ABSTRACT
During early mammalian embryogenesis, there is a wave of DNA demethylation postfertilization and de novo methylation around implantation. The paternal genome undergoes active DNA demethylation, whereas the maternal genome is passively demethylated after fertilization in most mammals except for sheep and rabbits. However, the emerging genome-wide DNA methylation landscape has revealed a regulatory and locus-specific DNA methylation reprogramming pattern in mammalian preimplantation embryos. Here we optimized a bisulfite sequencing protocol to draw base-resolution DNA methylation profiles of several selected genes in gametes, early embryos, and somatic tissue. We observed locus-specific DNA methylation reprogramming in early porcine embryos. First, some pluripotency genes (POU5F1 and NANOG) followed a typical wave of DNA demethylation and remethylation, whereas CpG-rich regions of SOX2 and CDX2 loci were hypomethylated throughout development. Second, a differentially methylated region of an imprint control region in the IGF2/H19 locus exhibited differential DNA methylation which was maintained in porcine early embryos. Third, a centromeric repeat element retained a moderate DNA methylation level in gametes, early embryos, and somatic tissue. The diverse DNA methylation reprogramming during early embryogenesis is thought to be possibly associated with the multiple functions of DNA methylation in transcriptional regulation, genome stability and genomic imprinting. The latest technology such as oxidative bisulfite sequencing to identify 5-hydroxymethylcytosine will further clarify the DNA methylation reprogramming during porcine embryonic development.
Keywords: CDX2, DNA methylation, NANOG, porcine preimplantation embryos, POU5F1, reprogramming, SOX2
DNA methylation reprogramming in early porcine embryos is locus-specific and associated with multiple functions of DNA methylation in transcriptional regulation, genomic imprinting, and genome stability.
INTRODUCTION
In mammals, both the maternal and paternal genomes are required for the completion of embryogenesis as they are not equivalent due to genomic imprinting [1, 2]. Upon fertilization, the parental genomes undergo dramatic epigenetic reprogramming to form the diploid genome. The paternal genome is actively demethylated within 6–8 hours after fertilization, before the onset of DNA replication, while the maternal genome is gradually demethylated until the blastocyst stage [3, 4]. The conservation of this active demethylation pattern in paternal genomes has been observed in many species such as human, rat, mice, cattle, and pigs [5, 6] but not in rabbits and sheep, where there was little DNA demethylation in the male pronucleus before DNA replication [7]. Nevertheless, genome-wide de novo methylation is initiated by the blastocyst stage and established by de novo DNA methyltransferases (DNMT3A and DNMT3B) [8]. The DNA methylation patterns are then faithfully retained by the maintenance DNA methyltransferase (DNMT1) during later development [9].
The typical wave of global DNA demethylation and remethylation in early mammalian embryos was revealed mainly by anti-5-methylcytosine (5mC) immunofluorescence staining [10] but has been widely accepted for the past decade [11]. Because transposon-related elements cover approximately 40% of mammalian genome and functional genes comprise only ∼1.5% of the entire genome [12, 13], most 5mC immunofluorescence signals are predicted to correspond to multiple copy repetitive regions [14]. However, genome-wide DNA methylation studies indicate a regulatory and genomic locus-specific DNA methylation reprogramming pattern during mammalian preimplantation development [15–18]. Accordingly, some differentially methylated regions (DMRs) at imprinted loci are resistant to this wave of active paternal and passive maternal DNA demethylation in the zygote and early preimplantation embryo [19]. Similarly, some repeat sequences, such as intracisternal A particle (IAP) elements are also exempted from complete DNA demethylation, although other repeat sequences (e.g., long interspersed elements [LINEs] and long terminal repeat [LTR] retroelements) are substantially demethylated during early embryonic development [20]. In addition, a number of promoter regions in nonimprinted genes also escape the global DNA methylation reprogramming in mouse preimplantation embryos [16]. Most CpG islands display incomplete DNA demethylation by the blastocyst stage although very few CpG islands are capable of resisting postfertilization methylation reprogramming [17], reflecting diverse DNA methylation options that are dependent upon genomic loci. Strikingly, in mouse gametes and early embryos, DNA methylation contributed by sperm in some retroelements remains unchanged and oocyte-contributed DMRs in many CpG island promoters retain their DNA methylation levels during early embryogenesis [15]. Furthermore, approximately half of germline differentially methylated regions between oocytes and sperm appears to resist genome-wide DNA demethylation in mouse preimplantation embryos [18]. Collectively, these studies suggest diverse DNA methylation reprogramming in preimplantation embryos, where DNA methylation in individual loci is mostly dynamic and stage-specific, possibly related to their functions in transcriptional regulation and genomic stability [21].
Pluripotency genes, such as POU5F1, NANOG, SOX2, and CDX2, are essential for the segregation and maintenance of embryonic and extraembryonic tissues. The POU family transcription factor Pou5f1 (also known as Oct3/4) is required for inner cell mass formation, pluripotency, and germ cell development in mice [22, 23]. Nanog is specifically localized in nascent epiblasts, thus demarcating the epiblast from the hypoblast, and is the gateway to the ground state of pluripotency in mouse embryos [24]. In addition, promoter DNA demethylation in Pou5f1 and Nanog gene loci is necessary for reprogramming somatic cells into induced pluripotent stem cells in mice [25]. Sox2 acts synergistically with Pou5f1 to maintain pluripotency and regulate germ layer cell fate determination in mouse embryonic stem cells [26, 27]. Cdx2 is required for placental formation by repressing Pou5f1 expression in trophectoderm and is essential for the maintenance of mouse trophoblast stem cell self-renewal [28, 29]. Transcriptional regulation of these pluripotency genes is considered to be governed by epigenetic modifications such as DNA methylation [30–33]. However, the dynamic DNA methylation profiles of pluripotency genes in vivo have been poorly understood due to the limited amounts of genomic DNA from preimplantation embryos. In mouse embryos, it appears that the regulatory regions of Sox2 and Cdx2 are never methylated, but Pou5f1 and Nanog loci have low levels of DNA methylation in zygotes but are completely demethylated in the inner cell mass (ICM) of blastocysts (Dr. Alexander Meissner, Broad Institute of MIT and Harvard, personal communication). Here we optimized a bisulfite sequencing protocol for small amounts of genomic DNA to address the dynamic DNA methylation reprogramming in pluripotency genes in early porcine embryos. We also examined DNA methylation profiles of a differentially methylated region in the IGF2/H19 imprinted locus and a centromeric repeat sequence. Intriguingly, we found diverse DNA methylation reprogramming patterns in porcine preimplantation embryos.
MATERIALS AND METHODS
Unless described elsewhere, all chemicals and reagents were purchased from Sigma (St. Louis, MO).
In Vitro Fertilization and Embryo Culture
Ovaries were collected from prepubertal gilts in a local Missouri slaughterhouse. Cumulus-oocyte complexes were aspirated and selected based on uniform cytoplasm and multiple layers of cumulus cells. Oocytes were cultured in in vitro maturation (IVM) medium covered with mineral oil for 40–44 h at 38.5°C in a humidified atmosphere of 5% CO2 in air. After maturation, cumulus cells were removed by vortexing in 0.1% (w/v) hyaluronidase in HEPES-buffered saline [34]. Denuded metaphase II (MII) oocytes with visible first polar body were then selected in oocyte manipulation medium (OMM) under a stereo microscope.
For in vitro fertilization (IVF), 30 MII oocytes were transferred to a 50-μl droplet of equilibrated modified Tris-buffered medium (mTBM) covered with mineral oil at 38.5°C in 5% CO2 in air. For each replication, a frozen semen pellet was thawed and washed twice by centrifugation at 1900 × g for 4 min. The number of spermatozoa was adjusted to 2 × 106 cells/ml, and 50 μl of resuspended sperm was added to each droplet containing MII oocytes. The sperm-oocyte-containing droplets were subsequently incubated at 38.5°C in 5% CO2 in air for 4–6 h. Then they were washed three times and cultured in porcine zygote medium-3 (PZM3) with 3 mg/ml bovine serum albumin (BSA) at 38.5°C in 5% CO2 in air. The 4-cell stage embryos were collected at approximately 36 h, and the blastocysts were collected on Day 7. Recipes for media IVM, OMM, mTBM, and PZM3 were assembled as previously described [34, 35].
Genomic DNA and RNA Isolation and Quantitative RT-PCR
A pool of 30–50 germinal vesicle (GV) oocyte, IVM MII oocytes, 4-cell IVF embryos, and blastocysts (BL) were produced according to the procedure described above. Zonae pellucidae were gently removed by 5 mg/ml pronase under a stereo microscope and immediately neutralized by polyvinyl alcohol, Tyrode lactate buffer with 0.1% BSA (w/v). The zona-free embryos were then washed three times in diethyl pyrocarbonate-treated PBS and then quickly frozen in liquid nitrogen before long-term storage at −80°C. Liver was taken from a postnatal 1-week old wild-type piglet. At least three biological replicates were collected for each stage sample. Genomic DNA and total RNA were isolated using AllPrep DNA/RNA Micro kit (Qiagen, Valencia, CA) following the manufacturer's instructions. First-strand complementary DNA (cDNA) was synthesized by using a QuantiTect reverse transcription kit (Qiagen) to remove any potential genomic DNA contamination. Real-time quantitative PCR (qRT-PCR) was performed by using iQ SYBR Green Supermix in an iCycler IQ single-color RT-PCR detection system (Bio-Rad, Hercules, CA). Melting curves were generated following RT-PCR to assess the specificity of the amplicons. Expression levels were analyzed by a relative standard curve method. Gradient dilutions (1/10×) of Ref cDNA [36] were used to create standard curves, and the YWHAG (a housekeeping gene encoding 14-3-3 protein gamma) was used as a calibrator gene. qRT-PCR data were obtained from three independent biological and two technical replicates and analyzed statistically by one-way analysis of variance (ANOVA).
Sperm Genomic DNA Extraction
Sperm were collected by centrifugation and incubated with Solution I (PBS-0.8% Triton X-100-0.8% SDS) for 10 min at room temperature to remove somatic cell contamination. After centrifugation for 5 min at 9000 × g, the supernatant was discarded. Sperm were then rinsed three times in STE buffer (100 mM NaCl-10 mM Tris-1 mM EDTA, pH 8.0) at 9000 × g for 5 min and subsequently resuspended in 675 μl of STE, followed with the orderly addition of 70 μl of 20% SDS (Thermo Fisher Scientific, Waltham, MA), 50 μl of 0.5 M dithiothreitol, and 5 μl of 20 mg/ml proteinase K (New England BioLabs, Ipswich, MA). Sperm were then incubated at 56°C overnight for digestion. The next day, genomic DNA was extracted by phenol-chloroform-isoamyl alcohol (25:24:1) combined with Maxtract high-density tubes (Qiagen). The upper phase containing DNA was further precipitated by 3 M sodium acetate (pH 5.5) and 100% ethanol and washed by 70% ethanol. The DNA pellet was dried completely to remove any trace of ethanol and resuspended with TE buffer (10 mM Tris-HCl-1 mM EDTA, pH 8.0). The quality and quantity of genomic DNA from sperm were evaluated and measured by using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific).
Bisulfite Sequencing
Genomic DNA was treated with sodium bisulfite and immediately cleaned up by using an imprint DNA modification kit (Sigma). Four replicates of bisulfite-treated genomic DNA from GV and MII oocytes, 4-cell embryos, and blastocysts were pooled for subsequent PCR amplification. However, three replicates of genomic DNA from sperm and liver were not pooled because of their abundance. The bisulfite primers were designed by using Methyl Primer Express version 1.0 (Life Technologies Corp., Grand Island, NY) and an online MethPrimer software (http://www.urogene.org/methprimer/index1.html). Large numbers of primer sets were selected and then tested by gradient PCRs with the template of bisulfite-treated liver DNA. Validated primer sequence information is summarized in Supplemental Table S1 (available online at www.biolreprod.org). The nested PCR primers for DMR3 in the IGF2/H19 locus were from a published study [37]. PCR was performed by using a GoTaq Green Master Mix (Promega, Madison, WI) with bisulfite-converted genomic DNA as the template. A typical PCR program (NANOG, POU5F1, CDX2, and SatRep) consisted of an initial denaturation step at 95°C for 4 min, followed by 45 cycles of denaturation at 95°C for 45 sec, annealing at 56°C for 1 min, and extension at 72°C for 45 sec. A final extension of 72°C for 15 min was also included. For SOX2 primers, two rounds of PCRs were performed using the same program. For DMR3 IGF2/H19 primers, the annealing temperature was 50°C for the outside primers and 56°C for the inner primers.
The PCR product was loaded in a 1.5% agarose gel, extracted, and purified by using a Wizard SV gel and PCR Clean Up System (Promega). Purified PCR fragments were then cloned into a pCR4-TOPO vector which was included in a TOPO TA cloning kit for sequencing (Invitrogen, Grand Island, NY). The TOPO cloning reaction was subsequently transformed into One Shot TOP10 chemically competent cells (Invitrogen) and grown on Luria-Bertani-kanamycin (50 mg/ml) agar plates overnight. For each transformation, 10–15 clones were randomly selected and plasmid DNA was isolated by using a PureLink Quick Plasmid Miniprep kit (Invitrogen). Plasmids were further screened by PCRs, and only positive clones were submitted to the DNA core at University of Missouri-Columbia for sequencing. The PCR amplifications and subsequent transformation were performed at least twice for each sample.
Data Interpretation
Sequencing data were aligned to the reference sequences by MacVector version 12.0 (MacVector Inc., Cary, NC). Reference sequences were created by replacing a “C” with a “T” in non-CpG sites but leaving the “C” in CpG sites intact. The bisulfite treatment converts an unmethylated “C” into a “U,” which will eventually turn into a “T” after multiple cycles of PCR amplification but has no effect on a methylated “C.” Therefore, for a fixed CpG site, if it is a “T” in the sample sequence, it means this CpG is unmethylated and is represented as an open circle. In contrast, if it is a “C,” it means this CpG is methylated and protected from bisulfite treatment, thus, is represented with a filled circle. The representative clones were carefully selected so that each one was at least one nucleotide (either in CpG or non-CpG site) different from another within the amplicon. The clones that had the same sequence information were only counted once. P values of pairwise comparisons were calculated by one-way ANOVA.
RESULTS
POU5F1 and NANOG Underwent Typical DNA Demethylation and Remethylation in Porcine Preimplantation Embryos
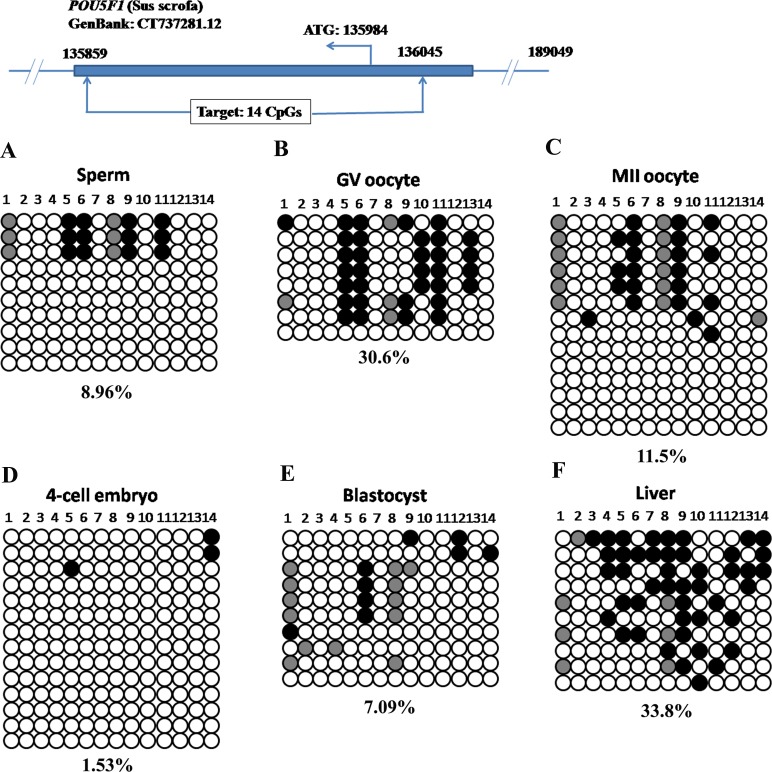
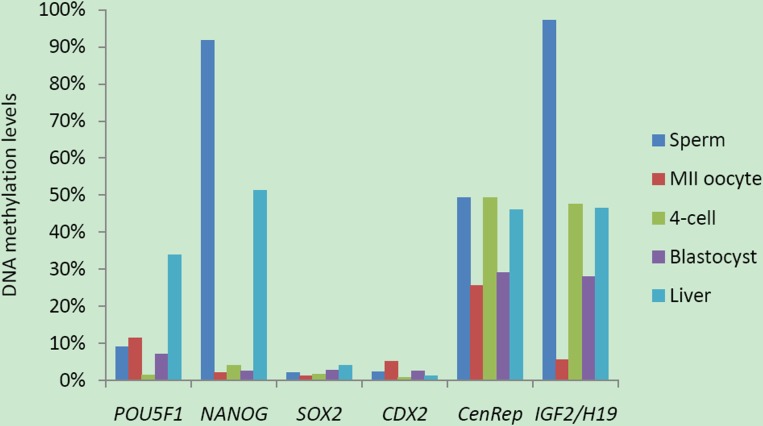
To examine the DNA methylation dynamics in preimplantation embryos, we started bisulfite sequencing with POU5F1 and NANOG. The POU5F1 amplicon spanned a CpG island and covered 14 CpG sites within 187 bp, whereas the NANOG promoter region had low CpG density and contained only 10 CpG sites within 500 bp (Supplemental Table S1). For the POU5F1 locus, there was a low level of DNA methylation in sperm (Fig. 1A, 8.96%). DNA methylation levels in GV oocytes (Fig. 1B, 30.6%) and MII oocytes (Fig. 1C, 11.5%) were not significantly different (GV vs. MII: P = 0.085). At 4-cell stage, the overall DNA methylation level decreased to 1.53% (Fig. 1D). When the blastocyst stage was reached, DNA methylation level was still low (Fig. 1E, 7.09%, 4-cell stage (4C) vs. blastocyst (BL): P = 0.117). This locus was moderately methylated in liver (Fig. 1F, 33.8%), implying a DNA remethylation event during postimplantation development (BL vs. liver: P = 0.0138).
FIG. 1.
Dynamic DNA methylation profiles in the POU5F1 locus in porcine gametes, preimplantation embryos and somatic tissue. Low DNA methylation levels were observed in sperm (A) and MII oocytes (C). However, GV oocytes were moderately methylated (B). After fertilization, the zygotic genome lost DNA methylation in 4-cell stage embryos (D). DNA methylation level was still low in blastocysts (E). A moderate level of DNA methylation was observed in liver (F). A closed circle shows methylated cytosine, whereas an open circle indicates unmethylated cytosine in each CpG site. A filled gray circle represents mutated and/or single nucleotide polymorphism (SNP) variation at certain CpG sites. The number below each panel denotes the percentage of methylated cytosines in observed total CpG sites, and each row of circles represents an individual clone which contains the inserted amplicon. The clones were arranged from the most methylated (top) to the least methylated (bottom). Note, the same legend was applied in Figures 2–6. The top diagram in each figure schematically denotes the genomic location of the target DNA methylation region. ATG, starting codon; TSS, transcription starting site.
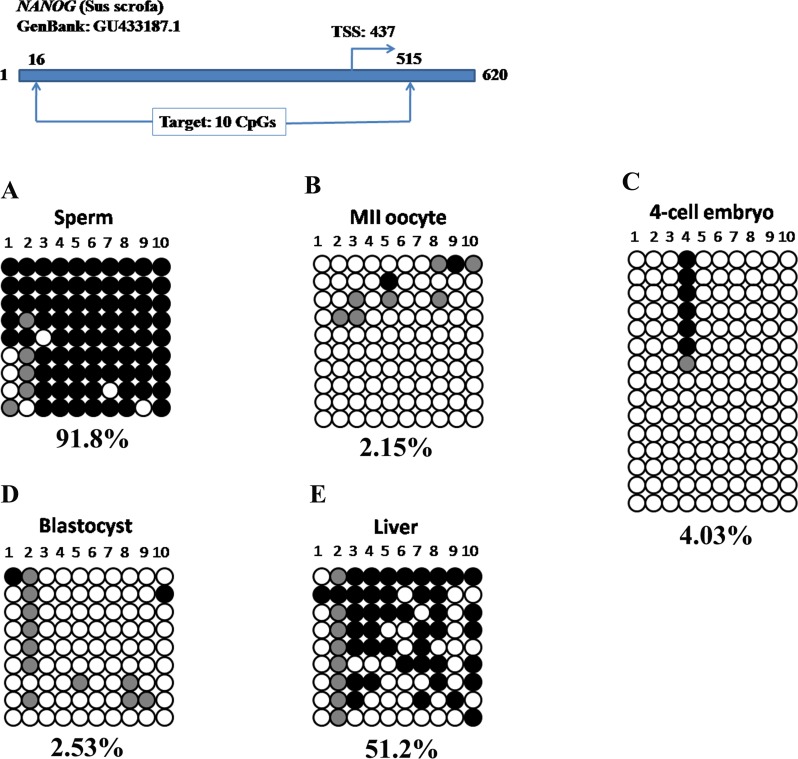
The NANOG promoter region was hypermethylated in sperm (Fig. 2A, 91.8%) but hypomethylated in MII oocytes (Fig. 2B, 2.15%). At the 4-cell stage, the NANOG promoter was hypomethylated with only 4.03% methylation (Fig. 2C). Similarly, DNA methylation level was also low in blastocysts (2.53%, Fig. 2D). The unmethylated NANOG promoter may be associated with its mRNA abundance in 4-cell embryos and blastocysts (Supplemental Fig. S1B). After implantation, de novo methylation occurred and 51.2% of methylated CpG sites were observed in the liver (Fig. 2E, BL vs. liver: P = 0.0054). In sum, porcine POU5F1 and NANOG underwent a typical wave of DNA demethylation and de novo methylation during porcine early embryogenesis.
FIG. 2.
Typical DNA demethylation and remethylation in the NANOG promoter during porcine early embryogenesis. The sperm DNA was highly methylated (A) whereas MII oocytes were hypomethylated (B). The paternal genome was actively demethylated with the overall 4.03% methylation at 4-cell stage (C). The DNA hypomethylation continued until the blastocyst stage (D). The subsequent de novo methylation rendered somatic tissue with a high level of DNA methylation (E).
SOX2 and CDX2 Loci Resisted DNA Methylation Reprogramming in Porcine Gametes, Early Embryos, and Somatic Tissue
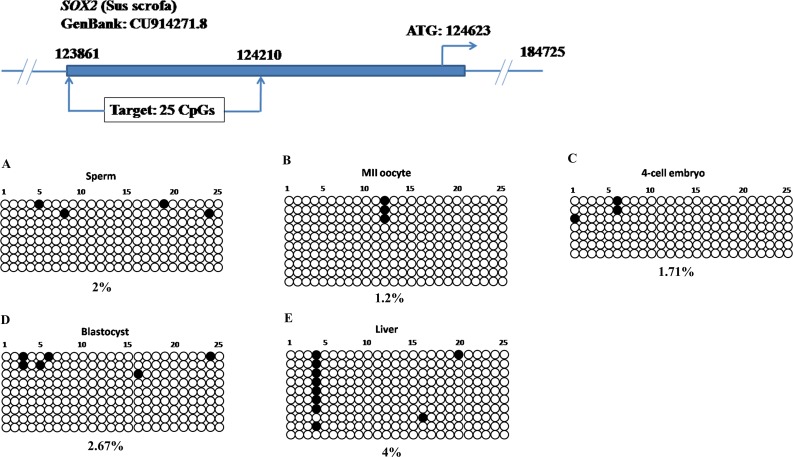
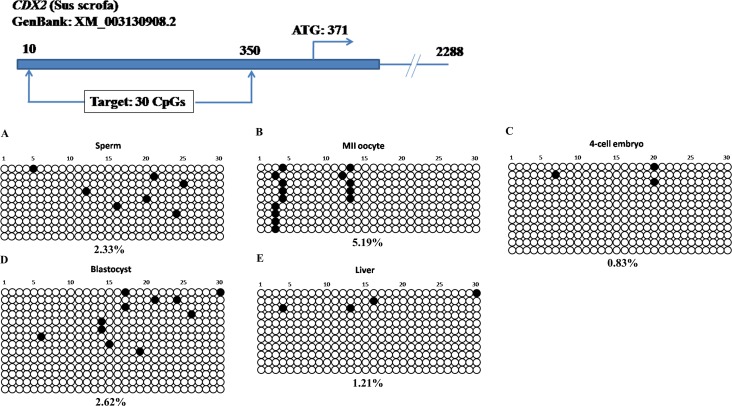
Next, we addressed the DNA methylation profiles in SOX2 and CDX2 loci. We designed bisulfite primers to target 5′ upstream regions of SOX2 and CDX2 genes. The SOX2 target sequence was 350 bp with 25 CpG sites upstream of transcription start site, and the CDX2 target was 341 bp with 30 CpGs upstream of the coding region (Supplemental Table S1). Both of them had CpG islands with a high density of CG contents. Strikingly, these CpG sites in SOX2 (Fig. 3) and CDX2 loci (Fig. 4) were hypomethylated in gametes, 4-cell stage embryos, blastocysts and somatic tissue. The DNA methylation levels were all below 6% and only a few sporadic methylated CpG sites were detected. Nevertheless, this DNA methylation pattern was different from that of POU5F1 which also contained a CpG island (Fig. 1).
FIG. 3.
The CpG island upstream of the SOX2 was unmethylated during porcine embryonic development. In porcine gametes (A and B) and early embryos (C and D), the CpG island with 25 CpG sites was generally hypomethylated. In addition, DNA methylation level was also low in the liver (E), implying that this locus was able to resist genome-wide de novo methylation after implantation.
FIG. 4.
CpG-rich region in the CDX2 locus maintained hypomethylation in gametes, early embryos, and somatic tissue. Hypomethylation (<6%) was observed in porcine sperm (A), MII oocytes (B), 4-cell stage embryos (C), blastocysts (D), and liver (E).
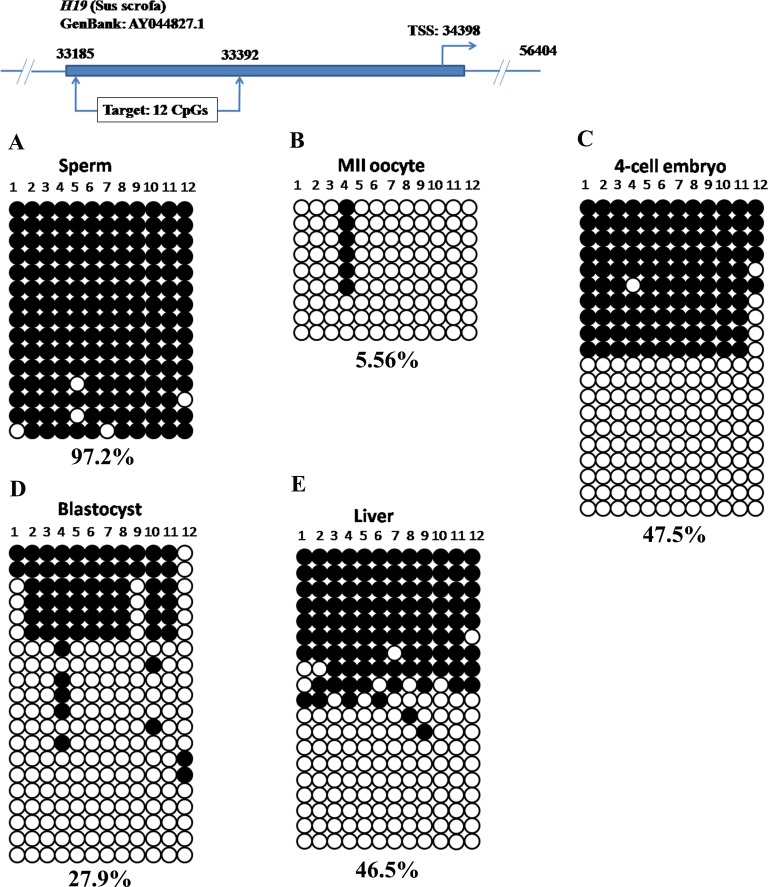
Differentially Methylated Region of the Imprinted IGF2/H19 Locus Displayed Differential DNA Methylation in Porcine Preimplantation Embryos
We selected the IGF2/H19 locus as a representative for imprinted genes. DNA methylation in the imprinting control region of the IGF2/H19 locus is thought to regulate their allele-specific expression by affecting the accessibility of CTCCC-binding factor (CTCF). We performed bisulfite sequencing to examine the DNA methylation profiles on the DMR3 of porcine IGF2/H19 gene locus [37]. The DMR3 was highly methylated in sperm (Fig. 5A, 97.2%) but hypomethylated in MII oocytes (Fig. 5B, 5.56%), indicating the paternal allele was methylated whereas the maternal allele was unmethylated. This differential DNA methylation pattern was also evident at 4-cell (Fig. 5C) and blastocyst stages (Fig. 5D). In somatic tissue, parental alleles were differentially methylated and nearly half were methylated (Fig. 5E). Together, the DMR in IGF2/H19 gene locus showed differential DNA methylation and resisted the genome-wide DNA demethylation in early porcine embryos.
FIG. 5.
Differential DNA methylation in the DMR of porcine IGF2/H19 locus. The paternal allele (sperm) was highly methylated (A) but the maternal allele (oocyte) was hypomethylated (B). This differential DNA methylation pattern was maintained in preimplantation embryos (C and D) and somatic tissue (E).
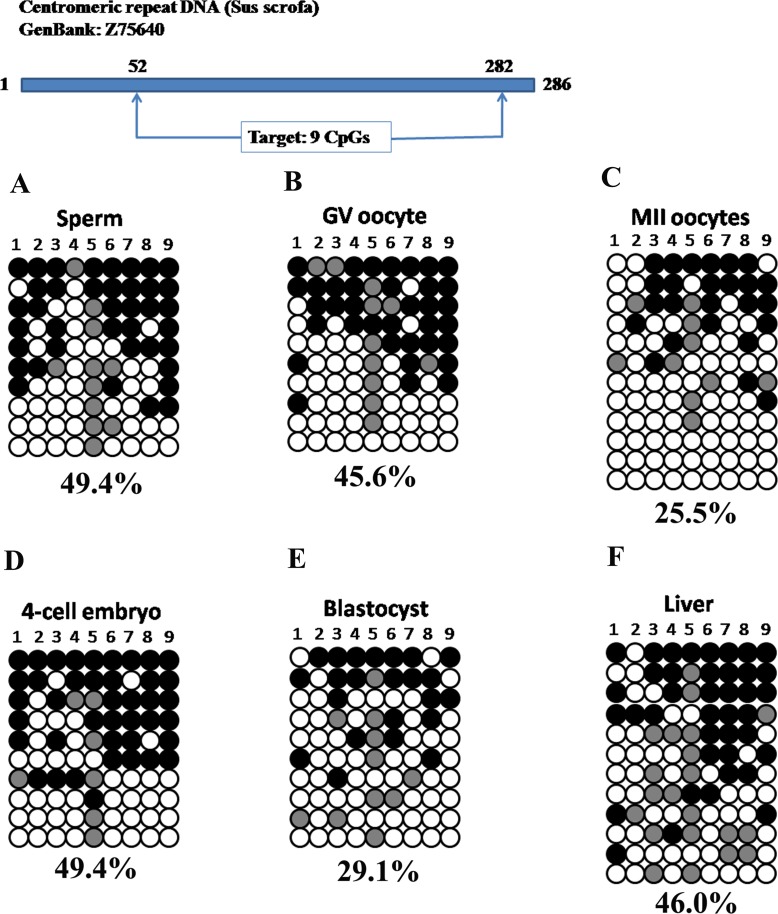
Centromeric Repeat Exhibited Moderate DNA Methylation Levels Throughout Embryonic Development
DNA methylation in repeat elements such as centromeres is essential for maintaining chromosome stability [21]. Previous studies by using immunofluorescence staining mainly reflected DNA methylation changes in repetitive gene families and transposable elements [14]. Thus, we selected a centromeric repeat to test whether it experienced DNA demethylation during early embryogenesis. We amplified a 231-bp fragment with 9 CpG sites (Supplemental Table S1). Bisulfite sequencing showed moderate DNA methylation levels in sperm (Fig. 6A, 49.4%), GV oocytes (Fig. 6B, 45.6%), and MII oocytes (Fig. 6C, 25.5%). Interestingly, this moderate DNA methylation persisted in 4-cell stage embryos (Fig. 6D, 49.4%) and decreased slightly at the blastocyst stage (Fig. 6E, 29.1%). Nevertheless, the DNA methylation variances during oocyte maturation (GV vs. MII: P = 0.357) and between 4-cell and blastocysts (4-cell vs. BL: P = 0.198) were not significantly different. Therefore, it is hard to presume that a DNA demethylation process took place by blastocyst stage. DNA methylation in somatic tissue (Fig. 6F, 46.0%) was not significantly different from that of blastocysts (BL vs. Liver: P = 0.190). On the whole, the centromeric repeat maintained moderate DNA methylation during porcine embryonic development.
FIG. 6.
Moderate DNA methylation in the centromeric repeat in porcine gametes, early embryos, and somatic tissue. DNA methylation level in sperm (A) was slightly higher than those of GV (B) and MII (C) oocytes. After fertilization, moderate DNA methylation was observed in 4-cell stage embryos (D) and blastocysts (E). The DNA methylation level rebounded to 46.0% in liver (F) post implantation.
DISCUSSION
Diverse DNA Methylation Reprogramming in Porcine Early Embryos
In mammals, DNA methylation plays an essential role in maintaining genomic imprinting, X-chromosome inactivation, transcriptional regulation, and suppression of transposable elements during normal development [21]. The concept that DNA methylation in certain genomic loci is dynamic rather than static is emerging from the latest DNA demethylation studies [38]. In mammalian preimplantation embryos, the overall DNA methylation level first decreases and then increases, following a typical pattern of demethylation and remethylation. In this study we found diverse DNA methylation patterns in early porcine embryos which are dependent upon genomic locus.
The pluripotency genes POU5F1 and NANOG follow a typical wave of DNA demethylation and de novo methylation during embryogenesis which fits well with the general DNA methylation reprogramming manner [11]. The low DNA methylation level in the POU5F1 locus throughout porcine preimplantation development was also seen in normal mouse embryos but not in cloned embryos which experienced gradual DNA demethylation starting from a higher methylation level [39]. In addition, the DNA demethylation pattern in the porcine NANOG promoter is similar to a mouse study which showed Nanog promoter methylation was erased by active and passive demethylation postfertilization [40]. DNA methylation dynamics in NANOG and POU5F1 may represent a general fashion of epigenetic reprogramming by which gamete-contributed methylation is removed in preimplantation embryos and then reestablished in somatic tissue.
It is generally thought that most CpG islands around the transcription start sites are exempted from DNA methylation when the entire genome undergoes de novo methylation [41]. However, some CpG islands which are associated with long-term silencing such as X chromosome inactivation and genomic imprinting are methylated during specific reprogramming events [21]. The CpG islands in the upstream region of SOX2 and CDX2 loci are hypomethylated and well protected from de novo DNA methylation during normal development. In contrast, the mRNA abundance of SOX2 (Supplemental Fig. S1C) was constantly high in gametes and early embryos relative to the reference gene YWHAG whereas CDX2 (Supplemental Fig. S1D) was only highly expressed in blastocysts. Therefore, the constant DNA hypomethylation in SOX2 and CDX2 loci is not likely to directly modulate their gene expression. Instead, chromatin modifications such as histone methylation and acetylation may be more significantly involved in the transcriptional regulation of SOX2 and CDX2 in porcine early embryos [42].
The imprinted H19 gene is expressed only from the maternal allele, whereas IGF2 is expressed only from the paternal allele [19]. IGF2 and H19 share a common enhancer downstream of H19. The imprint control region in the paternal allele is methylated and thus prevents CTCF binding so that the enhancer can interact with the IGF2 promoter which eventually initiates the transcription of the IGF2 gene. Simultaneously, DNA methylation also silences H19 transcription from the paternal allele. On the contrary, the imprint control region in the maternal allele is hypomethylated and attracts CTCF binding which abolishes the downstream enhancer activity on the IGF2 promoter, leading to the transcriptional silencing of IGF2 [43, 44]. The allele-specific imprinting by DNA methylation is established during germ cell formation but is maintained during preimplantation embryogenesis [45]. Several bisulfite sequencing studies support the idea that allele-specific DNA methylation of DMR upstream of the H19 gene is faithfully replicated during mouse preimplantation development [46, 47]. However, Park et al. [37] argued that porcine DMR of the IGF2/H19 locus was demethylated at the 8-cell stage but was then remethylated in morulae, and suggested dynamic DNA methylation changes in imprinted genes in porcine embryos. In this study, we observed that the differential DNA methylation pattern in the IGF2/H19 locus was well maintained during embryonic development. However, we did not check DNA methylation levels of this particular DMR at porcine 8-cell, 16-cell, and morula stages. Therefore, there might be dynamic DNA methylation changes between 8-cell embryos and morulae because DMRs in imprinted genes were not always faithfully protected from epigenetic reprogramming events during mouse preimplantation embryogenesis [48].
In view of the overall DNA methylation levels in the centromeric repeat, there was no sharp demethylation process postfertilization. Instead, partial DNA demethylation appeared after the 4-cell stage but was not statistically significant. The dynamic DNA methylation profile is similar to a previous report [49] except a relatively higher methylation in blastocysts in our study. It is suggested that repetitive sequences show diverse DNA methylation profile during preimplantation development: LINEs and LTRs transposable elements lose methylation dramatically after fertilization, whereas IAPs retain DNA methylation through the blastocysts stage [15, 20]. Collectively, the nontypical DNA methylation programming in the repeat elements underlies a diverse methylation program during early mammalian development.
Modified DNA Methylation Reprogramming Model in Preimplantation Embryos
Genome-scale DNA methylation studies [15–18], together with our observations (Fig. 7), suggest a diverse DNA methylation reprogramming model during mammalian early embryogenesis. First, most of the differentially methylated regions in imprinted genes retain differential DNA methylation pattern throughout early embryonic development although a small number of gametic DMRs show dynamic stage-specific changes and are not fully protected from DNA methylation reprogramming [48]. Second, repetitive elements exhibit bimodal DNA methylation: some elements (LINE 1) obey DNA demethylation and remethylation behaviors whereas others (IAPs and centromeric repeat) maintain moderate DNA methylation throughout embryonic development. Third, some genomic loci (NANOG and POU5F1) are methylated in either sperm or oocytes and undergo DNA demethylation during preimplantation development. Fourth, the CpG islands located in 5′ upstream region (SOX2 and CDX2) or at housekeeping promoters are generally unmethylated in gametes, early embryos, and somatic tissues. The diverse DNA methylation reprogramming patterns in various genomic loci may be associated with their functions in transcriptional regulation, genomic imprinting and maintaining genome stability.
FIG. 7.
Locus-specific DNA methylation reprogramming in porcine early embryos. The dynamic DNA methylation patterns are generally diverse in porcine early embryos. The pluripotency genes, such as POU5F1 and NANOG, display regular DNA demethylation and de novo methylation process, while the others, such as SOX2 and CDX2, retain hypomethylation in their CpG islands during porcine embryogenesis. The DMR in imprinted gene (IGF2/H19) exhibits differential DNA methylation during development. Additionally, centromeric repeats (multiple copies) maintain moderate DNA methylation during porcine embryonic development.
Provocatively, recent identification of several intermediates such as 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine during active DNA demethylation has further confounded our understanding of the diverse DNA methylation reprogramming in preimplantation embryos [50–52]. Because conventional bisulfite sequencing is not able to distinguish between 5-methylcytosine and 5-hydroxymethylcytosine [53], it is conceivable that some 5-hydroxymethylcytosine may be located in the filled circles shown as methylated in this study. The new oxidative bisulfite sequencing to map 5-hydroxymethylcytosine at single-base resolution will further clarify DNA methylation reprogramming during early porcine embryogenesis [54].
Supplementary Material
ACKNOWLEDGMENT
We would like to thank Dr. Kristin M. Whitworth and Lee D. Spate for technical assistance in embryo culture and bisulfite sequencing.
Footnotes
Supported by National Institutes of Health grant R01 RR013438 and Food for the 21st Century at the University of Missouri.
REFERENCES
- Barton SC, Surani MA, Norris ML. Role of paternal and maternal genomes in mouse development. Nature 1984; 311: 374 376 [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 1984; 37: 179 183 [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature 2000; 403: 501 502 [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 2000; 10: 475 478 [DOI] [PubMed] [Google Scholar]
- Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci U S A 2001; 98: 13734 13738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulka J, Fulka H, Slavik T, Okada K, Fulka J., Jr. DNA methylation pattern in pig in vivo produced embryos. Histochem Cell Biol 2006; 126: 213 217 [DOI] [PubMed] [Google Scholar]
- Young LE, Beaujean N. DNA methylation in the preimplantation embryo: the differing stories of the mouse and sheep. Anim Reprod Sci 2004; 82–83: 61 78 [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999; 99: 247 257 [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 2010; 11: 607 620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 2002; 241: 172 182 [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science 2001; 293: 1089 1093 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D. et al. Initial sequencing and analysis of the human genome. Nature 2001; 409: 860 921 [DOI] [PubMed] [Google Scholar]
- Lander ES. Initial impact of the sequencing of the human genome. Nature 2011; 470: 187 197 [DOI] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 2012; 139: 15 31 [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012; 484: 339 344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, Targets Weber M. and dynamics of promoter DNA methylation during early mouse development. Nat Genet 2010; 42: 1093 1100 [DOI] [PubMed] [Google Scholar]
- Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet 2011; 43: 811 814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, Suzuki Y, Kono T. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet 2012; 8: e1002440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol 2011; 3: a002592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 2003; 35: 88 93 [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012; 13: 484 492 [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998; 95: 379 391 [DOI] [PubMed] [Google Scholar]
- Kehler J, Tolkunova E, Koschorz B, Pesce M, Gentile L, Boiani M, Lomeli H, Nagy A, McLaughlin KJ, Scholer HR, Tomilin A. Oct4 is required for primordial germ cell survival. EMBO Rep 2004; 5: 1078 1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell 2009; 138: 722 737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 2007; 1: 55 70 [DOI] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol 2007; 9: 625 635 [DOI] [PubMed] [Google Scholar]
- Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 2011; 145: 875 889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 2005; 123: 917 929 [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 2005; 132: 2093 2102 [DOI] [PubMed] [Google Scholar]
- Liu T, Zhang X, So CK, Wang S, Wang P, Yan L, Myers R, Chen Z, Patterson AP, Yang CS, Chen X. Regulation of Cdx2 expression by promoter methylation, and effects of Cdx2 transfection on morphology and gene expression of human esophageal epithelial cells. Carcinogenesis 2007; 28: 488 496 [DOI] [PubMed] [Google Scholar]
- Hattori N, Imao Y, Nishino K, Ohgane J, Yagi S, Tanaka S, Shiota K. Epigenetic regulation of Nanog gene in embryonic stem and trophoblast stem cells. Genes Cells 2007; 12: 387 396 [DOI] [PubMed] [Google Scholar]
- Hattori N, Nishino K, Ko YG, Ohgane J, Tanaka S, Shiota K. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J Biol Chem 2004; 279: 17063 17069 [DOI] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol 2009; 10: 526 537 [DOI] [PubMed] [Google Scholar]
- Lai L, Prather RS. Production of cloned pigs by using somatic cells as donors. Cloning Stem Cells 2003; 5: 233 241 [DOI] [PubMed] [Google Scholar]
- Hao Y, Mathialagan N, Walters E, Mao J, Lai L, Becker D, Li W, Critser J, Prather RS. Osteopontin reduces polyspermy during in vitro fertilization of porcine oocytes. Biol Reprod 2006; 75: 726 733 [DOI] [PubMed] [Google Scholar]
- Zhao MT, Whitworth KM, Lin H, Zhang X, Isom SC, Dobbs KB, Bauer B, Zhang Y, Prather RS. Porcine skin-derived progenitor (SKP) spheres and neurospheres: distinct “stemness” identified by microarray analysis. Cellular Reprogramming 2010; 12: 329 345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Kim HS, Lee SG, Lee CK. Methylation status of differentially methylated regions at Igf2/H19 locus in porcine gametes and preimplantation embryos. Genomics 2009; 93: 179 186 [DOI] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell 2011; 146: 866 872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Fujita TC, Low EW, Alarcon VB, Yanagimachi R, Marikawa Y. Gradual DNA demethylation of the Oct4 promoter in cloned mouse embryos. Mol Reprod Dev 2006; 73: 180 188 [DOI] [PubMed] [Google Scholar]
- Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet 2008; 4: e1000116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem 2012; 81: 97 117 [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M, Morris SA, Bruce AW. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet 2009; 10: 467 477 [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 2000; 405: 482 485 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 2000; 405: 486 489 [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet 2001; 2: 21 32 [DOI] [PubMed] [Google Scholar]
- Warnecke PM, Mann JR, Frommer M, Clark SJ. Bisulfite sequencing in preimplantation embryos: DNA methylation profile of the upstream region of the mouse imprinted H19 gene. Genomics 1998; 51: 182 190 [DOI] [PubMed] [Google Scholar]
- Olek A, Walter J. The pre-implantation ontogeny of the H19 methylation imprint. Nat Genet 1997; 17: 275 276 [DOI] [PubMed] [Google Scholar]
- Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 2011; 138: 811 820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YK, Koo DB, Park JS, Choi YH, Kim HN, Chang WK, Lee KK, Han YM. Typical demethylation events in cloned pig embryos. Clues on species-specific differences in epigenetic reprogramming of a cloned donor genome. J Biol Chem 2001; 276: 39980 39984 [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324: 930 935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011; 333: 1303 1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011; 333: 1300 1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 2010; 5: e8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 2012; 336: 934 937 [DOI] [PubMed] [Google Scholar]
- Zhao M, Isom SC, Lin H, Hao Y, Zhang Y, Zhao J, Whyte JJ, Dobbs KB, Prather RS. Tracing the stemness of porcine skin-derived progenitors (pSKP) back to specific marker gene expression. Cloning Stem Cells 2009; 11: 111 122 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.