Abstract
This study demonstrates the effects of simulated microgravity on E. coli K 12 MG1655 grown on LB medium supplemented with glycerol. Global gene expression analysis indicated that the expressions of hundred genes were significantly altered in simulated microgravity conditions compared to that of normal gravity conditions. Under these conditions genes coding for adaptation to stress are up regulated (sufE and ssrA) and simultaneously genes coding for membrane transporters (ompC, exbB, actP, mgtA, cysW and nikB) and carbohydrate catabolic processes (ldcC, ptsA, rhaD and rhaS) are down regulated. The enhanced growth in simulated gravity conditions may be because of the adequate supply of energy/reducing equivalents and up regulation of genes involved in DNA replication (srmB) and repression of the genes encoding for nucleoside metabolism (dfp, pyrD and spoT). In addition, E. coli cultured in LB medium supplemented with glycerol (so as to protect the cells from freezing temperatures) do not exhibit multiple stress responses that are normally observed when cells are exposed to microgravity in LB medium without glycerol.
Introduction
Microbes have the ability to sense and respond to environmental changes occurring in their vicinity. This adaptability confers on them the capacity to thrive under various extreme environmental niches including microgravity. When exposed to microgravity cells experience reduced gravity resulting in a relative lack of sedimentation, low shear stress and low turbulence [1]. These physical effects of microgravity may influence the growth and also induce other physiological changes. Studies have indicated that bacteria exposed to reduced gravity exhibit up regulation of genes involved in starvation response, acid stress, osmotic stress, oxidative stress, biofilm formation, curli biosynthesis and lipid biosynthesis [1]–[3]. Resistance towards antibiotics and increase in virulence was also reported in different bacteria exposed to microgravity [4]–[6]. The above effects of microgravity on bacteria are possibly dependent on the specific media used for culturing the bacteria [7]–[9].
Functional genomic technologies [10] using Escherichia coli strains as a model organism have provided insights in to the effects of microgravity at the genome level. Tucker et al., [11] demonstrated that the expression of genes in E. coli under microgravity conditions varied depending on whether the culture was grown on minimal medium or rich medium like Luria-Bertani (LB) broth [11]. Subsequently, Vukanti et al., [1] using DNA microarrays and E. coli K 12 identified a number of genes that were significantly altered in expression under microgravity conditions [1]. In two recent studies again in Escherichia coli it was clearly demonstrated that microgravity induced expression of multiple stress genes depends on the nutritional status and nutrient resources modulate the responses [12]–[13]. Further, in Salmonella typhimurium microgravity decreased the virulence of the bacteria if LB medium was supplemented with phosphate ions [6]. Taken together all these studies substantiate that media composition influences the changes occurring in gene expression under microgravity condition.
In this study, a clinostat that mimics microgravity conditions was used to investigate the effects of microgravity on E. coli K12 MG1655 grown in LB medium supplemented with glycerol to monitor the effects on growth and global gene expression. The need for supplementing LB with glycerol was crucial because the incubator in which the bacteria is proposed to be grown at 30°C in space, under microgravity conditions, in a space recovery capsule due to unforeseen reasons may fail and the incubation temperature may decrease to below 0°C. Such unforeseen failures are not very uncommon in space flights. Haga and Saleh [14] analysed 168 satellites that were launched during 1995 to 2009 and recorded a total of 170 failure events which included 62 minor, 105 major and 3 fatal failure events. Thus, it may be essential to culture the bacterium in the presence of glycerol to protect the cultures from any kind of power failure in the spacecraft resulting in freezing temperatures.
The present study differs from the earlier studies in that the LB medium used in the present study is supplemented with glycerol. There is a reasonable chance to assume that the response of the bacterium to microgravity may vary with respect to growth and expression of genes. These assumptions are based on the earlier findings that media components influence the growth and expression of genes of bacteria exposed to microgravity [11]–[13]. In addition it would be interesting to see how glycerol supplementation in the medium would help to overcome the nutrient limiting conditions imposed due to a nutrient depletion zone formed when cells are grown under microgravity conditions [1], [9].
The results of the present study is in accordance with earlier studies that had indicated that E. coli exhibits enhanced growth rate and that several genes are differentially expressed when exposed to microgravity [1], [12]–[13]. But, the genes that were differentially regulated were not identical. For instance Vukanti et al., [1] observed that many of the genes that were up regulated were identified as stress-inducible genes and included starvation-inducible genes, genes associated with multiple stress responses, genes involved in biofilm formation, curli biosynthesis and lipid biosynthesis [1]. None of these genes were up regulated in the current study implying that the presence of glycerol in both the control and microgravity exposed cultures nullified the up regulation of stress-inducible genes.
Further, the genes that facilitate the formation of the nutrient depletion zone in cells grown under microgravity conditions such as up regulation of genes coding for nutrient transport and metabolic enzymes and simultaneously down regulation of genes associated with translation apparatus, DNA replication and cell division [1] were not differentially expressed in the presence of glycerol in the medium. Thus implying that glycerol may not favour the formation of a nutrient depletion zone since it could be utilized as a carbon source. In fact the study also indicates that the enhanced growth of E. coli under microgravity may be because of adequate supply of energy/reducing equivalents and increase in transcripts for DNA replication.
This study also for the first time assigns the differentially regulated genes to sixteen different functional pathways such as several metabolic pathways (purine, pyrimidine, carbohydrate, amino acid etc.), ABC transporter systems, two component systems etc., by KEGG (Kyoto Encyclopedia of Genes and Genomes). In addition, DAVID (Database for Annotation, Visualization and Integrated Discovery) was used for geneontology annotations and the analysis yielded up regulated genes involved in DNA transcription and regulation of transcription, genes coding for integral membrane proteins, ion binding and DNA binding etc. The 21 down regulated genes were implicated in nitrogen and amine compound biosynthetic process, ion transport, carbohydrate catabolic process and nucleoside metabolic process. DAVID analysis also annotated genes involved in cellular components such as cell wall, cell membrane and organelle membrane and seven hypothetical protein coding genes namely yfjD, ydcQ, ynfA, ybdJ, yniB, ygaQ and hokE were assigned to various functional processes in response to microgravity conditions in E. coli.
Materials and Methods
Bacterial Strain and Growth Conditions
In this study, Escherichia coli K12 MG1655 was grown in LB broth (containing 10 g peptone, 10 g NaCl and 5 g yeast extract in 1000 ml distilled water) which was supplemented with 10% (v/v) glycerol (hereafter LB) at 30°C. To develop the inoculums, a single colony of E. coli K 12 MG1655 from the LB agar plate was inoculated into LB broth and incubated for growth under shaking at 150 rpm and at 30°C. E. coli growth was monitored by measuring absorbance at 600 nm. E. coli grown to a OD of 0.8 (106 CFU/ml) was used as inoculum for all the experiments unless otherwise mentioned.
Growth of E. coli K12 MG1655 in a Clinostat
A three-dimensional (3D) clinostat is an apparatus that nullifies the effect of gravity, and it has been used to evaluate the effects of microgravity on cells [1], [15]. A 3D clinostat is capable of randomizing motion and thus cancels the uniform gravity influence and as a consequence an object is subjected to weightlessness which is referred commonly as simulated microgravity. The formula for microgravity (g′) is g′ = ω2R/g where g = 9.8 m/s2, R = radius from the centre of rotation, ω = constant angular velocity (ω) where angular velocity is equal to angular displacement in radians/time taken (θ/τ). The angular velocity obtained using the clinostat was 2 rads/s. At this angular velocity, the simulated microgravity is 1×10−3 [16]. The microgravity in the present study was 1×10−3.
To 300 ml of LB medium 3.0 ml of logarithmically growing E. coli culture (0.8 OD) was added and 15 ml of this inoculum was distributed into 15 ml separate vessels and grown in clinostat at 30°C. Under these conditions the g was 1×10−3 g. After every one hour interval one vessel was detached from the instrument and OD600nm of the culture was determined and used to construct the growth curves. E. coli culture that was setup as above and grown in an incubator shaker at 30°C under normal gravity conditions served as control. Data of three experiments that were conducted under similar conditions separately was considered in plotting the growth curves.
For simulated microgravity studies, 50 ml of the LB broth was inoculated with 500 µl of logarithmically growing E. coli culture and grown in a 50 ml vessel in a clinostat at 30°C. A control was set up as above and grown under normal gravity conditions in an incubator shaker at 30°C. Three separate vessels were maintained under each condition. When the OD600nm reached 0.8, 10 ml of the culture collected from each of these vessels was suspended in 3 volumes of RNAlater solution (Ambion Inc., USA) in their respective sample bottles and stored at 4°C. From the stored culture 3 ml was used for the isolation of RNA.
RNA Extraction
Qiagen RNeasy mini-prep kit was used for the extraction of RNA from about 3 ml of E. coli culture. The culture was pelleted, suspended in 200 µl Tris-lysozymemixture at room temperature for 5 min and then subjected to lysis with 700 µl of lysis buffer containing 1% β-mercaptoethanol. Lysis was facilitated by vigorous vortexing the suspension of cells for 5 min. Absolute ethanol (500 µl) was added and the mixture was transferred to a mini-spin column placed in a 2 ml centrifuge tube and centrifuged at 13,000 rpm for 30 s. At this stage, 10 µl of DNase prepared in 90 µl of buffer was added to the columns and the columns were kept at room temperature for 10 min. The columns were then washed at 13,000 rpm for 30 s with 500 µl of wash buffers RW1 and purified RNA was collected in 20 µl of RNase-free water after centrifugation at 13,000 rpm for 2 min. Quality of the purified RNA was assessed following gel electrophoresis and the OD 260/280 ratio of the purified RNA ranged from 1.8–2.0. Quantification of the RNA was done using a Nanodrop spectrophotometer (Nano Drop Technologies, USA).
cDNA Synthesis
RNA (5–10 µg) extracted as above was used for cDNA synthesis using the first strand cDNA synthesis kit from Invitrogen (Invitrogen Bioservices India Pvt. Ltd., Bangalore). Annealing of the primers with the purified RNA was accomplished by incubating the RNA and primer reaction mix at 70°C for 10 min followed by incubation at 25°C for 10 min. cDNA was then synthesized by adding superscript III and incubating the reaction mixture at 25°C for 10 min, 37°C for 60 min, 42°C for 60 min and 70°C for 10 min. The cDNA was fragmented with DNAse 1 (Promega Corporation, Madison, USA) and then labelled with biotin at the 3′ end using the labelling reagent from Affymetrix (CA, USA) and Terminal transferase enzyme (Promega).
DNA Microarray Analysis
E.coli Genome 2.0 gene chip arrays were purchased from Affymetrix. The chip contained the complete genome of four E. coli strains (viz., non-pathogenic E. coli K12 MG1655, uropathogenic E. coli strain CFT073 and enterohemorragic E. coli O157:H7 strains EDL 933 and Sakai). The gene chip consists of approximately 10,000 probe sets for the 20,366 genes of all the four strains of E. coli. DNA micro array chips were hybridized with the labelled cDNA using the Affymetrix protocol (www.affymetrix.com). Microarray slides were then scanned using Affymetrix 428 Array Scanner and GCOS software to obtain images of the chips and further processed to get intensity cell files for the probe sets. The intensity cell files were then imported normalized for background correction and data analysed using Gene Spring 11.5 software. Genes that exhibited ≥1.5 fold increase or decrease (treated versus control) in expression and P≤0.05 were considered as differentially regulated due to microgravity.
The microarray data was submitted to Gene Expression Omnibus (GEO) web deposit of National Centre for Biotechnology Information (NCBI) with an accession number GSE40648.
Real Time PCR
The RT-PCR reactions (10 µl) were performed in triplicate with 2.5 pM primer (Table 1) and SYBER Green PCR Master Mix (Applied Biosystems, CA, USA). Template was pre-incubated at 50°C for 2 min, denatured at 95°C for 10 min and subjected to 40 cycles under the following thermal conditions: 95°C (15 s) and 52°C (30 s). Relative expression of genes was calculated by ΔΔCT method which is based on product cycle threshold (CT). Expression of 16S rRNA gene was used as an internal standard for RT-PCR. All values reported represent the mean of at least three independent experiments.
Table 1. Primers used for Real-Time PCR analysis of a few genes of Escherichia coli.
| Sl.No. | Gene | Primer | Sequence (5′-3′) |
| 1 | hyaE | hyaE-rt F | CTTGACGACTGGCTTACG |
| hyaE-rt R | GCCACCTGCCATGTATAG | ||
| 2 | mdtD | mdt-rt F | TTATCGTCGGGTACTGGTAG |
| mdtD -rt R | GAGTTGACCATCCCTTGTAA | ||
| 3 | srmB | srmB-rt F | AACATATTGCTGGCGAAA |
| srmB-rt R | GAGGGATTGGCAGAAACT | ||
| 4 | pyrD | pyrD-rt F | GAAGAATTGATCCAGGTTGC |
| pyrD-rt R | TCCCTGAACAAGAGAACGAT | ||
| 5 | rhaD | rhaD-rt F | GCGAATGTTTTTGCATCTCT |
| rhaD -rt R | CGAGTGTCTGGTGGTATTC | ||
| 6 | yicL | yicL-rt F | CGTCGCAGTTTTTGACTATG |
| yicL -rt R | AAAAATCAGCAGGCTAATGG | ||
| 7 | ldcC | ldcC-rt F | TGTTGATGCCTGGAGAAA |
| ldcC-rt R | TCAAAACCGGGGTAATGT | ||
| 8 | 16S | 16S-rtF | GTGCAATATTCCCCACTGCT |
| 16S-rtR | CGATCCCTAGCTGGTCTGAG |
Assignment of the Differentially Regulated Genes to Functional Pathways by KEGG and DAVID
Genes identified by the microarray analysis were analyzed to identify relevant functional pathways by KEGG (Kyoto Encyclopedia of Genes and Genomes) and DAVID (Database for Annotation, Visualization and Integrated Discovery). A cutoff p value of 0.05 was used for enriched KEGG pathways and geneontology functions by DAVID. Out of 100 genes that were differentially regulated only 57 genes were considered for analysis using KEGG and DAVID. The remaining 43 genes that coded for unknown or intergenic regions were not included in the analysis.
Results
Growth of E. coli in the Clinostat
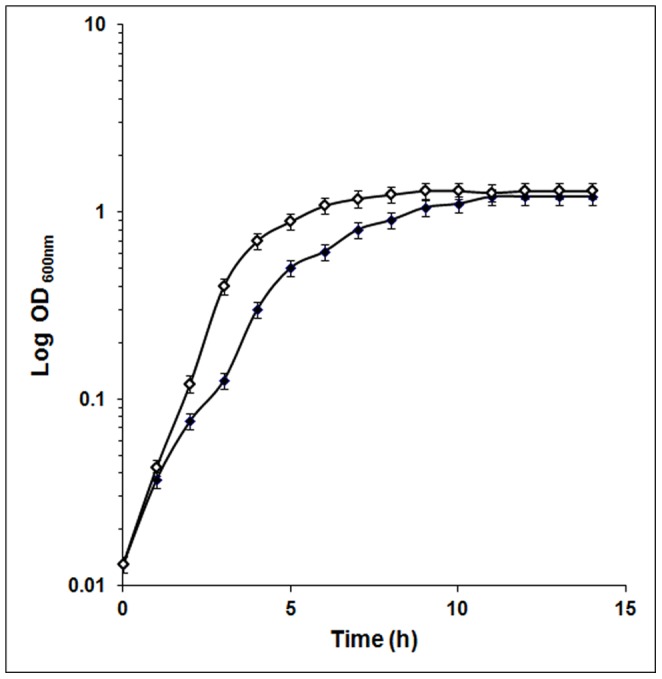
Growth of E. coli was monitored under both simulated microgravity conditions and normal gravity conditions. Cultures grown in the clinostat exhibited enhanced growth rate and reached the stationary phase earlier than the culture grown at normal gravity (Fig.1).
Figure 1. Growth of E. coli at 30°C under microgravity conditions in a clinostat (□) and under normal gravity (▪) conditions.
Expression of Genes in E. coli Grown in a Clinostat
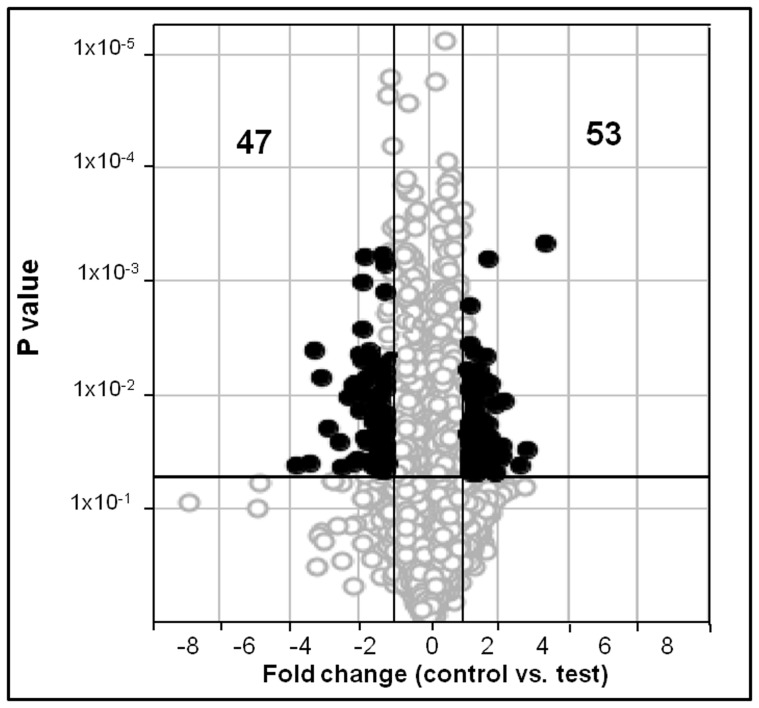
In E. coli grown under simulated microgravity conditions hundred genes were differentially expressed with a fold change ≥1.5 (P≤0.05). Three chips were used for control and experimental conditions respectively. Fifty three genes were up regulated while forty seven genes were down regulated under simulated microgravity conditions (Tables 2 and 3 and Fig. 2). The up regulated genes were identified as genes coding for ATP-dependent DNA helicase (srmB), multidrug efflux system protein (mdtD/yegB), cysteine desulfuration protein involved in oxidative stress (sufE/ynhA), periplasmic proteins involved in metal detoxification (cusF), chaperone for hydrogenase isoenzyme (hyaE), a DNA binding transcriptional regulator (IlvY), non coding RNA genes (ryjA and ssrA), one pseudo gene (intK), twelve hypothetical proteins (ymgG, yaiB, ybdJ, hokE, mokB, ydcQ, ynfA, yniB, ECs3519, ybaM, ydbL and ydfO), thirty one intergenic region genes and one gene coding for an unknown protein (Table 2).
Table 2. Microgravity-induced up regulation of genes in Escherichia coli.
| Probe set ID | Fold change* | Gene | Gene function |
| 1761737_at | 1.5 | srmB | ATP-dependent RNA helicase |
| 1764160_s_at | 1.5 | mdtD/yegB | multidrug efflux system protein |
| 1759467_s_at | 1.7 | sufE/ynhA | cysteine desufuration protein SufE |
| 1761044_at | 1.6 | cusF | periplasmic copper-binding protein |
| 1761315_s_at | 1.5 | hyaE | hydrogenase-1 operon protein HyaE |
| 1768123_s_at | 1.6 | ilvY | DNA-binding transcriptional regulator IlvY |
| 1767538_at | 1.5 | intK | pseudo |
| 1760679_at | 2.1 | ryjA | ncRNA |
| 1762568_s_at | 1.7 | ssrA | misc_RNA/tmRNA |
| 1763277_s_at | 1.5 | ymgG | hypothetical protein |
| 1769259_s_at | 1.5 | yaiB | hypothetical protein |
| 1765637_s_at | 1.5 | ybdJ | hypothetical protein |
| 1764857_s_at | 1.8 | hokE | hypothetical protein |
| 1762609_s_at | 1.7 | mokB | hypothetical protein |
| 1764733_s_at | 1.5 | ydcQ | hypothetical protein |
| 1760864_s_at | 1.5 | ynfA | hypothetical protein |
| 1766918_s_at | 1.6 | yniB | hypothetical protein |
| 1763411_s_at | 1.8 | ECs3519 | hypothetical protein |
| 1760935_s_at | 1.8 | ybaM | hypothetical protein |
| 1766171_s_at | 1.8 | ydbL | hypothetical protein |
| 1763909_s_at | 1.5 | ydfO | hypothetical protein |
| 1767707_at | 1.7 | – | Unknown |
| 1764171_s_at | 1.6 | – | intergenic region |
| 1767139_s_at | 1.5 | – | intergenic region |
| 1762045_s_at | 1.5 | – | intergenic region |
| 1765005_s_at | 1.6 | – | intergenic region |
| 1768087_s_at | 1.6 | – | intergenic region |
| 1765131_s_at | 1.9 | – | intergenic region |
| 1763703_s_at | 1.7 | – | intergenic region |
| 1761358_s_at | 1.8 | – | intergenic region |
| 1761890_s_at | 1.6 | – | intergenic region |
| 1762015_s_at | 2.1 | – | intergenic region |
| 1761719_s_at | 1.7 | – | intergenic region |
| 1760269_s_at | 1.5 | – | intergenic region |
| 1761431_s_at | 1.6 | – | intergenic region |
| 1767216_s_at | 2.1 | – | intergenic region |
| 1760296_s_at | 1.5 | – | intergenic region |
| 1763899_s_at | 1.5 | – | intergenic region |
| 1759771_s_at | 2 | – | intergenic region |
| Probe set ID | Fold change * | Gene | Gene function |
| 1765387_s_at | 1.8 | – | intergenic region |
| 1765961_s_at | 1.5 | – | intergenic region |
| 1762257_s_at | 2 | – | intergenic region |
| 1767591_s_at | 2 | – | intergenic region |
| 1761987_s_at | 1.6 | – | intergenic region |
| 1765027_s_at | 1.8 | – | intergenic region |
| 1765126_s_at | 1.7 | – | intergenic region |
| 1762765_s_at | 1.5 | – | intergenic region |
| 1767249_s_at | 1.6 | – | intergenic region |
| 1764126_s_at | 2.5 | – | intergenic region |
| 1767857_s_at | 1.8 | – | intergenic region |
| 1766406_s_at | 1.8 | – | intergenic region |
| 1764615_s_at | 1.6 | – | intergenic region |
| 1763140_s_at | 2 | – | intergenic region |
Genes that showed fold change greater than 1.5 (P<0.05).
Table 3. Microgravity-induced down regulation of genes in Escherichia coli.
| Probe set ID | Fold change* | Gene | Gene function |
| 1768937_at | 1.6 | ompC | outer membrane porin protein C |
| 1767282_s_at | 1.5 | hisA | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase |
| 1766205_x_at | 2.4 | hisL | his operon leader peptide |
| 1763370_s_at | 1.5 | insA | KpLE2 phage-like element; IS1 repressor protein InsA |
| 1763159_s_at | 2 | ldcC | lysine decarboxylase, constitutive |
| 1761146_s_at | 1.5 | livG | leucine/isoleucine/valine transporter ATP-binding subunit |
| 1765703_at | 1.9 | aroP | aromatic amino acid transporter |
| 1767440_s_at | 1.5 | cysW | sulfate/thiosulfate transporter subunit |
| 1759826_s_at | 2.7 | cysH | 3′-phosphoadenosine 5′-phosphosulfate reductase |
| 1761464_at | 1.6 | nupC | nucleoside (except guanosine) transporter |
| 1763832_at | 1.7 | actP | acetate transporter |
| 1759775_at | 1.6 | ptsA | fused predicted PTS enzymes: Hpr component/enzyme I component/enzyme IIA component |
| 1764279_s_at | 1.5 | mgtA | magnesium-transporting ATPase MgtA |
| 1766765_s_at | 1.5 | nikB | nickel transporter permease NikB |
| 1768074_s_at | 1.5 | gst | glutathionine S-transferase |
| 1769236_s_at | 2.1 | rhaD | rhamnulose-1-phosphate aldolase |
| 1768302_s_at | 1.8 | rhaS | transcriptional activator RhaS |
| 1764298_s_at | 1.5 | ydcR | putative transcriptional regulator |
| 1759346_s_at | 1.6 | hiuH | transthyretin-like protein precursor |
| 1763816_s_at | 1.6 | dfp | bifunctional phosphopantothenoylcysteine decarboxylase/phosphopantothenate synthase |
| 1766833_s_at | 1.6 | spoT | bifunctional (p)ppGpp synthetase II/guanosine-3′,5′-bis pyrophosphate 3′-pyrophosphohydrolase |
| 1759907_s_at | 2 | pyrD | dihydro-orotate oxidase, FMN-linked |
| 1767020_s_at | 1.5 | pldB | lysophospholipase L2 |
| 1767318_s_at | 2 | exbB | biopolymer transport protein ExbB |
| 1763278_at | 1.5 | pbl | pseudo |
| 1761419_s_at | 2 | speB | agmatinase |
| 1766645_s_at | 2.1 | yicI | alpha-xylosidase YicI |
| 1767015_at | 1.7 | yfjV | pseudo |
| 1763267_s_at | 1.6 | yeaM | putative ARAC-type regulatory protein |
| 1759710_at | 1.5 | ybeR | predicted protein |
| 1763523_s_at | 1.7 | ypjA | hypothetical protein |
| 1765691_s_at | 1.6 | yfaT | hypothetical protein |
| 1759640_s_at | 1.6 | yfjD | hypothetical protein |
| Probe set ID | Fold change * | Gene | Gene function |
| 1768572_s_at | 1.8 | yehP | hypothetical protein |
| 1768317_s_at | 1.6 | yjbG | hypothetical protein |
| 1765663_s_at | 1.8 | ykiA | hypothetical protein |
| 1762026_at | 1.9 | – | Unknown |
| 1768646_s_at | 1.6 | – | intergenic region |
| 1762447_s_at | 1.6 | – | intergenic region |
| 1761029_s_at | 1.5 | – | intergenic region |
| 1760120_s_at | 1.6 | – | intergenic region |
| 1765833_s_at | 1.7 | – | intergenic region |
| 1766353_s_at | 1.6 | – | intergenic region |
| 1762591_s_at | 3.7 | – | intergenic region |
| 1763867_s_at | 1.5 | – | intergenic region |
| 1766044_s_at | 2.4 | – | intergenic region |
| 1767764_s_at | 2.2 | – | intergenic region |
Genes that showed fold change greater than 1.5 (P<0.05).
Figure 2. DNA microarray analysis of clinostat-induced gene expression in E. coli.
The Volcano plot depicts gene expression in E. coli culture at 0.8 OD (OD600 nm) cultured in the presence of 10% glycerol under microgravity conditions compared to the control. Genes that are represented on the right side of the volcano-axis are up regulated and those that are on left side of the axis are down regulated. Out of the 4377 genes (O) analysed, 53genes were upregulated (•) and 47were down regulated (•).Only those genes that showed more than 1.5 fold change in expression and a P value <0.05 were identified as either up- or down-regulated. The x-axis represents the fold change and the dark vertical lines represent cut-offs at 1.5 fold decrease and increase. The y-axis represents the p-values and the dark horizontal line indicates a p value cut-off of 0.05.
The genes that were down regulated included a gene coding for outer membrane porin protein C (ompC), his operon genes (hisL, hisA), insertion element repressor protein (insA), lysine decarboxylase (ldcC), transporter proteins (actP, aroP, exbB, livG, nupC and cysW), 3′-phosphoadenosine 5′-phosphosulphate reductase (cysH,), metal transport proteins (mgtA and nikB), fused predicted PTS system enzyme (ptsA), glutathionine S-transferase (gst), rhamulose-1-phosphate aldolase (rhaD), transcriptional activator (rhaS), putative transcriptional regulatory protein (ydcR and yeaM), transthyretin-like protein (hiuH), a bifunctional synthase (dfp), bifunctional (p)ppGpp synthetase II/guanosine-3′,5′-bis pyrophosphate 3,-pyrophosphohydrolase (spoT), dihydro-orotate oxidase (pyrD), lysophospholipase L2 (pldB), agmatinase (speB) and xylosidase (yicL). The other down regulated genes include two pseudo genes (pbl, yfjV), seven hypothetical protein coding genes (ybeR, ypjA, yfaT, yfjD, yehP, yjbG and ykiA), one unknown gene and ten intergenic region coding genes (Table 3).
Validation of the Expression of Genes by RT-PCR
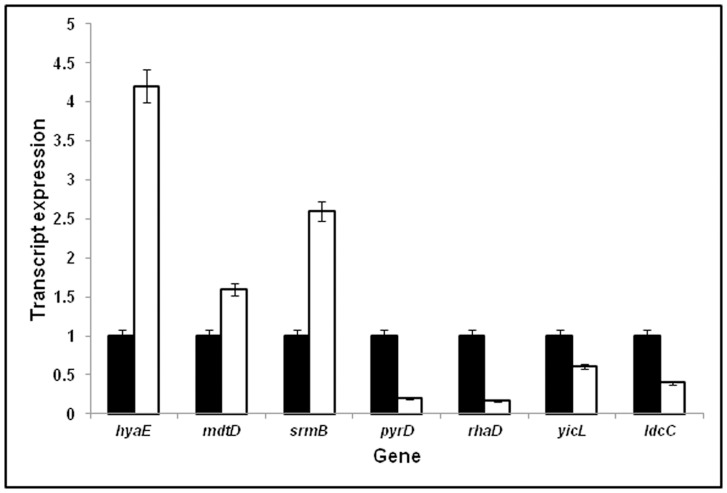
RT-PCR was carried out on seven genes (hyaE, mdtD, srmB, pyrD, rhaD, yicL and ldcC) and the results indicated that, pyrD and rhaD (P<0.001), yicL, (P<0.05), hyaE, ldcC, mdtD and srmB (P<0.1) were either significantly up regulated or down regulated in accordance with the DNA microarray results (Tables 2 and 3) thus validating the DNA microarray results in E. coli exposed to simulated microgravity (Fig. 3). The expression of the genes was calculated based on the product Cycle threshold (CT) value. The data was analysed using ANOVA (Prism 3.0 software).
Figure 3. Effect of microgravity on the expression of genes hyaE, mdtD, srmB, pyrD, rhaD, yicL and ldcC using RNA from E. coli grown in a clinostat (▪)and compared with E. coli grown in normal gravity conditions (□).
The OD of the the cultures was 0.8 (OD600 nm) The P values were pyrD, rhaD (P<0.001), yicL (p<0.05), srmB, hyaE, mdtD and ldcC (P<0.1) respectively.
Assignment of the Differentially Regulated Genes to Functional Pathways by KEGG and DAVID
Out of 100 genes that were differentially regulated only 57 genes were considered for analysis using KEGG and DAVID. The remaining 43 genes that coded for 2 unknown and 41 Intergenic regions were not included in the analysis. Out of 57 genes that were differentially regulated only 14 down regulated and one up regulated gene could be associated with a particular pathway when analysed by KEGG pathways. Hyperlinks have been provided to these genes. These fifteen genes belonged to sixteen different pathways like metabolic pathways (cysH, dfp, hisA, hiuH, ldcC, pyrD and speB), the two-component system (cusF and ompC), purine metabolism (hiuH and spoT), microbial metabolism in diverse environments (cysH, and hiuH), biosynthesis of secondary metabolites (hisA and ldcC), ABC transporters (cysW and nikB), glycerophospholipid metabolism (pldB), phosphotransferase system (ptsA), pantothenate and CoA biosynthesis (dfp), pyrimidine metabolism (pyrD), fructose and mannose metabolism (rhaD), pentose and glucuronate interconversion (rhaD), arginine and proline metabolism (speB), histidine metabolism (hisA), lysine degradation (ldcC) and sulfur metabolism (cysH). The KEGG pathway analysis also indicated that certain genes like cysH, hisA, hiuH, ldcC, pyrD, speB, and rhaD were associated with more than one pathway.
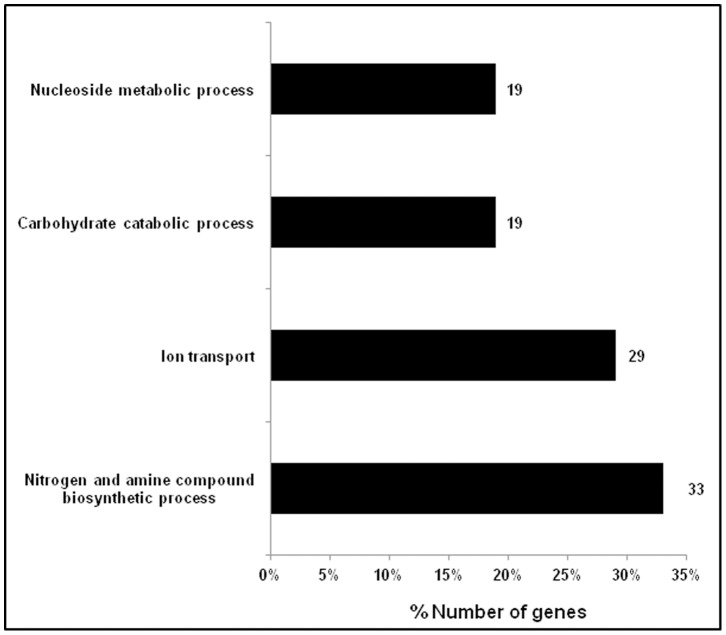
DAVID was also used for geneontology annotations and term enrichments for various biological processes, molecular functioning and cellular components. Out of 57 genes that were differentially regulated and analysed by DAVID only 21 down regulated and 9 up regulated genes yielded term enrichments. Hyperlinks have been provided to these genes. The analysis yielded that the 9 upregulated genes were involved in two biological processes namely DNA transcription and regulation of transcription (ilvY and ydcQ), one cellular component which included a number of genes coding for integral membrane proteins (hokE, mdtD, ynfA, ybdJ and yniB) and three molecular functioning terms including ion binding (cusF and ygaQ) and DNA binding (ilvY and ydcQ). The 21 down regulated genes were implicated in four biological processes (Fig. 4) such as genes involved in nitrogen and amine compound biosynthetic process (hisA, speB, aroP, pyrD, livG, mgtA and cysH), ion transport (exbB, actP, mgtA, ompC, cysW and nikB), carbohydrate catabolic process (rhaS, ldcC, ptsA and rhaD) and nucleoside metabolic process (dfp, pyrD, spoT and nupC). DAVID analysis also annotated genes involved in cellular components such as cell wall, cell membrane and organelle membrane (aroP, exbB, actP, pyrD, pldB, mgtA, nikB, nupC, ompC, cysW and yfjD). Similarly GO terms enriched for molecular functioning included genes coding for proteins involved in metal-ion binding, cation-binding and ion-binding (speB, actP, dfp, spoT, mgtA, ptsA, nikB and rhaD).
Figure 4. Distribution of down regulated genes (%) based on biological process classification reported by Gene ontology term functional categories using DAVID version 2.0 software.
Discussion
Bacteria under reduced gravity conditions exhibit a number of distinct physiological changes such as changes in growth characteristics, increase in resistance to acidic, osmotic, and thermal stress [2], [17], greater tolerance to ethanol exposure [18], enhanced attachment abilities [19], [20], increase in virulence, ability to use substrates more effectively [21] and have altered gene expression [22]. Such studies are best done in a spacecraft in orbit. But due to logistic reasons and practical limitations, studies conducted in ground-based systems that mimic microgravity conditions like using a clinostat, could help to generate data and provide clues to important effects of microgravity on life systems [23]–[25]. A clinostat creates microgravity conditions characterized by reduced sedimentation, low shear and low turbulence as in space flight [25], [26]. Thus studies have used clinostat as a device to subject microbes to reduced gravity conditions and monitor the effects on the growth, physiology and expression of genes in bacteria [27], [28].
Earlier studies demonstrated that E. coli under simulated microgravity conditions exhibits decreased lag phase and an increase in exponential phase compared to normal gravity [1], [11], [29], [30]. Our study on growth of E. coli on LB medium in a clinostat also showed increased growth rate compared to normal gravity conditions. On the contrary Bouloc and D’Ari [31] showed no difference in growth of E. coli in the orbiting Biocosmos 2044 satellite compared to its ground based controls. Studies have indicated that the effects of reduced gravity on bacterial viability and growth is dependent on whether the bacteria were cultured in rich or dilute media. In S. aureus growth characteristics were similar in LB under reduced and normal gravity conditions but in 1/10 LB, the growth of S. aureus was significantly increased under microgravity [13]. The authors have also suggested that the physiological responses to microgravity conditions vary with growth medium and growth phase. However, recent studies by Berry et al [32] on the effect of hypobaric pressures similar to those of the surface of mars (0.69 kPa global average) on the growth of E.coli revealed slight reduction in growth.
In the present study a fold change of ≥1.5 was considered to analyse the data on differential gene expression in simulated microgravity conditions. Several other studies also used a fold change ≥1.5 as a cut-off for the analysis of differential gene expression [33], [34]. In this study using a fold change ≥1.5 cut-off a total of 100 genes were differentially regulated in E. coli under simulated microgravity conditions, out of which fifty three were upregulated and forty seven down regulated compared to normal gravity grown culture (Tables 2 and 3).
Wilson et al. [22] were the first to monitor global gene expression in Salmonella in response to microgravity. A total of 163 genes were differentially regulated by microgravity. These differentially regulated genes included genes coding for transcriptional regulators, virulence factors, lipopolysaccharide biosynthetic enzymes, iron-utilization enzymes and several genes with no homology to other genes in the current databases. Subsequent studies on Salmonella also led to the discovery of novel virulence mechanisms involving a ferric uptake regulator under reduced gravity conditions [3], [6], [22]. Recent studies also revealed that P. aeruginosa responded to spaceflight conditions through differential regulation of 167 genes and 28 proteins, with Hfq as a global transcriptional regulator [35]. Vukanti et al. [1] using E. coli K12 demonstrated that 430 genes were significantly altered in expression under modeled reduced gravity conditions. Up regulated genes included starvation inducible genes (csiD, cspD, ygaF, gabDTP, ygiG, fliY and cysK), genes associated with multiple stress responses (asr, yhiW, yehZYW, katE and btuDE), genes involved in biofilm formation (lldR, lamB, yneA, fadB and ydeY), curli biosynthesis (csgDEF), and lipid biosynthesis (yfbEFG). None of these genes were up regulated in the current study. The only difference between the present study and that of Vukanti et al. [1] being that E. coli K12 MG1655 in the present study was grown in LB containing glycerol. Therefore the effect of glycerol on gene expression was studied (GEO accession number is GSE34275). It was observed that in E. coli K12 MG1655 grown in the presence of glycerol 103 genes were up regulated whereas 209 were down regulated (Supplementary Tables S1 and S2). The results also indicated that the presence of glycerol in both the control and microgravity exposed cultures nullified the up regulation of starvation induced genes (cbpA, uspB and wrbA), genes associated with multiple stress responses (hdeA, hdeB, hdeD, gadA, gadB, gadC, gadW, cadB, dhaL, dhaK, ompW, tar and tnaL), genes involved in curli biosynthesis (fliC, fimA), and lipid biosynthesis (ybjP) as observed by Vukanti et al. [1]. The mechanism or the reason for the difference observed still remains unknown. A few of the differentially regulated genes were also validated for their expression by RT-PCR and found to be consistent with the DNA microarray results (Tables 2 and 3 and Fig. 2).
Earlier studies demonstrated that cells grown under microgravity conditions develop a nutrient depletion zone around the cells [24]. As a consequence, genes coding for nutrient transport and salvage systems, multiple stress resistance and metabolic enzymes are up regulated and simultaneously down regulation of genes associated with translation apparatus, DNA replication and cell division were observed [1]. However, surprisingly in the present study the above genes were not differentially expressed implying that the presence of glycerol in the medium may not favour the formation of a nutrient depletion zone. In fact this may be so because glycerol can enter the cell through simple diffusion and thus get utilized as a carbon source and generate acidic metabolic products. This may be the reason as to why E. coli grown in the presence of glycerol exhibits up regulation of genes related to acid stress (Supplementary Table S1and S2). Recently, Vukanti and Leff [12] also demonstrated that general stress response of the bacteria grown under microgravity can be reversed by supplementing a nutrient rich medium.
Comparison of genes identified in the present study to genes reported in previous studies [1] revealed that only a few differentially expressed genes were common. These genes include hyaE, livG and pyrD. Gene hyaE encodes for HyaE protein of hydrogenase-1 operon which comprises six genes hyaABCDEF. In Vukanti et al. [1] all the six genes of the operon were up regulated whereas in the present study only hyaE was upregulated. pyrD which encodes dihydroorotate dehydrogenase and involved in pyrimidine biosynthesis is down regulated in both the studies. In addition, in the paper of Vukanti et al. [1] few other genes of the pyr operon (pyrF, carA, pyrB, pyrG, pyrC and carB) were also down regulated. Gene livG that belongs to livFGHMK operon that is involved in high-affinity branched-chain amino acid transport system is down regulated. While ilvGMEDA operon an amino acid biosynthetic operon required for the synthesis of the branched chain aminoacids isoleucine, leucine and valine is down regulated in previous studies [1].
In the present study cysW involved in sulfate/thiosulfate transport, cysH involved in cystein biosysnthesis and gene actP that is involved in acetate transport are down regulated, so as to possibly maintain pH homeostasis in the acidified media conditions. This is in contrast to the previous observations that genes pertaining to acetate metabolism acs and actP and cysAV involved in sulfate/thiosulfate transport as well as genes involved in cysteine biosynthesis cysCDIJKN were upregulated [1], to overcome oxidative stress due to microgravity conditions [1].
In the present study 100 genes were either up or down regulated when E. coli was cultured under microgravity conditions. Analysis based on the DAVID geneontology (GO) term enrichments included the following up regulated genes belonging to DNA transcription (srmB) and those involved in regulation of transcription (ilVY and ydcQ). Gene srmB encodes for DNA helicase. Upregulation of helicases is interesting as these plays a vital role in DNA repair, recombination and RNA transcription [36]. At the same time DAVID analysis also revealed enrichment of GO terms such as nucleoside metabolic process for the down regulated genes dfp, pyrD, spoT. Gene dfp is an essential gene required for DNA synthesis [37] and gene pyrD involved in pyrimidine biosynthesis [38] are down regulated. Gene ydcR that belongs to one of the transcriptional regulator family GntR is also repressed. The low abundance of these genes may be because of the active growth of E. coli culture under microgravity conditions. The spoT gene which codes for ppGpp in response to nutrients limitation [39], [40] is also repressed implying that cells are not in a nutrients limiting condition in the clinostat. Based on DAVID analysis, genes involved in carbohydrate catabolic process (rhaS, rhaD, ldcC and ptsA) are also down regulated suggesting that energy/reducing equivalents are adequately present. Gene ilvY is involved in the transcriptional regulation of the ilvA gene of the ilvGMEDA operon encoding for biosynthesis of branched chain aminoacids leucine, isoleucine and valine [41] is up regulated. It is also observed that DAVID analysis enriched GO terms nitrogen compound and amine biosynthetic processes for down regulated genes hisA, speB, aroP, pyrD, livG, mgtG and cysH. These results imply that cells growing under microgravity conditions are not deprived of nitrogenous compounds.
The other up regulated genes enriched include those coding for cation-binding (cusF and ygaQ) and DNA-binding (ydcQ). Tucker et al., [1] reported down regulation of cusF when E. coli was grown in MOPS medium under simulated microgravity conditions. cusF is a periplasmic chaperone and exports copper and silver ions both from cytoplasm and periplasm to the external environment to maintain cell homeostasis. Enrichment of DAVID GO terms for ion transport, metal ion transport and cation transport for the down regulated genes (ompC, exbB, actP, mgtA, cysW and nikB) is also indicated. These transport systems assist in uptake of essential nutrients, regulate the flow of physiologically relevant chemicals, and release substances such as signalling molecules. Gene ompC coding for major outer membrane protein functions as a porin, allows free diffusion of small hydrophilic molecules across the membrane [42] is down regulated. Apart from these ion transporters, detoxification genes like gst encoding for glutathione transferase that protects cells by detoxification of metals Hg2+ and Cd2+ [43] is also down regulated may be because the cells are in an environment free of metal ions.
Based on DAVID enrichment terms the up regulation of genes integral to membrane include mdtD along with hypothetical protein coding genes hokE, ynfA, ybdJ and yniB. Gene mdtD that encodes for multidrug efflux system [44] may indicate acquisition of drug resistance by E. coli cultured under microgravity conditions. This observation is in accordance with the observations of Leys et al. [45] that bacteria growing in space require greater concentrations of various antibiotics to inhibit their growth. However, Tucker et al. [11] did not observe any significant change in the antibiotic resistance in E. coli exposed to microgravity. The up regulation of sufE implies that in the prevailing conditions iron was limiting since suf genes are induced under iron limitation conditions [46]. Up regulation of gene ssrA that encodes for small RNA, is required for adaptation to environmental changes and growth under stress conditions [47] could be an advantage to the growth of the bacterium under microgravity conditions.
On the whole in this study a number of genes that were not reported earlier when E. coli is grown on LB media under microgravity conditions appeared when 10% v/v glycerol is additionally supplemented to LB medium. The up regulated genes comprised those that are related to transcription and integral to membrane. Among the down regulated genes, those involved in nitrogen, nucleoside and carbohydrate metabolic process and membrane related transporters were present. We did not observe elevation of multiple stress genes in response to nutrient depletion zone in our study, may be because of the reversal of this stress with glycerol in the medium. Using DAVID seven hypothetical protein coding genes namely yfjD, ydcQ, ynfA, ybdJ, yniB, ygaQ and hokE were assigned to various functional processes in response to microgravity conditions in E. coli. A number of hypothetical and Intergenic region genes were also differentially expressed.
Conclusions
The results imply that E. coli in the presence of glycerol under simulated microgravity conditions grows better than compared to the normal gravity control. The enhanced growth may be because of adequate supply of energy/reducing equivalents and increase in transcripts for DNA replication. Further, glycerol supplementation in the medium helps in overcoming multiple stressors exerted in microgravity conditions due to nutrient limitations.
Supporting Information
Glycerol-induced up regulation of genes in Escherichia coli. DNA microarray analysis of E. coli grown in the presence of 10% glycerol showed up regulation of 103 genes with a fold change >1.5 (P<0.05).
(DOCX)
Glycerol-induced down regulation of genes in Escherichia coli . DNA microarray analysis of E. coli grown in the presence of 10% glycerol showed down regulation of 209 genes with a fold change >1.5 (P<0.05).
(DOCX)
Acknowledgments
We would like to thank Santosh Bhaskaran, Vascular Biology Lab, AU-KBC research centre, Anna University, Chennai for deducing gravity calculations of the study. We would also like to thank Dr. Mohammed Idris for help in using DAVID software and P. Ramesh and M.B. Madhavi for microarray data analysis all from the Centre for Cellular and Molecular Biology, Hyderabad.
Funding Statement
The project was funded by ISRO, India up to March 2012. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vukanti R, Mintz E, Leff L (2008) Changes in gene expression of E. coli under conditions of modelled reduced gravity. Microgravity Sci Technol 20: 41–57. [Google Scholar]
- 2. Nickerson CA, Ott CM, Mister SJ, Orrow BJ, Burns-Keliher L, et al. (2000) Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect Immun 68: 3147–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wilson JW, Ott CM, Bentrup KH, Ramamurthy R, Quicka L, et al. (2007) Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc Natl Acad Sci 104: 16299–16304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rosenzweig JA, Abogunde O, Thomas K, Lawal A, Y-Uyen Nguyen, et al. (2010) Spaceflight and modeled microgravity effects on microbial growth and virulence. Appl Microbiol Biotechnol 85: 885–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chopra VA, Fadl AA, Sha J, Chopra S, Galindo CL, et al. (2006) Alterations in the virulence potential of enteric pathogens and bacterial–host cell interactions under simulated microgravity conditions. J Toxicol Environ Health A 69: 1345–1370. [DOI] [PubMed] [Google Scholar]
- 6. Wilson JW, Ott CM, Quick L, Davis R, Höner zu Bentrup K (2008) Media ion composition controls regulatory and virulence response of Salmonella in spaceflight.PLoS ONE. 3: e3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baker PW, Leff LG (2004) The effect of simulated microgravity on bacteria from the Mir space station. Microgravity Sci Technol 15: 35–41. [DOI] [PubMed] [Google Scholar]
- 8. Baker PW, Leff LG (2006) Mir space station bacteria responses to modeled reduced gravity under starvation conditions. Adv Space Res 38: 1152–1158. [Google Scholar]
- 9. Benoit MR, Klaus DM (2007) Microgravity, bacteria and the influence of motility. Adv Space Res 39: 1225–1232. [Google Scholar]
- 10. Simon C, Daniel R (2011) Metagenomic analyses; Past anad future trends. Appl Environ Microbiol 77: 1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tucker DL, Ott CM, Huff S, Fofanov Y, Pierson DL, et al. (2007) Characterization of Escherichia coli MG1655 grown in a low-shear modeled microgravity environment. BMC Microbiol 7: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vukanti R, Leff LG (2012) Expression of multiple stress response genes by Escherichia coli under modelled reduced gravity. Microgravity Sci Technol 24: 267–279. [Google Scholar]
- 13. Vukanti R, Model MA, Leff LG (2012) Effect of modelled reduced gravity conditions on bacterial morphology and physiology. BMC Microbiol 12: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haga RA, Saleh JH (2011) “Epidemiology of satellite anomalies and failures: A subsystem-centric approach,” Aerospace Conference, 2011 IEEE, 5–12.
- 15. Gruener R, Roberts R, Reitstetter R (1994) Reduced receptor aggregation and altered cytoskeleton in cultured myocytes after space-flight. Biol Sci Space 8: 79–93. [DOI] [PubMed] [Google Scholar]
- 16.Huijser RH (2000) Desktop RPM: new small size microgravity simulator for the bioscience laboratory. FS-MG-0017, Fokker Space.
- 17. Wilson JW, Ott CM, Ramamurthy R, Porwollik S, McClelland M, et al. (2002) Low-Shear modeled microgravity alters the Salmonella enterica serovar Typhimurium stress response in an RpoS-independent manner. Appl Environ Microbiol 68: 5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao Q, Fang A, Pierson DL, Mishra SK, Demain AL (2001) Shear stress enhances microcin B17 production in a rotating wall bioreactor, but ethanol stress does not. Appl Microbiol Biotechnol 56: 384–387. [DOI] [PubMed] [Google Scholar]
- 19. Baker PW, Leff LG (2005) Attachment to stainless steel by Mir Space Station bacteria growing under modeled reduced gravity at varying nutrient concentrations. Biofilms 2: 1–7. [Google Scholar]
- 20. McLean RJ, Cassanto JM, Barnes MB, Koo JH (2001) Bacterial biofilm formation under microgravity conditions. FEMS Microbiol Lett 195: 115–119. [DOI] [PubMed] [Google Scholar]
- 21. Brown RB, Klaus D, Todd P (2002) Effects of space flight, clinorotation and centrifugation on the substrate utilization efficiency of E. coli . Microgravity Sci Technol 13: 24–29. [DOI] [PubMed] [Google Scholar]
- 22. Wilson JW, Ramamurthy R, Porwollik S, McClelland M, Hammond T, et al. (2002) Microarray analysis identifies Salmonella genes belonging to the low-shear modeled microgravity regulon. Proc Natl Acad Sci 99: 13807–13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao H, Ayyaswamy PS, Ducheyne P (1997) Dynamics of a microcarrier particle in the simulated microgravity environment of a rotating wall vessel. Microgravity Sci Technol 10: 154–165. [PubMed] [Google Scholar]
- 24. Hammond TG, Hammond JM (2001) Optimized suspension culture: the rotating-wall vessel. Am J Physiol Renal Physiol 281: F12–25. [DOI] [PubMed] [Google Scholar]
- 25. Nickerson CA, Ott CM, Wilson JW, Ramamurthy R, LeBlanc CL, et al. (2003) Lowshear modeled microgravity: a global environmental regulatory signal affecting bacterial gene expression, physiology, and pathogenesis. J Microbiol Methods 54: 1–11. [DOI] [PubMed] [Google Scholar]
- 26. Nickerson CA, Ott CM, Wilson JW, Ramamurthy R, Pierson DL (2004) Microbial responses to microgravity and other low-shear environments. Microbiol Mol Biol Rev 68: 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yamazaki T, Yoshimoto M, Nishiyama Y, Okubo Y, Makimura K (2012) Phenotypic characterization of Aspergillus niger and Candida albicans grown under simulated microgravity using a three-dimensional clinostat. Microbiol Immunol 56: 441–446. [DOI] [PubMed] [Google Scholar]
- 28. Benoit MR, Klaus DM (2005) Can genetically modified Escherichia coli with neutral buoyancy induced by gas vesicles be used as an alternative method to clinorotation for microgravity studies?. Microbiology 151: 69–74. [DOI] [PubMed] [Google Scholar]
- 29. Baker PW, Leff LG (2004) The effect of simulated microgravity on bacteria from the Mir space station. Microgravity Sci Technol 15: 35–41. [DOI] [PubMed] [Google Scholar]
- 30. Klaus D, Simske S, Todd P, Stodieck L (1997) Investigation of space flight effects on Escherichia coli and a proposed model of underlying physical mechanisms. Microbiology 143: 449. [DOI] [PubMed] [Google Scholar]
- 31. Bouloc P (1991) D’Ari (1991) Escherichia coli metabolism in space. J Gen Microbiol 137: 2839–2843. [DOI] [PubMed] [Google Scholar]
- 32. Berry BJ, Jenkins DG, Schuerger AC (2010) Effects of simulated mars conditions on the survival and growth of Escherichia coli and Serratia liquefaciens. Appl Environ microbiol 76: 2377–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yuan L, Hillman JD, Progulske-Fox A (2005) Microarray Analysis of Quorum-Sensing-Regulated Genes in Porphyromonas gingivalis. . Infect Immun 73: 4146–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dalman MR, Deeter A, Nimishaka G, Duan Z-H (2012) Fold change and p-value cutoffs significantly alter microarray interpretations. BMC Bioinformatics 13 Suppl 2 S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crabbe A, Schurr MJ, Monsieurs P, Morici L, Schurr J, et al. (2011) Transcriptional and proteomic responses of Pseudomonas aeruginosa PA01 to spaceflight conditions involve Hfq regulation and reveal a role for oxygen. Appl Environ Microbiol 77: 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jagessar KL, Jain C (2010) Functional and molecular analysis of Escherichia coli strains lacking multiple DEAD-box helicases. RNA 16: 1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spitzer ED, Jimenez-Billini HE, Weiss B (1988) 1-Alanine auxotrophy associated with dfp, a locus affecting DNA synthesis in Escherichia coli . J Baceriol 170: 872–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilson HR, Turnbough JRCL (1990) Role of the purine repressor in the regulation of pyrimidine gene expression in Escherichia coli K-12. J Bacteriol 172: 3208–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mittenhuber G (2001) Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT Proteins). J Mol Microbiol Biotechnol 3: 585–600. [PubMed] [Google Scholar]
- 40. Mechold U, Cashel M, Steiner K, Gentry D, Malke H (1996) Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. . J Bacteriol 178: 1401–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parekh BS, Hatfield GW (1997) Growth rate-related regulation of the ilvGMEDA Operon of Escherichia coli K-12 Is a consequence of the polar frame shift mutation in the ilvG gene of this strain. J Bacteriol 179: 2086–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ikenaka SK, Ramakrishnan G, Inouye M, Tsung K, Inouye M (1986) Regulation of the ompC gene of Escherichia coli. Involvement of three tandem promoters. J biologic chem 261: 9316–9320. [PubMed] [Google Scholar]
- 43. Chen S, Wilson DB (1997) Construction and characterization of Escherichia coli genetically engineered for bioremediation of Hg21-contaminated environments. Appl Env Microbiol 63: 2442–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nagakub S, Nishino K, Hirata T, Yamaguchi A (2002) The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J Bacteriol 184: 4161–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leys N, Hendrickx L, De Bover P, Baatout S, Mergeay M (2004) Space flight effects on bacterial physiology. J Biol Regul Homeost Agents. 18: 193–199. [PubMed] [Google Scholar]
- 46. Nachin L, El Hassouni M, Loiseau L, Expert D, Barras F (2001) SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol Microbiol 39: 960–972. [DOI] [PubMed] [Google Scholar]
- 47. Ranquet C, Gottesman S (2007) Translational regulation of the Escherichia coli stress factor RpoS: a role for SsrA and Lon J Bacteriol. 189: 4872–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Glycerol-induced up regulation of genes in Escherichia coli. DNA microarray analysis of E. coli grown in the presence of 10% glycerol showed up regulation of 103 genes with a fold change >1.5 (P<0.05).
(DOCX)
Glycerol-induced down regulation of genes in Escherichia coli . DNA microarray analysis of E. coli grown in the presence of 10% glycerol showed down regulation of 209 genes with a fold change >1.5 (P<0.05).
(DOCX)