Abstract
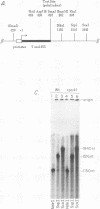
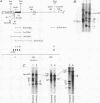
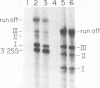
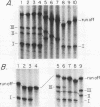
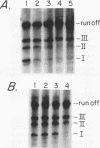
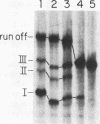
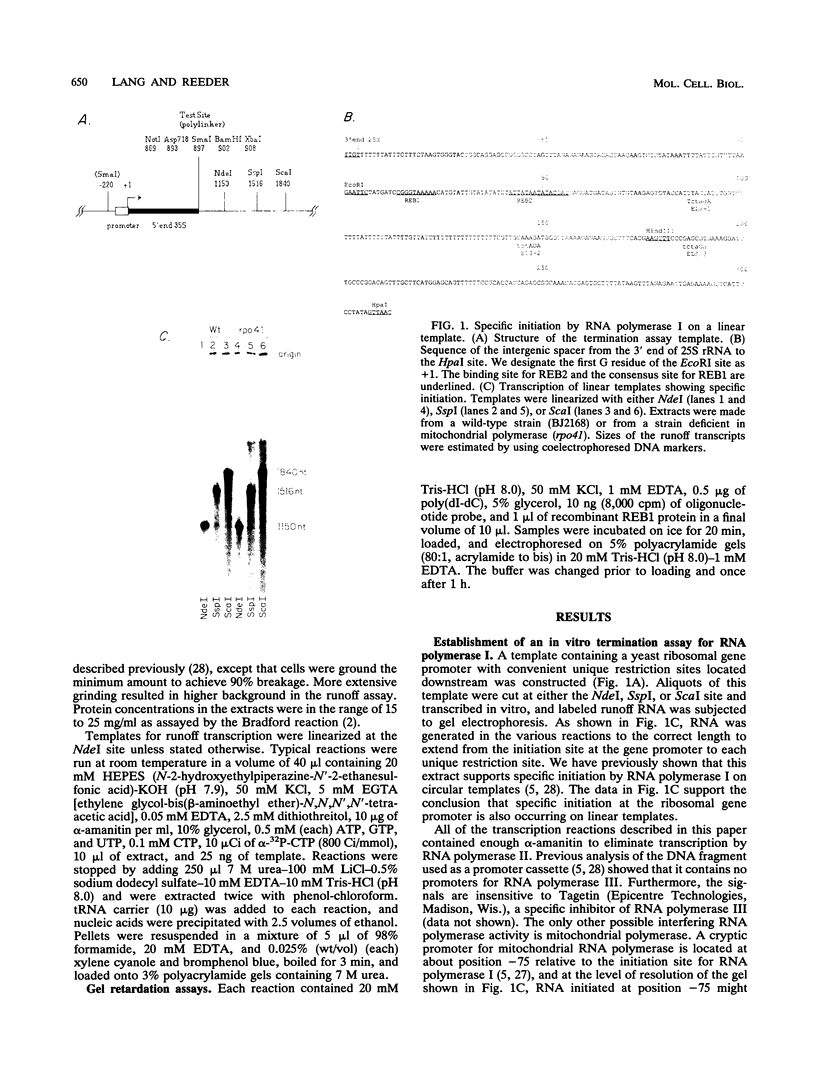
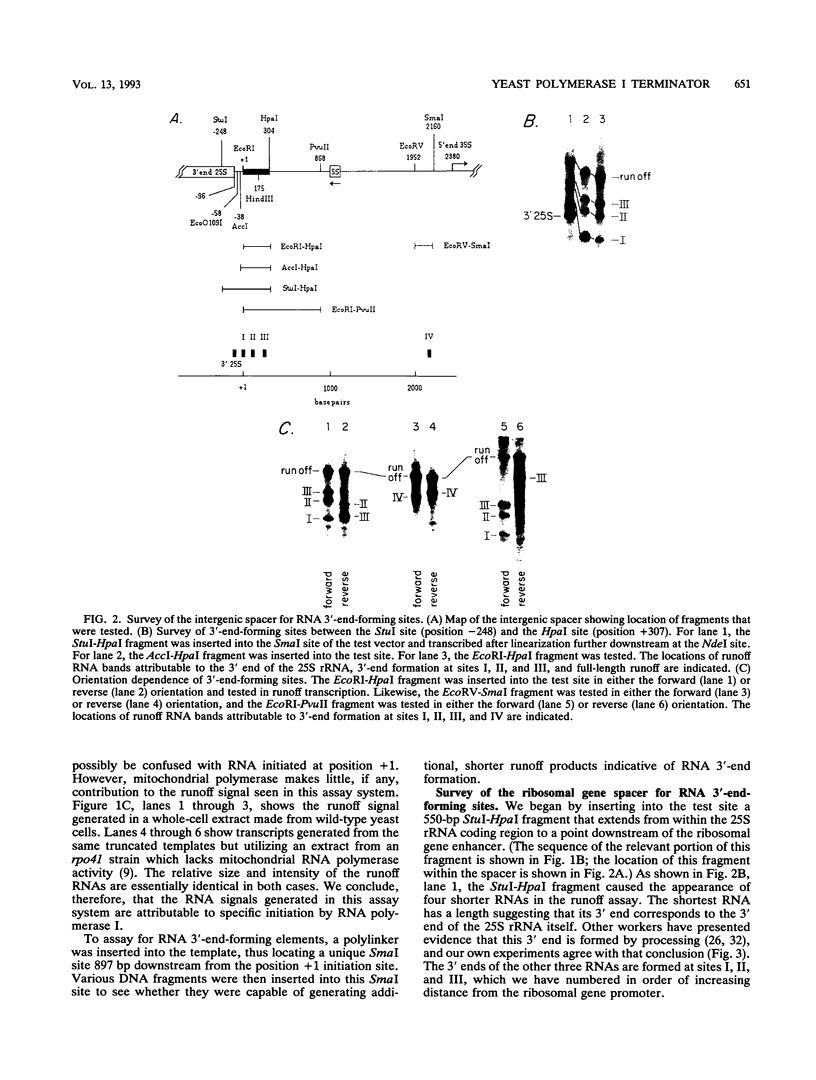
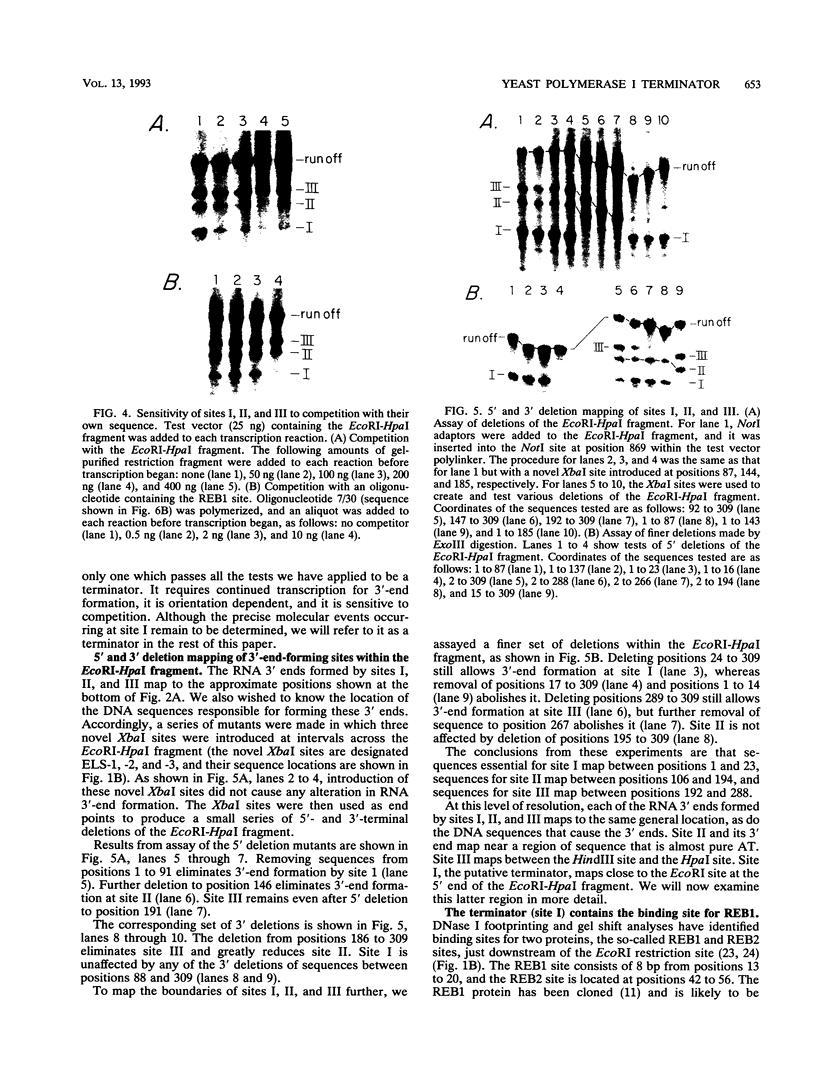
We have identified a terminator for transcription by RNA polymerase I in the genes coding for rRNA of the yeast Saccharomyces cerevisiae. The terminator is located 108 bp downstream of the 3' end of the mature 25S rRNA and shares several characteristics with previously studied polymerase I terminators in the vertebrates. For example, the yeast terminator is orientation dependent, is inhibited by its own sequence, and forms RNA 3' ends 17 +/- 2 bp upstream of an essential protein binding site. The recognition sequence for binding of the previously cloned REB1 protein (Q. Ju, B. E. Morrow, and J. R. Warner, Mol. Cell. Biol. 10:5226-5234, 1990) is an essential component of the terminator. In addition, the efficiency of termination depends upon sequence context extending at least 12 bp upstream of the REB1 site.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kornberg R. D. A yeast ARS-binding protein activates transcription synergistically in combination with other weak activating factors. Mol Cell Biol. 1990 Mar;10(3):887–897. doi: 10.1128/mcb.10.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D. I., Lue N. F., Buchman A. R., LaPointe J. W., Lorch Y., Kornberg R. D. A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev. 1990 Apr;4(4):503–514. doi: 10.1101/gad.4.4.503. [DOI] [PubMed] [Google Scholar]
- Choe S. Y., Schultz M. C., Reeder R. H. In vitro definition of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1992 Jan 25;20(2):279–285. doi: 10.1093/nar/20.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle U., Hipskind R. A., Bujard H. RNA polymerase II transcription blocked by Escherichia coli lac repressor. Science. 1990 Apr 27;248(4954):480–483. doi: 10.1126/science.2158670. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Lue N. F., Kornberg R. D. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988 Nov 5;204(1):109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Greenleaf A. L., Kelly J. L., Lehman I. R. Yeast RPO41 gene product is required for transcription and maintenance of the mitochondrial genome. Proc Natl Acad Sci U S A. 1986 May;83(10):3391–3394. doi: 10.1073/pnas.83.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Rosenbauer H., Niedermeyer I., Maier U., Ohrlein A. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell. 1986 Jun 20;45(6):837–846. doi: 10.1016/0092-8674(86)90558-1. [DOI] [PubMed] [Google Scholar]
- Ju Q. D., Morrow B. E., Warner J. R. REB1, a yeast DNA-binding protein with many targets, is essential for growth and bears some resemblance to the oncogene myb. Mol Cell Biol. 1990 Oct;10(10):5226–5234. doi: 10.1128/mcb.10.10.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Oliemans J., Offenberg H., Dekker A. F., Piper P. W., Planta R. J., Klootwijk J. 3'-End formation of transcripts from the yeast rRNA operon. EMBO J. 1986 Oct;5(10):2703–2710. doi: 10.1002/j.1460-2075.1986.tb04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Hidaka M., Nishizawa M., Horiuchi T. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol Gen Genet. 1992 Jun;233(3):355–362. doi: 10.1007/BF00265431. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Bartsch I., Grummt I. Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature. 1990 Apr 5;344(6266):559–562. doi: 10.1038/344559a0. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Grummt I. 3'-end formation of mouse pre-rRNA involves both transcription termination and a specific processing reaction. Genes Dev. 1989 Feb;3(2):224–231. doi: 10.1101/gad.3.2.224. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Normann A., Bartsch I., Grummt I. The mouse ribosomal gene terminator consists of three functionally separable sequence elements. EMBO J. 1988 May;7(5):1497–1502. doi: 10.1002/j.1460-2075.1988.tb02968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. A 12-base-pair sequence is an essential element of the ribosomal gene terminator in Xenopus laevis. Mol Cell Biol. 1987 May;7(5):1900–1905. doi: 10.1128/mcb.7.5.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. A point mutation uncouples RNA 3'-end formation and termination during ribosomal gene transcription in Xenopus laevis. Genes Dev. 1990 Feb;4(2):269–276. doi: 10.1101/gad.4.2.269. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Ribosomal precursor 3' end formation requires a conserved element upstream of the promoter. Cell. 1987 Jul 3;50(1):51–57. doi: 10.1016/0092-8674(87)90661-1. [DOI] [PubMed] [Google Scholar]
- Lorch Y., Lue N. F., Kornberg R. D. Interchangeable RNA polymerase I and II enhancers. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8202–8206. doi: 10.1073/pnas.87.21.8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A DNA-binding protein is required for termination of transcription by RNA polymerase I in Xenopus laevis. Mol Cell Biol. 1990 Jun;10(6):2793–2800. doi: 10.1128/mcb.10.6.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestel R., Yip M., Holland J. P., Wang E., Kang J., Holland M. J. Sequences within the spacer region of yeast rRNA cistrons that stimulate 35S rRNA synthesis in vivo mediate RNA polymerase I-dependent promoter and terminator activities. Mol Cell Biol. 1989 Mar;9(3):1243–1254. doi: 10.1128/mcb.9.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow B. E., Johnson S. P., Warner J. R. Proteins that bind to the yeast rDNA enhancer. J Biol Chem. 1989 May 25;264(15):9061–9068. [PubMed] [Google Scholar]
- Morrow B. E., Ju Q., Warner J. R. Purification and characterization of the yeast rDNA binding protein REB1. J Biol Chem. 1990 Dec 5;265(34):20778–20783. [PubMed] [Google Scholar]
- Müller E., Neuhaus H., Tobler H., Müller F. Unusual transcription termination of the ribosomal RNA genes in Ascaris lumbricoides. EMBO J. 1990 Sep;9(9):2849–2856. doi: 10.1002/j.1460-2075.1990.tb07474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P. W., Bellatin J. A., Lockheart A. Altered maturation of sequences at the 3' terminus of 5S gene transcripts in a Saccharomyces cerevisiae mutant that lacks a RNA processing endonuclease. EMBO J. 1983;2(3):353–359. doi: 10.1002/j.1460-2075.1983.tb01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs D. L., Nomura M. Specific transcription of Saccharomyces cerevisiae 35 S rDNA by RNA polymerase I in vitro. J Biol Chem. 1990 May 5;265(13):7596–7603. [PubMed] [Google Scholar]
- Schultz M. C., Choe S. Y., Reeder R. H. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S. E., Roeder G. S. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Aug;9(8):3464–3472. doi: 10.1128/mcb.9.8.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Nicholson P. R., Stillman D. J. Identification of a Saccharomyces cerevisiae DNA-binding protein involved in transcriptional regulation. Mol Cell Biol. 1990 Apr;10(4):1743–1753. doi: 10.1128/mcb.10.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip M. T., Holland M. J. In vitro RNA processing generates mature 3' termini of yeast 35 and 25 S ribosomal RNAs. J Biol Chem. 1989 Mar 5;264(7):4045–4051. [PubMed] [Google Scholar]
- van der Sande C. A., Kulkens T., Kramer A. B., de Wijs I. J., van Heerikhuizen H., Klootwijk J., Planta R. J. Termination of transcription by yeast RNA polymerase I. Nucleic Acids Res. 1989 Nov 25;17(22):9127–9146. doi: 10.1093/nar/17.22.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]