Abstract
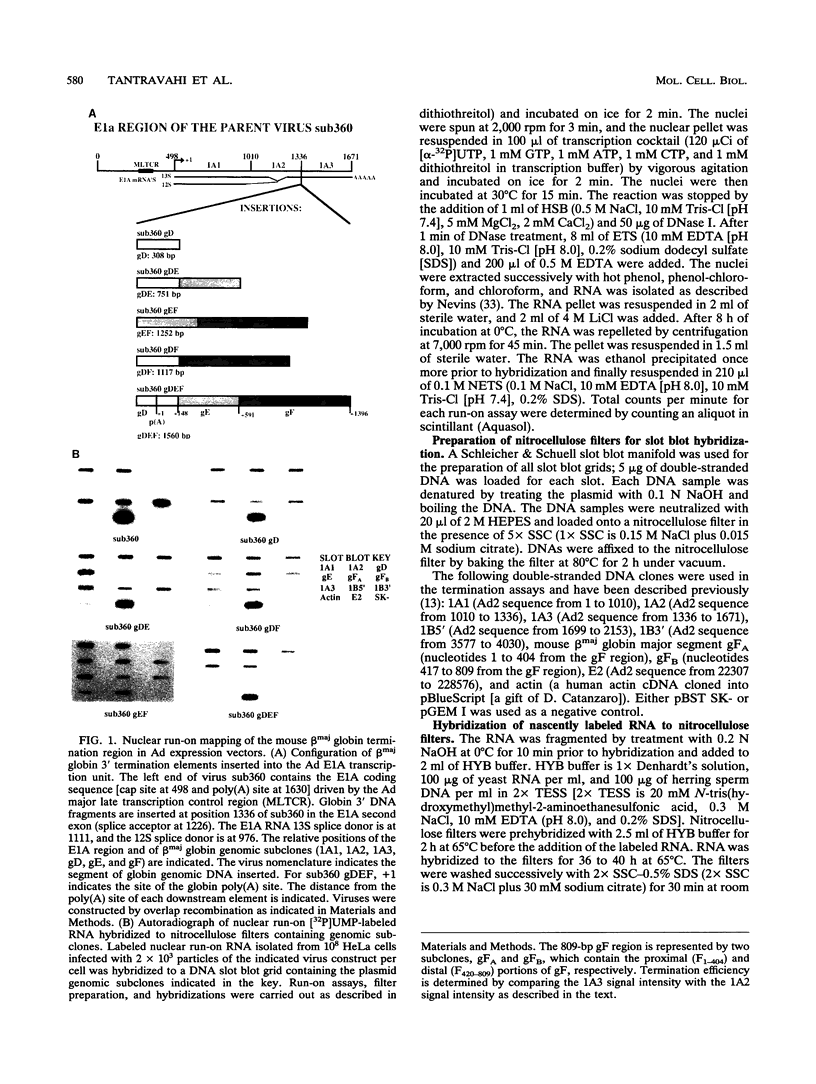
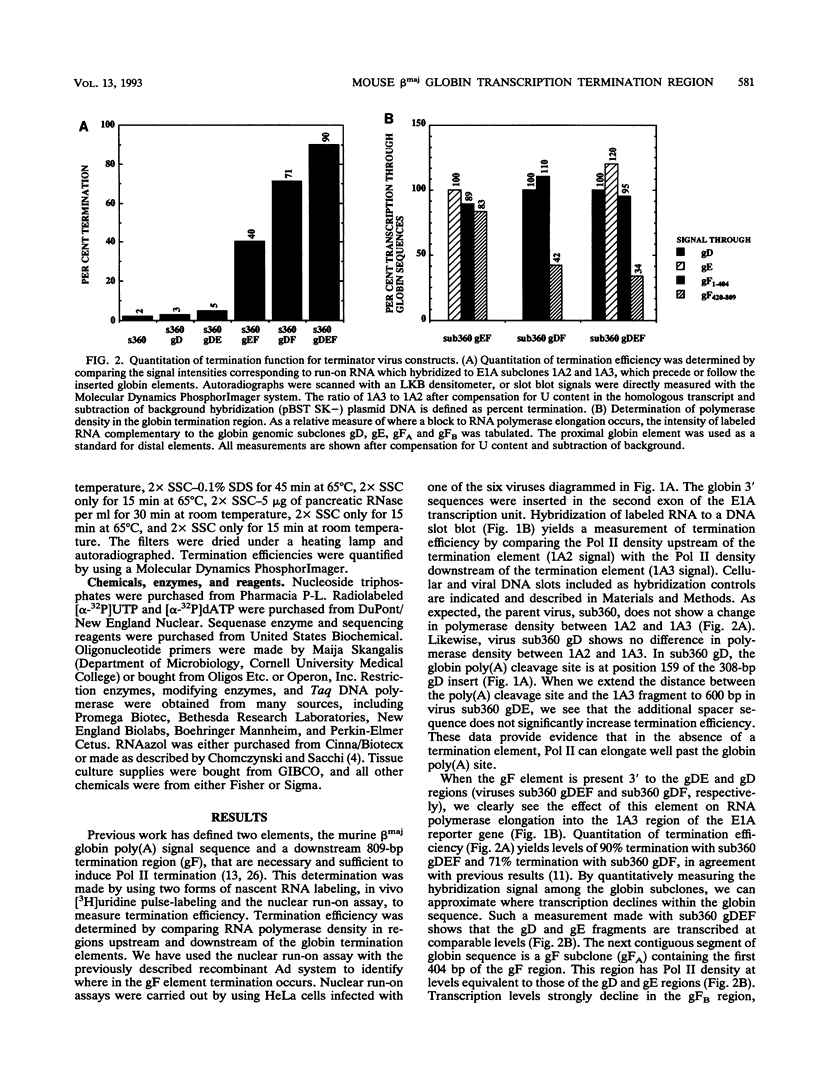
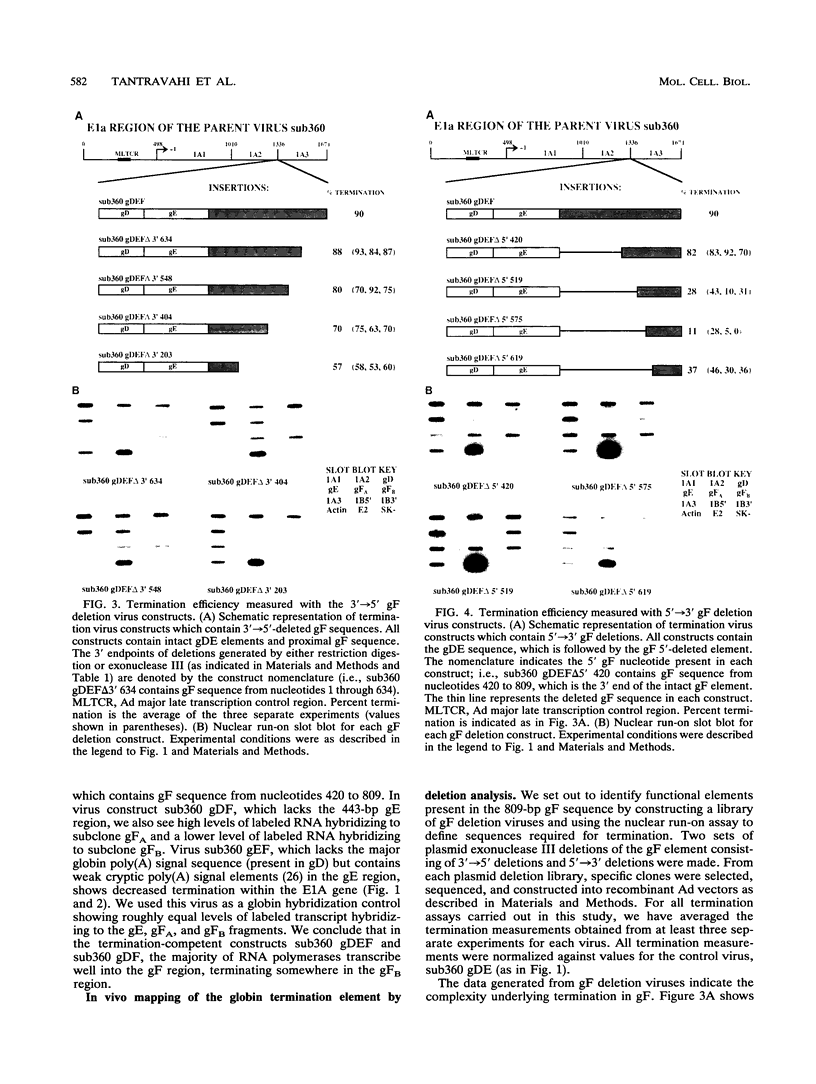
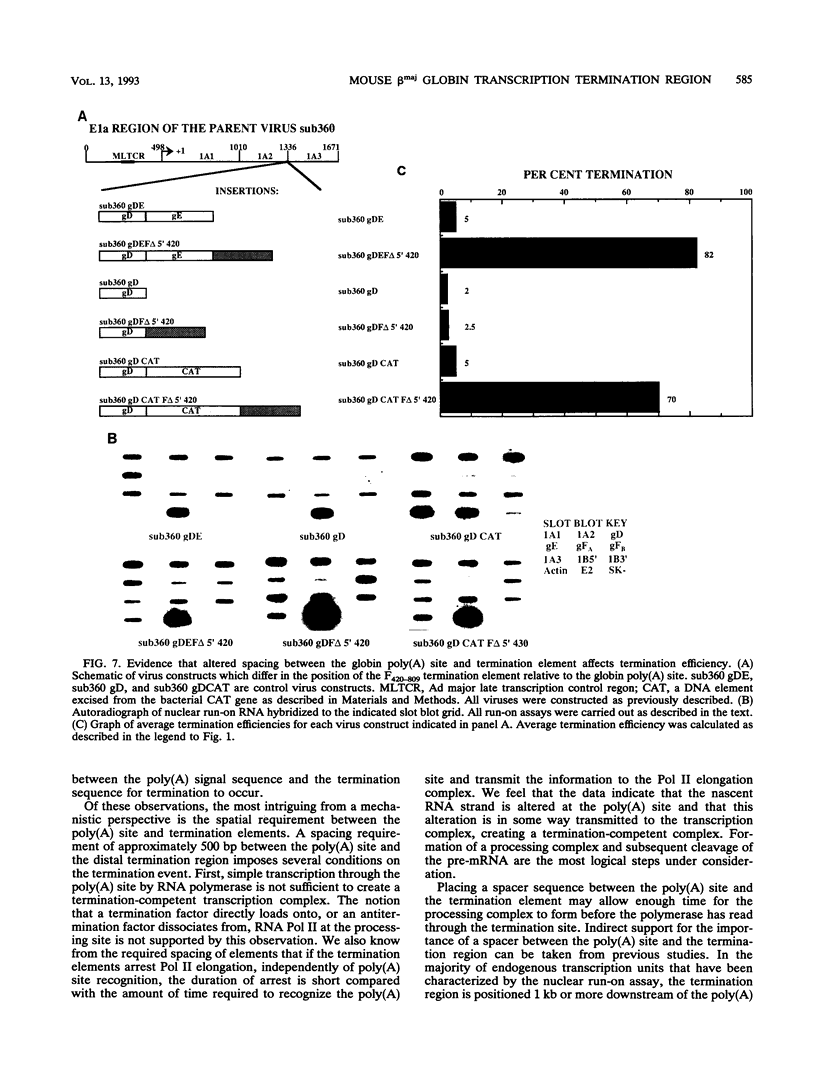
For the majority of mRNA encoding eukaryotic transcription units, there is little or no knowledge of the elements responsible for transcription termination or how they may interact with RNA polymerase. In this report, we have used recombinant adenovirus reporter vectors to characterize the mouse beta maj globin sequence elements that cause transcription termination. Within the globin 3' termination region, we have identified at least three sequence elements which induce significant levels of transcription termination (> 50%). The smallest functionally active element (64% termination) is 69 bp in length. The natural arrangement of these elements results in a cumulative termination which is greater than 90%. Recognition of the termination elements by RNA polymerase II depends on the presence of a functional poly(A) signal sequence. We demonstrate that efficient transcription termination depends on appropriate spacing between the poly(A) signal sequence and the termination element.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birchmeier C., Schümperli D., Sconzo G., Birnstiel M. L. 3' editing of mRNAs: sequence requirements and involvement of a 60-nucleotide RNA in maturation of histone mRNA precursors. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1057–1061. doi: 10.1073/pnas.81.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Chodchoy N., Pandey N. B., Marzluff W. F. An intact histone 3'-processing site is required for transcription termination in a mouse histone H2a gene. Mol Cell Biol. 1991 Jan;11(1):497–509. doi: 10.1128/mcb.11.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Dathan N., Frank R., Philipson L., Mattaj I. W. Formation of the 3' end on U snRNAs requires at least three sequence elements. EMBO J. 1986 Nov;5(11):2931–2937. doi: 10.1002/j.1460-2075.1986.tb04589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron B., Falck-Pedersen E., Salditt-Georgieff M., Darnell J. E., Jr Transcription termination occurs within a 1000 base pair region downstream from the poly(A) site of the mouse beta-globin (major) gene. Nucleic Acids Res. 1984 Nov 26;12(22):8723–8731. doi: 10.1093/nar/12.22.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S., Manley J. L. A CCAAT box sequence in the adenovirus major late promoter functions as part of an RNA polymerase II termination signal. Cell. 1989 May 19;57(4):561–571. doi: 10.1016/0092-8674(89)90126-8. [DOI] [PubMed] [Google Scholar]
- Connelly S., Manley J. L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988 Apr;2(4):440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- Connelly S., Manley J. L. RNA polymerase II transcription termination is mediated specifically by protein binding to a CCAAT box sequence. Mol Cell Biol. 1989 Nov;9(11):5254–5259. doi: 10.1128/mcb.9.11.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. R., Darnell J. E., Jr Cycloheximide stimulates early adenovirus transcription if early gene expression is allowed before treatment. J Virol. 1983 Feb;45(2):683–692. doi: 10.1128/jvi.45.2.683-692.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Harris P., Levitt N., Briggs D., Proudfoot N. J. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 1991 Jul;10(7):1833–1842. doi: 10.1002/j.1460-2075.1991.tb07709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisst S., Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992 Jan 11;20(1):3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck-Pedersen E., Logan J., Shenk T., Darnell J. E., Jr Transcription termination within the E1A gene of adenovirus induced by insertion of the mouse beta-major globin terminator element. Cell. 1985 Apr;40(4):897–905. doi: 10.1016/0092-8674(85)90349-6. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I., Imperiale M. J., Adhya S. L. RNA 3' end formation in the control of gene expression. Annu Rev Genet. 1987;21:453–488. doi: 10.1146/annurev.ge.21.120187.002321. [DOI] [PubMed] [Google Scholar]
- Georgiev O., Birnstiel M. L. The conserved CAAGAAAGA spacer sequence is an essential element for the formation of 3' termini of the sea urchin H3 histone mRNA by RNA processing. EMBO J. 1985 Feb;4(2):481–489. doi: 10.1002/j.1460-2075.1985.tb03654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., Smiley J., Russell W. C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977 Jul;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Grummt I., Maier U., Ohrlein A., Hassouna N., Bachellerie J. P. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985 Dec;43(3 Pt 2):801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Wellauer P. K., Cribbs D. L., Schibler U. Termination of transcription in the mouse alpha-amylase gene Amy-2a occurs at multiple sites downstream of the polyadenylation site. Cell. 1984 Oct;38(3):737–744. doi: 10.1016/0092-8674(84)90269-1. [DOI] [PubMed] [Google Scholar]
- Hernandez N. Formation of the 3' end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985 Jul;4(7):1827–1837. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel G. R., Pederson T. Transcription boundaries of U1 small nuclear RNA. Mol Cell Biol. 1985 Sep;5(9):2332–2340. doi: 10.1128/mcb.5.9.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoix J., Acheson N. H. A rabbit beta-globin polyadenylation signal directs efficient termination of transcription of polyomavirus DNA. EMBO J. 1988 Aug;7(8):2515–2522. doi: 10.1002/j.1460-2075.1988.tb03099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L. F., Roberts J. W., Wu R. RNA polymerase pausing and transcript release at the lambda tR1 terminator in vitro. J Biol Chem. 1983 Aug 10;258(15):9391–9397. [PubMed] [Google Scholar]
- LeMeur M. A., Galliot B., Gerlinger P. Termination of the ovalbumin gene transcription. EMBO J. 1984 Dec 1;3(12):2779–2786. doi: 10.1002/j.1460-2075.1984.tb02209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo S. M., Marzluff W. F. Synthesis of U1 RNA in isolated mouse cell nuclei: initiation and 3'-end formation. Mol Cell Biol. 1987 Dec;7(12):4290–4296. doi: 10.1128/mcb.7.12.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan J., Falck-Pedersen E., Darnell J. E., Jr, Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan J., Shenk T. Adenovirus tripartite leader sequence enhances translation of mRNAs late after infection. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3655–3659. doi: 10.1073/pnas.81.12.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maa M. C., Chinsky J. M., Ramamurthy V., Martin B. D., Kellems R. E. Identification of transcription stop sites at the 5' and 3' ends of the murine adenosine deaminase gene. J Biol Chem. 1990 Jul 25;265(21):12513–12519. [PubMed] [Google Scholar]
- Manley J. L. Polyadenylation of mRNA precursors. Biochim Biophys Acta. 1988 May 6;950(1):1–12. doi: 10.1016/0167-4781(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Morgan W. D., Bear D. G., von Hippel P. H. Rho-dependent termination of transcription. II. Kinetics of mRNA elongation during transcription from the bacteriophage lambda PR promoter. J Biol Chem. 1983 Aug 10;258(15):9565–9574. [PubMed] [Google Scholar]
- Mowry K. L., Oh R., Steitz J. A. Each of the conserved sequence elements flanking the cleavage site of mammalian histone pre-mRNAs has a distinct role in the 3'-end processing reaction. Mol Cell Biol. 1989 Jul;9(7):3105–3108. doi: 10.1128/mcb.9.7.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribyl T. M., Martinson H. G. Transcription termination at the chicken beta H-globin gene. Mol Cell Biol. 1988 Dec;8(12):5369–5377. doi: 10.1128/mcb.8.12.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci. 1989 Mar;14(3):105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991 Feb 22;64(4):671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Rohrbaugh M. L., Johnson J. E., 3rd, James M. D., Hardison R. C. Transcription unit of the rabbit beta 1 globin gene. Mol Cell Biol. 1985 Jan;5(1):147–160. doi: 10.1128/mcb.5.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal E. R., Calvo J. M. Transcription termination sites at the distal end of the leu operon of Salmonella typhimurium. J Mol Biol. 1987 Apr 5;194(3):443–452. doi: 10.1016/0022-2836(87)90673-5. [DOI] [PubMed] [Google Scholar]
- Sato K., Ito R., Baek K. H., Agarwal K. A specific DNA sequence controls termination of transcription in the gastrin gene. Mol Cell Biol. 1986 Apr;6(4):1032–1043. doi: 10.1128/mcb.6.4.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasserot A. P., Schaufele F. J., Birnstiel M. L. Conserved terminal hairpin sequences of histone mRNA precursors are not involved in duplex formation with the U7 RNA but act as a target site for a distinct processing factor. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4345–4349. doi: 10.1073/pnas.86.12.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E., Keller W. The biochemistry of 3'-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- Whitelaw E., Proudfoot N. Alpha-thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3' end processing in the human alpha 2 globin gene. EMBO J. 1986 Nov;5(11):2915–2922. doi: 10.1002/j.1460-2075.1986.tb04587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A. M., Christie G. E., Platt T. Tandem termination sites in the tryptophan operon of Escherichia coli. Proc Natl Acad Sci U S A. 1981 May;78(5):2913–2917. doi: 10.1073/pnas.78.5.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuo C. Y., Ares M., Jr, Weiner A. M. Sequences required for 3' end formation of human U2 small nuclear RNA. Cell. 1985 Aug;42(1):193–202. doi: 10.1016/s0092-8674(85)80115-x. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]









