing studies. Red: putative and established tumor suppressors; green: oncogenes; brown: other relevant genes/proteins; blue: viral proteins. Loss of TP53 and CDKN2A, amplification of CCND1, and loss of TGFBR2/SMAD4 permit abnormal proliferation and decrease apoptosis. However, abnormal cell cycling may still be restrained by intact differentiation and apoptotic programs. Loss of NOTCH1 and/or abnormal expression of TP63, together with alterations in “survival” genes (e.g., CASP8, PIK3CA, EGFR), may remove additional barriers to tumor cell proliferation and survival. Loss of cell adhesion genes (e.g., FAT1) could permit release of cells from the mucosal lining, while invasion through the basement membrane is promoted by TGFB1 (and SMAD3). (C) Schematic of HNSCC hallmarks. The precise order of acquisition of distinct alterations is not clear. In addition, several genes (e.g., TP53, TP63, NOTCH1) may contribute to more than one hallmark.
Cellular proliferation and p53/Rb/CDKN2A/CCND1
Mutation of the TP53 tumor suppressor gene is the most common and among the earliest identified genetic alterations in HNSCC, occurring in more than half of all cases (2). As in other human cancers, missense mutations primarily within the DNA binding domain account for 75% of all mutations in the TP53 gene and confer both dominant negative and poorly understood gain-of-function properties (12–15). In many of the remaining HNSCC tumors in which p53 is wild-type, p53 function may be inactivated by other mechanisms. These include expression of the HPV E6 protein (which binds p53 and targets it for proteasomal degradation), overexpression/amplification of MDM2 (which also mediates p53 proteasomal degradation), and deletion of CDKN2A, which may eliminate p14/ARF, a negative regulator of MDM2 (16–19). Overall, the data suggest that the p53 pathway is downregulated in at least 80% of HNSCCs (2).
The finding that TP53 is mutated in both leukoplakia (a histologically recognizable precursor lesion) and benign-appearing mucosa has led to a “patch-field” progression model of HNSCC development, in which the index squamous carcinoma (as well as subsequent tumors) develops from a field of genetically abnormal mucosa, itself the result of expansion of a clonal patch arising from a putative stem cell containing a mutated TP53 gene (20). Interestingly, in some cases the TP53 mutations found in the tumor and adjacent mucosa are different, implying a distinct clonal origin for multiple patches and suggesting that metachronous tumors from the same patient (e.g., primary versus locally recurrent) could in fact develop from unique clones through independent acquisition of additional alterations (21). In addition to tumor initiation, TP53 inactivation also contributes to the clinical behavior of tumors, at least in part independent of an influence on the response to genotoxic therapy. Thus, truncating and function-disrupting mutations of TP53 are significantly associated with decreased survival — after primary surgery with or without postoperative radiotherapy — compared with either non-disruptive mutations or no mutation at all (22, 23).
The essential role of the retinoblastoma (Rb) pathway is evidenced by the finding of inactivation of CDKN2A, encoding the cell cycle regulators p16/INK4A and p14/Arf/INK4B, in HNSCC. CDKN2A mutations were found in approximately 7% of tumors by exome sequencing, with copy number losses in another 20%–30% (10, 11). It has been previously shown that CDKN2A inactivation by mutation is significantly more rare than deletion or epigenetic inactivation, which together account for inactivation of the gene in up to 75% of HNSCCs (24–26). Although p16/INK4A loss (whether genetic or functional) has been repeatedly demonstrated to correlate with indicators of worse prognosis, data on p14/Arf/INK4B loss (e.g., by methylation, when the genomic locus itself is not deleted) is conflicting, with one study suggesting worsened prognosis, while two others suggested improved prognosis, perhaps a result of increased radiation sensitivity (27–29). In the case of HPV+ HNSCC, inactivation of the Rb pathway is achieved through expression of the HPV E7 protein, which binds RB1 and abrogates the requirement for p16/INK4A silencing. As a result, assaying p16 protein expression in tumor cells by immunohistochemistry (IHC) is of clinical value in determining HPV+ status (30).
Amplification of a discrete, approximately 5-Mb region of chromosome 11q13 containing the CCND1 gene (encoding cyclin D1) occurs in approximately one-third of HNSCCs, and perhaps even more frequently in HPV-negative tumors (4, 31). Furthermore, overexpression of cyclin D1 has been observed in up to 80% of HNSCCs (2). This high frequency is remarkable given that CDKN2A loss or CCND1 amplification would seem to be redundant mechanisms to promote cell cycle progression through activation of G1 phase cyclin-dependent kinases (CDKs) 4 and 6. Nevertheless, these two genetic events are not mutually exclusive in HNSCC, potentially reflecting either qualitatively or quantitatively different effects. For example, cyclin D1 may indirectly stimulate CDK2 activity by sequestering the CDK2 inhibitors p21 and p27, and alternatively, cyclin D1 may function as a cofactor independent of its role in cell cycle regulation, through binding to transcription factors (e.g., PPARγ) or DNA repair proteins (e.g., BRCA2, Rad51) (refs. 32, 33; reviewed in ref. 34). In keeping with their distinct contributions, loss of p16 expression and overexpression of cyclin D1 are independent predictors of death from tongue cancer, and the loss of p16 together with overexpression of cyclin D1 confers significantly worse 5 year survival than either condition observation alone (35).
Terminal differentiation and the Notch/p63 axis
Perhaps the most novel finding to emerge from the whole-exome sequencing studies of HNSCC is the discovery of mutations within the NOTCH1 gene in 12%–15% of cases, and within additional NOTCH family members in 3%–5% (10, 11). Although Notch signaling had previously been implicated as pro-tumorigenic — by virtue of activating mutations and translocations observed in the genes for Notch receptors or their regulators in T cell acute lymphoblastic leukemia, chronic lymphocytic leukemia, and diffuse large B-cell lymphoma (36–40) — several of the NOTCH family mutations in HNSCC (and in chronic myelomonocytic leukemia, a rare myeloproliferative disease) encode inactivating mutations, suggesting a tumor suppressor function (41). The physiologic relevance of these findings is supported by animal models in which NOTCH activation in hematopoietic cells leads to T cell leukemias and inactivation in squamous epidermis promotes skin tumorigenesis (refs. 42–44; reviewed in ref. 45).
Notch signaling has been linked to multiple biological functions, including regulation of self-renewal capacity, cell cycle exit (in part through upregulation of p21/CDKN1A expression), and cell survival (46–48). In the stratified epithelium, Notch has a central role in promoting terminal differentiation (48, 49), which is mediated through both direct effects (e.g., on activation of suprabasal keratins) and indirect effects on the Wnt, hedgehog, and interferon response pathways (43, 47, 50, 51). Additionally, Notch activity has been linked to suppression of HPV E6 and E7 protein expression, potentially providing additional selective pressure for loss of Notch in HPV+ HNSCC (52, 53).
Further supporting a role for Notch in squamous epithelial differentiation is its control by the p53-related transcription factor p63, a master regulator of proliferative potential, lineage specification, and differentiation in stratified epithelia. Constitutive knockout of Tp63 in mouse models results in complete failure of normal epidermal development (54, 55). In mature epithelium, expression of p63 is highest in basal epithelial cells, where it functions as an inhibitor of NOTCH1 expression, and becomes downregulated during terminal differentiation coincident with NOTCH1 upregulation (Figure 1A and ref. 56). Reactivation of p63 expression is observed in the suprabasal layers of dysplastic mucosa, and overexpression and/or genomic amplification of the TP63 locus is observed in the majority of invasive HNSCCs (57, 58). TP63 gives rise to two major isoform classes, TAp63 and ΔNp63, which differ in the presence and absence, respectively, of an N-terminal transcriptional transactivation domain. Although tumor incidence data from Tp63-heterozygous mice are conflicting, ΔNp63 isoforms, which are selectively overexpressed in HNSCC, are likely oncogenic (59, 60). Importantly, TP63 was found to be mutated or amplified in 8% of samples in one of the sequencing studies (11). Consistent with a contribution of ΔNp63 in these tumors, two of the mutations uncovered are predicted to alter the function of TAp63 (including a nonsense mutation) but not ΔNp63. In addition to its contribution through Notch suppression, ΔNp63 has been demonstrated to control other key tumor-relevant pathways, including cell survival (in part through suppression of the proapoptotic p53-related protein p73), senescence suppression (through suppression of p16/INK4A expression), and growth factor signaling (through induction of EGFR) (61–64).
Cell survival through EGFR/Ras/PIK3CA/PTEN/CASP8
The PI3K signaling pathway is commonly activated in HNSCC, as evidenced by recurrent alterations of two central regulators: PTEN, encoding a negative regulator, and PIK3CA, encoding a positive regulator of this pathway. PTEN is subject to frequent loss of heterozygosity in a variety of cancers, including up to 40% of HNSCCs, although biallelic inactivation occurs less frequently (65–68). Loss of just a single PTEN allele in the remaining samples may contribute to tumorigenesis, however, since recent data suggesting a gene dosage effect for PTEN (69). Activating mutations in two “hot spot” regions of the PIK3CA gene occur in 6%–11% of HNSCCs, with a potential enrichment in tumors originating from the pharynx (70, 71). The latter finding is particularly noteworthy given the increased incidence of PIK3CA mutations in HPV-related versus non-HPV-related tumors observed in both exome sequencing studies. This observation suggests that PIK3CA mutations may cooperate with HPV E6 and E7 proteins in the development of invasive OPSCC, as has been suggested for cervical carcinomas (72). The prominent role of the PI3K pathway in HNSCC has potentially important clinical implications, given that numerous targeted inhibitors of this pathway are currently being evaluated in clinical trials (reviewed in ref. 73).
Activating missense mutations causing single amino acid substitutions in one of three positions (codon 12, codon 13, and codon 61) in the HRAS gene were uncovered in 3%–5% of samples in both whole-exome sequencing studies. While it is currently unknown whether HRAS-dependent signals function in collaboration with or independently of PI3K activation in HNSCC, several findings underscore the importance of this particular RAS family member to the pathogenesis of the disease. These include the more frequent occurrence of HRAS than KRAS mutations in HNSCC, particularly in relationship to tobacco history, whereas the reverse is true for several other malignancies (74, 75); the presence of HRAS mutations in HPV-driven tumors, suggesting potential cooperativity in tumor promotion (76); and the more frequent association of HRAS versus KRAS mutation in squamous cell carcinomas arising in the setting of tobacco exposure in humans and chemical carcinogen exposure in mice (77). Although Ras proteins themselves have proven difficult to target directly, therapeutic strategies that target downstream effectors of Ras proteins or the synthetic lethal dependencies that result from their mutational activation have already been successful in preclinical models (78–80).
Upstream signaling to both Ras and PI3K pathways may occur through activation or overexpression of receptor tyrosine kinases (RTKs) including EGFR. Although it is often considered to be among the most important therapeutic targets in HNSCC (81), our understanding of the role of EGFR is evolving with the appreciation that EGFR activating mutations are rare in HNSCC and that the reported frequency of EGFR gene amplification in HNSCC varies widely, in part due to varying definitions of the degree and size of copy number gain that constitute amplification (82, 83). Furthermore, although copy number gain of EGFR has been suggested to correlate with poor prognosis in HNSCC (83, 84), in general gain of EGFR has not been clearly demonstrated to predict improved outcomes following EGFR-directed therapy (85, 86). Similarly, therapeutic agents that inhibit EGFR, including the small molecule inhibitors gefitinib and erlotinib and the therapeutic antibody cetuximab, have modest activity in HNSCC, with little or no correlation with EGFR status (87–91).
Two other genetic abnormalities affecting RTK signaling have received less attention but have potential near-term clinical impact. Expression of EGFRvIII, a variant EGFR protein that results from the in-frame genomic deletion of exons 2–7 and is present in gliomas and lung squamous cancers, was recently reported in 42% of HNSCCs (92). Importantly, an antibody thought to be specific for EGFRvIII (e.g., rather than full-length EGFR) was used to initially identify cases; this finding was not reproduced in another study that sequenced the full-length EGFR cDNA (93). It will be important to resolve whether EGFRvIII is expressed with any appreciable frequency in HNSCC, as EGFR kinase inhibitors have demonstrated clinical activity against tumors expressing this variant (94). Mutation or amplification of the MET (c-Met) RTK gene has been reported in some HNSCC cases (95). This finding is of clinical interest both because MET amplification is thought to confer resistance to EGFR-directed therapy (96) and because the small molecule therapeutic crizotinib, which inhibits both the MET and ALK kinases, has recently been FDA approved for use in lung cancers harboring ALK translocations (97).
While each of the above genes and pathways are associated with activities that may indirectly prevent programmed cell death, several constituents of the apoptotic signaling cascade may also have an important role in HNSCC. These include caspase-8 (CASP8), encoding the critical proapoptotic enzyme that initiates a cascade of proteolysis responsible for executing apoptosis and found to be mutated in 8% of samples in one exome sequencing study (11, 98, 99); and BCL2, encoding a key antiapoptotic regulator reported to be overexpressed in a fraction of HNSCC cell lines, particularly those with reduced expression of p63 (63).
Adhesion and invasion signaling through TGF-β/SMAD and FAT1
Inactivation of TGF-β signaling components is well established in human cancer, including HNSCC, most commonly through loss of TGF-β receptor (TGFBR2) and SMAD genes as a result of chromosome 18q deletion (100). Notably, although missense mutations in TGFBR2 have been previously described in primary HNSCCs (101), and SMAD2 and SMAD4 mutations have been reported in HNSCC cell lines (102), no point mutations in these genes were found through exome sequencing, perhaps due to the low frequency of these events. The TGF-β pathway is a pleiotropic regulator in human cancer, as mutational inactivation of its signaling components is associated with tumor initiation, while activation of the pathway is known to promote metastasis. Thus, genetic loss of TGF-β pathway factors would at first glance seem at odds with a contribution of this pathway to invasion and metastasis in HNSCC. Recent mouse models, however, suggest a more complex interaction. Conditional deletion of Smad4 in the mouse stratified epithelium led to HNSCC in association with increased genomic instability and increased inflammation, the latter attributed in part to elevation of TGF-β1 and activation of other SMADs in stroma, mucosal epithelia, and tumor cells (103). In addition, Tgfbr2 deletion within the mouse head and neck epithelia is insufficient to cause HNSCC, but cooperates with activated Kras to promote squamous carcinomas that metastasize to local lymph nodes (104). TGF-β1 itself has also been associated with epithelial-mesenchymal transition (EMT) and metastasis, the latter in the absence of functional TGF-βRII (105).
New insight into potential mechanisms of HNSCC invasion and metastasis was provided by the identification of mutations in the FAT1 gene in nine HNSCC samples (12%) in one of the two exome sequencing studies (11). Six nonsense mutations and a seventh frameshift were noted, suggesting FAT1 may function as a tumor suppressor. Notably, focal, intragenic homozygous deletions of FAT1 have previously been described in oral cancer (106). As it is a member of the cadherin superfamily of cell membrane proteins that have demonstrated roles in the establishment of cell polarity and mediate cell-cell contacts, loss of FAT1 might be predicted to permit loosening of the adhesions that normally restrain growth and/or migration of cells in an epithelial sheet. Similarly, mutations in genes encoding other membrane-associated proteins with a role in the establishment of polarity and cell adhesion have been described in HNSCC (107).
Additional genes/pathways
Several genes with unclear roles in HNSCC were found to be mutated at appreciable frequencies in at least one of the exome sequencing studies. Although the functional significance of the identified missense mutations is not clear (and some may merely represent passenger mutations), recurrent inactivating mutations were observed in several additional genes, suggestive of tumor suppressor activity. These include MLL2 (11) and NSD1 (11), both encoding histone methyltransferases, and SYNE1 (10, 11), a nuclear envelope protein. Mutations within MLL2 have recently been described in non-Hodgkin’s lymphoma, and mutations within several other histone-modifying enzymes have been identified in renal cell carcinoma and diverse human cancers, suggesting a role for chromatin-mediated gene expression deregulation in cancer pathogenesis (108–110). Although SYNE1 loss has been previously described in ovarian cancers and gliomas, this genomic locus spans more than 0.5 Mb (the longest isoform comprises 146 exons) and is subject to copy number variation in normal tissues. As such, this locus could be expected to accumulate relatively frequent passenger mutations, resulting in an overestimate of the significance of mutations if gene size is not taken into account (111–113).
Implications for therapy
The majority of patients with HNSCC come to clinical attention before the development of metastases, and as a result they have the potential to be cured with aggressive multimodality therapy (e.g., surgery, radiation, and/or chemotherapy). However, although improvements in surgical techniques, chemotherapy and radiation delivery, and supportive care have improved quality of life for patients with HNSCC, survival as a whole has not markedly improved (9).
As noted above, activating mutations in tyrosine kinases are relatively rare in HNSCC, and thus only one kinase-targeted therapy, cetuximab, is FDA approved for patients with HNSCC. This agent has generally modest efficacy, and efforts to identify a subset of patients whose tumors are “addicted” to EGFR signaling have not been successful (86, 87). One intriguing possibility is that production of EGFR activating ligands by tumor cells or tumor-associated stroma may increase sensitivity to EGFR inhibitors in HNSCC (114).
The recognition of HPV-related OPSCC as a distinct clinical entity with a better prognosis has led to the hypothesis that less-intensive therapy might mitigate toxicity without affecting survival. Clinical trials to test this hypothesis are now underway (115), but it should be weighed against the possibility that the primary survival benefit of intensive therapy may lie with HPV-positive patients (116). Also relevant is the demonstration that human cancer cell lines derived from HPV-positive HNSCCs may be addicted to the continuous activity of the viral E6 and E7 proteins, suggesting that the latter represent important therapeutic targets. In addition, endogenous expression of E6 and E7 depends in part on the activity of transcription factors (including Notch), which themselves are controlled by potentially targetable upstream signaling pathways (52, 53).
The convergence of many HNSCC cancer genes on the activity of the G1 cyclin-dependent kinases (e.g., CDK2, -4, and -6) suggests that targeting these kinases could have potent efficacy. A number of direct CDK inhibitors are currently being evaluated in clinical trials; several appear particularly effective in combination with cytotoxic agents with established efficacy in HNSCC (e.g., cisplatin, taxol; reviewed in ref. 34). A potentially novel approach to targeting the cell cycle in HSNCC has been suggested by the finding that inhibitors of the deubiquitinating enzyme USP2 could promote the proteasomal degradation of cyclin D1 (117, 118).
The ability to selectively target tumors with decreased p53 activity could have major implications for the majority of HNSCC patients. Unfortunately, the majority of p53-selective compounds currently in early phase clinical trials are designed to induce the activation of wild-type p53 in cells in which p53 activity is downmodulated by alterations in endogenous p53 regulators. These agents are unlikely to be effective in tumors in which p53 activity is abrogated primarily through mutation. An exception could be the large class of structural mutations that affect p53 protein stability; in these cases, decreasing p53 degradation could lead to restoration of some p53 activity (16). In addition, a variety of creative preclinical strategies have been undertaken to target tumor cells expressing mutated p53. These include the development of compounds that stabilize p53 through allosteric binding and synthetic lethal screens to identify candidate agents (including polo-like kinase inhibitors, some of which are in clinical trials) that selectively induce toxicity in p53-deficient cells (119).
Conclusions
As noted above, the relative lack of readily targeted oncogenes in these tumors has challenged traditional approaches to drug development for HNSCC. Nevertheless, the increasingly refined knowledge of epithelial biology and HNSCC genetics, combined with newer approaches aimed at targeting proteins previously thought undruggable, could lead to novel therapies with greater potency and less toxicity for these cancers (120). These conceptual advances will allow the development of ever-more-sophisticated animal models, which in turn will be essential for testing the emerging host of candidate targeted therapeutics, and for determining which of these may have the most clinical viability. A strong biologic rationale will be essential if progress against this disease is to be accelerated in the near term.
Acknowledgments
The authors wish to thank Lori Wirth and members of the Ellisen laboratory for helpful discussions and review of the manuscript. This work was supported by NIH R01 DE-015945 (to L.W. Ellisen) and by NIH K08 DE-020139 (to S.M. Rothenberg).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2012;122(6):1951–1957. doi:10.1172/JCI59889.
References
- 1.Hermsen M, et al. New chromosomal regions with high-level amplifications in squamous cell carcinomas of the larynx and pharynx, identified by comparative genomic hybridization. J Pathol. 2001;194(2):177–182. doi: 10.1002/path.862. [DOI] [PubMed] [Google Scholar]
- 2.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 3.Slebos RJ, et al. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12(3 pt 1):701–709. doi: 10.1158/1078-0432.CCR-05-2017. [DOI] [PubMed] [Google Scholar]
- 4.Smeets SJ, et al. Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene. 2006;25(17):2558–2564. doi: 10.1038/sj.onc.1209275. [DOI] [PubMed] [Google Scholar]
- 5.Fakhry C, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 6.Gillison ML, et al. Survival outcomes by tumor human papillomavirus (HPV) status in stage III-IV oropharyngeal cancer (OPC) in RTOG 0129. J Clin Oncol. 2009;27(suppl):15S. doi: 10.1200/JCO.2008.21.7695. [DOI] [Google Scholar]
- 7.Licitra L, et al. High-risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J Clin Oncol. 2006;24(36):5630–5636. doi: 10.1200/JCO.2005.04.6136. [DOI] [PubMed] [Google Scholar]
- 8.Posner MR, et al. Cisplatin and fluorouracil alone or with docetaxel in head and neck cancer. N Engl J Med. 2007;357(17):1705–1715. doi: 10.1056/NEJMoa070956. [DOI] [PubMed] [Google Scholar]
- 9.Chaturvedi AK, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119(6):861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119(6):847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2(1):a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petitjean A, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Human Mutation. 2007;28(6):622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 16.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9(12):862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 17.Millon R, et al. Loss of MDM2 expression in human head and neck squamous cell carcinomas and clinical significance. Oral Oncol. 2001;37(8):620–631. doi: 10.1016/S1368-8375(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 18.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 19.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248(4951):76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 20.Braakhuis BJ, Leemans CR, Brakenhoff RH. A genetic progression model of oral cancer: current evidence and clinical implications. J Oral Pathol Med. 2004;33(6):317–322. doi: 10.1111/j.1600-0714.2004.00225.x. [DOI] [PubMed] [Google Scholar]
- 21.Nees M, et al. Expression of mutated p53 occurs in tumor-distant epithelia of head and neck cancer patients: a possible molecular basis for the development of multiple tumors. Cancer Res. 1993;53(18):4189–4196. [PubMed] [Google Scholar]
- 22.Lindenbergh-van der Plas M, et al. Prognostic significance of truncating TP53 mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2009;17(11):3733–3741. doi: 10.1158/1078-0432.CCR-11-0183. [DOI] [PubMed] [Google Scholar]
- 23.Poeta ML, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357(25):2552–2561. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Sayans M, Suarez-Penaranda JM, Gayoso-Diz P, Barros-Angueira F, Gandara-Rey JM, Garcia-Garcia A. p16(INK4a)/CDKN2 expression and its relationship with oral squamous cell carcinoma is our current knowledge enough? Cancer Lett. 2009;306(2):134–141. doi: 10.1016/j.canlet.2011.02.039. [DOI] [PubMed] [Google Scholar]
- 25.Reed AL, et al. High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1996;56(16):3630–3633. [PubMed] [Google Scholar]
- 26.Smeets SJ, et al. Genetic classification of oral and oropharyngeal carcinomas identifies subgroups with a different prognosis. Cell Oncol. 2009;31(4):291–300. doi: 10.3233/CLO-2009-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominguez G, et al. Prevalence of aberrant methylation of p14ARF over p16INK4a in some human primary tumors. Mutat Res. 2003;530(1–2):9–17. doi: 10.1016/s0027-5107(03)00133-7. [DOI] [PubMed] [Google Scholar]
- 28.Ogi K, et al. Aberrant methylation of multiple genes and clinicopathological features in oral squamous cell carcinoma. Clin Cancer Res. 2002;8(10):3164–3171. [PubMed] [Google Scholar]
- 29.Sailasree R, Abhilash A, Sathyan KM, Nalinakumari KR, Thomas S, Kannan S. Differential roles of p16INK4A and p14ARF genes in prognosis of oral carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17(2):414–420. doi: 10.1158/1055-9965.EPI-07-0284. [DOI] [PubMed] [Google Scholar]
- 30.Schache AG, et al. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin Cancer Res. 2011;17(19):6262–6271. doi: 10.1158/1078-0432.CCR-11-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheu JJ, et al. Functional genomic analysis identified epidermal growth factor receptor activation as the most common genetic event in oral squamous cell carcinoma. Cancer Res. 2009;69(6):2568–2576. doi: 10.1158/0008-5472.CAN-08-3199. [DOI] [PubMed] [Google Scholar]
- 32.Fu M, et al. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J Biol Chem. 2005;280(17):16934–16941. doi: 10.1074/jbc.M500403200. [DOI] [PubMed] [Google Scholar]
- 33.Jirawatnotai S, et al. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474(7350):230–234. doi: 10.1038/nature10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11(8):558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 35.Bova RJ, et al. Cyclin D1 and p16INK4A expression predict reduced survival in carcinoma of the anterior tongue. Clin Cancer Res. 1999;5(10):2810–2819. [PubMed] [Google Scholar]
- 36.Ellisen LW, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66(4):649–661. doi: 10.1016/0092-8674(91)90111-B. [DOI] [PubMed] [Google Scholar]
- 37.Lee SY, et al. Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer Sci. 2009;100(5):920–926. doi: 10.1111/j.1349-7006.2009.01130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malecki MJ, et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26(12):4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 41.Klinakis A, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473(7346):230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Jette C, Kanki JP, Aster JC, Look AT, Griffin JD. NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia. 2007;21(3):462–471. doi: 10.1038/sj.leu.2404546. [DOI] [PubMed] [Google Scholar]
- 43.Nicolas M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33(3):416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 44.Pear WS, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183(5):2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dotto GP. Notch tumor suppressor function. Oncogene. 2008;27(38):5115–5123. doi: 10.1038/onc.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devgan V, Mammucari C, Millar SE, Brisken C, Dotto GP. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 2005;19(12):1485–1495. doi: 10.1101/gad.341405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niimi H, Pardali K, Vanlandewijck M, Heldin CH, Moustakas A. Notch signaling is necessary for epithelial growth arrest by TGF-beta. J Cell Biol. 2007;176(5):695–707. doi: 10.1083/jcb.200612129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rangarajan A, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20(13):3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nickoloff BJ, Qin JZ, Chaturvedi V, Denning MF, Bonish B, Miele L. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-kappaB and PPARgamma. Cell Death Differ. 2002;9(8):842–855. doi: 10.1038/sj.cdd.4401036. [DOI] [PubMed] [Google Scholar]
- 50.Blokzijl A, et al. Cross-talk between the Notch and TGF-beta signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J Cell Biol. 2003;163(4):723–728. doi: 10.1083/jcb.200305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duan L, Yao J, Wu X, Fan M. Growth suppression induced by Notch1 activation involves Wnt-beta-catenin down-regulation in human tongue carcinoma cells. Biol Cell. 2006;98(8):479–490. doi: 10.1042/BC20060020. [DOI] [PubMed] [Google Scholar]
- 52.Talora C, et al. Constitutively active Notch1 induces growth arrest of HPV-positive cervical cancer cells via separate signaling pathways. Exp Cell Res. 2005;305(2):343–354. doi: 10.1016/j.yexcr.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 53.Talora C, Sgroi DC, Crum CP, Dotto GP. Specific down-modulation of Notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of malignant transformation. Genes Dev. 2002;16(17):2252–2263. doi: 10.1101/gad.988902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 55.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 56.Yugawa T, Handa K, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol Cell Biol. 2007;27(10):3732–3742. doi: 10.1128/MCB.02119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dotto GP. Crosstalk of Notch with p53 and p63 in cancer growth control. Nat Rev Cancer. 2009;9(8):587–595. doi: 10.1038/nrc2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reis-Filho JS, Simpson PT, Martins A, Preto A, Gartner F, Schmitt FC. Distribution of p63, cytokeratins 5/6 and cytokeratin 14 in 51 normal and 400 neoplastic human tissue samples using TARP-4 multi-tumor tissue microarray. Virchows Arch. 2003;443(2):122–132. doi: 10.1007/s00428-003-0859-2. [DOI] [PubMed] [Google Scholar]
- 59.Flores ER, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7(4):363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 60.Keyes WM, et al. p63 heterozygous mutant mice are not prone to spontaneous or chemically induced tumors. Proc Natl Acad Sci U S A. 2006;103(22):8435–8440. doi: 10.1073/pnas.0602477103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Danilov AV, et al. DeltaNp63alpha-mediated induction of epidermal growth factor receptor promotes pancreatic cancer cell growth and chemoresistance. PLoS One. 2011;6(10):e26815. doi: 10.1371/journal.pone.0026815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rocco JW, Ellisen LW. p63 and p73: life and death in squamous cell carcinoma. Cell Cycle. 2006;5(9):936–940. doi: 10.4161/cc.5.9.2716. [DOI] [PubMed] [Google Scholar]
- 63.Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9(1):45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 64.Su X, Cho MS, Gi YJ, Ayanga BA, Sherr CJ, Flores ER. Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf. EMBO J. 2009;28(13):1904–1915. doi: 10.1038/emboj.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 66.Steck PA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 67.Okami K, et al. Analysis of PTEN/MMAC1 alterations in aerodigestive tract tumors. Cancer Res. 1998;58(3):509–511. [PubMed] [Google Scholar]
- 68.Shao X, et al. Mutational analysis of the PTEN gene in head and neck squamous cell carcinoma. Int J Cancer. 1998;77(5):684–688. doi: 10.1002/(SICI)1097-0215(19980831)77:5<684::AID-IJC4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 69.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476(7359):163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qiu W, et al. PIK3CA mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12(5):1441–1446. doi: 10.1158/1078-0432.CCR-05-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qiu W, Tong GX, Manolidis S, Close LG, Assaad AM, Su GH. Novel mutant-enriched sequencing identified high frequency of PIK3CA mutations in pharyngeal cancer. Int J Cancer. 2008;122(5):1189–1194. doi: 10.1002/ijc.23217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Henken FE, et al. PIK3CA-mediated PI3-kinase signalling is essential for HPV-induced transformation in vitro. Mol Cancer. 2011;10:71. doi: 10.1186/1476-4598-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9(8):550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 74.Saranath D, et al. High frequency mutation in codons 12 and 61 of H-ras oncogene in chewing tobacco-related human oral carcinoma in India. Br J Cancer. 1991;63(4):573–578. doi: 10.1038/bjc.1991.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anderson JA, Irish JC, Ngan BY. Prevalence of RAS oncogene mutation in head and neck carcinomas. J Otolaryngol. 1992;21(5):321–326. [PubMed] [Google Scholar]
- 76.Anderson JA, Irish JC, McLachlin CM, Ngan BY. H-ras oncogene mutation and human papillomavirus infection in oral carcinomas. Arch Otolaryngol Head Neck Surg. 1994;120(7):755–760. doi: 10.1001/archotol.1994.01880310059011. [DOI] [PubMed] [Google Scholar]
- 77.Lu S-L, Herrington H, Wang X-J. Mouse models for human head and neck squamous cell carcinomas. Head Neck. 2006;28(10):945–954. doi: 10.1002/hed.20397. [DOI] [PubMed] [Google Scholar]
- 78.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137(5):821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 80.Singh A, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15(6):489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharafinski ME, Ferris RL, Ferrone S, Grandis JR. Epidermal growth factor receptor targeted therapy of squamous cell carcinoma of the head and neck. Head Neck. 2010;32(10):1412–1421. doi: 10.1002/hed.21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taoudi Benchekroun M, et al. Epidermal growth factor receptor expression and gene copy number in the risk of oral cancer. Cancer Prev Res (Phila). 2010;3(7):800–809. doi: 10.1158/1940-6207.CAPR-09-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Temam S, et al. Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J Clin Oncol. 2007;25(16):2164–2170. doi: 10.1200/JCO.2006.06.6605. [DOI] [PubMed] [Google Scholar]
- 84.Chung CH, et al. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol. 2006;24(25):4170–4176. doi: 10.1200/JCO.2006.07.2587. [DOI] [PubMed] [Google Scholar]
- 85.Bonner JA, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354(6):567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 86.Licitra L, et al. Evaluation of EGFR gene copy number as a predictive biomarker for the efficacy of cetuximab in combination with chemotherapy in the first-line treatment of recurrent and/or metastatic squamous cell carcinoma of the head and neck: EXTREME study. Ann Oncol. 2011;22(5):1078–1087. doi: 10.1093/annonc/mdq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cohen EE, et al. Response of some head and neck cancers to epidermal growth factor receptor tyrosine kinase inhibitors may be linked to mutation of ERBB2 rather than EGFR. Clin Cancer Res. 2005;11(22):8105–8108. doi: 10.1158/1078-0432.CCR-05-0926. [DOI] [PubMed] [Google Scholar]
- 88.Cohen EE, et al. Phase II trial of ZD1839 in recurrent or metastatic squamous cell carcinoma of the head and neck. J Clin Oncol. 2003;21(10):1980–1987. doi: 10.1200/JCO.2003.10.051. [DOI] [PubMed] [Google Scholar]
- 89.Kirby AM, et al. Gefitinib (ZD1839, Iressa) as palliative treatment in recurrent or metastatic head and neck cancer. Br J Cancer. 2006;94(5):631–636. doi: 10.1038/sj.bjc.6602999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Soulieres D, Senzer NN, Vokes EE, Hidalgo M, Agarwala SS, Siu LL. Multicenter phase II study of erlotinib, an oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with recurrent or metastatic squamous cell cancer of the head and neck. J Clin Oncol. 2004;22(1):77–85. doi: 10.1200/JCO.2004.06.075. [DOI] [PubMed] [Google Scholar]
- 91.Wirth LJ, et al. Phase I study of gefitinib plus celecoxib in recurrent or metastatic squamous cell carcinoma of the head and neck. J Clin Oncol. 2005;23(28):6976–6981. doi: 10.1200/JCO.2005.02.4182. [DOI] [PubMed] [Google Scholar]
- 92.Sok JC, et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res. 2006;12(17):5064–5073. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- 93.Hama T, et al. Prognostic significance of epidermal growth factor receptor phosphorylation and mutation in head and neck squamous cell carcinoma. Oncologist. 2009;14(9):900–908. doi: 10.1634/theoncologist.2009-0058. [DOI] [PubMed] [Google Scholar]
- 94.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353(19):2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 95.Seiwert TY, et al. The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma. Cancer Res. 2009;69(7):3021–3031. doi: 10.1158/0008-5472.CAN-08-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 97.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim HS, et al. Inactivating mutations of caspase-8 gene in colorectal carcinomas. Gastroenterology. 2003;125(3):708–715. doi: 10.1016/S0016-5085(03)01059-X. [DOI] [PubMed] [Google Scholar]
- 99.Soung YH, et al. CASPASE-8 gene is inactivated by somatic mutations in gastric carcinomas. Cancer Res. 2005;65(3):815–821. [PubMed] [Google Scholar]
- 100.Levy L, Hill CS. Alterations in components of the TGF–beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17(1–2):41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 101.Wang D, Song H, Evans JA, Lang JC, Schuller DE, Weghorst CM. Mutation and downregulation of the transforming growth factor beta type II receptor gene in primary squamous cell carcinomas of the head and neck. Carcinogenesis. 1997;18(11):2285–2290. doi: 10.1093/carcin/18.11.2285. [DOI] [PubMed] [Google Scholar]
- 102.Qiu W, Schonleben F, Li X, Su GH. Disruption of transforming growth factor beta–Smad signaling pathway in head and neck squamous cell carcinoma as evidenced by mutations of SMAD2 and SMAD4. Cancer Lett. 2007;245(1–2):163–170. doi: 10.1016/j.canlet.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bornstein S, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119(11):3408–3419. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lu SL, et al. Loss of transforming growth factor-beta type II receptor promotes metastatic head-and-neck squamous cell carcinoma. Genes Dev. 2006;20(10):1331–1342. doi: 10.1101/gad.1413306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Han G, et al. Distinct mechanisms of TGF-beta1-mediated epithelial-to-mesenchymal transition and metastasis during skin carcinogenesis. J Clin Invest. 2005;115(7):1714–1723. doi: 10.1172/JCI24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nakaya K, et al. Identification of homozygous deletions of tumor suppressor gene FAT in oral cancer using CGH-array. Oncogene. 2007;26(36):5300–5308. doi: 10.1038/sj.onc.1210330. [DOI] [PubMed] [Google Scholar]
- 107.Rothenberg SM, et al. A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers. Cancer Res. 2010;70(6):2158–2164. doi: 10.1158/0008-5472.CAN-09-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41(5):521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 112.Marme A, et al. Loss of Drop1 expression already at early tumor stages in a wide range of human carcinomas. Int J Cancer. 2008;123(9):2048–2056. doi: 10.1002/ijc.23763. [DOI] [PubMed] [Google Scholar]
- 113.Masica DL, Karchin R. Correlation of somatic mutation and expression identifies genes important in human glioblastoma progression and survival. Cancer Res. 2011;71(13):4550–4561. doi: 10.1158/0008-5472.CAN-11-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yonesaka K, et al. Autocrine production of amphiregulin predicts sensitivity to both gefitinib and cetuximab in EGFR wild-type cancers. Clin Cancer Res. 2008;14(21):6963–6973. doi: 10.1158/1078-0432.CCR-08-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Radiation Therapy Oncology Group. Phase III randomized study of radiotherapy with cisplatin or cetuximab in patients with human papilloma virus–associated oropharyngeal cancer 2011. National Cancer Institute at the NIH Web site.http://www.cancer.gov/clinicaltrials/search/view?cdrid=695731&version=HealthProfessional . Updated March 15, 2012. Accessed March 20, 2012.
- 116.Posner MR, et al. Survival and human papillomavirus in oropharynx cancer in TAX 324: a subset analysis from an international phase III trial. Ann Oncol. 2011;22(5):1071–1077. doi: 10.1093/annonc/mdr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dragnev KH, et al. Bexarotene and erlotinib for aerodigestive tract cancer. J Clin Oncol. 2005;23(34):8757–8764. doi: 10.1200/JCO.2005.01.9521. [DOI] [PubMed] [Google Scholar]
- 118.Shan J, Zhao W, Gu W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol Cell. 2009;36(3):469–476. doi: 10.1016/j.molcel.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lambert JM, et al. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15(5):376–388. doi: 10.1016/j.ccr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 120.Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin Cancer Res. 2007;13(24):7264–7270. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]