Abstract
Pluripotent stem cells hold enomous potential for therapuetic applications in tissue replacement therapy. Reprogramming somatic cells from a patient donor to generate pluripotent stem cells involves both ethical concerns inherent in the use of embryonic and oocyte-derived stem cells, as well as issues of histocompatibility. Among the various pluripotent stem cells, induced pluripotent stem cells (iPSC)—derived by ectopic expression of four reprogramming factors in donor somatic cells—are superior in terms of ethical use, histocompatibility, and derivation method. However, iPSC also show genetic and epigenetic differences that limit their differentiation potential, functionality, safety, and potential clinical utility. Here, we discuss the unique characteristics of iPSC and approaches that are being taken to overcome these limitations.
Keywords: iPSC, ntESC, ESC, pESC, Stem cell, Epigenetic, Differentiation, Reprogramming
1. Introduction
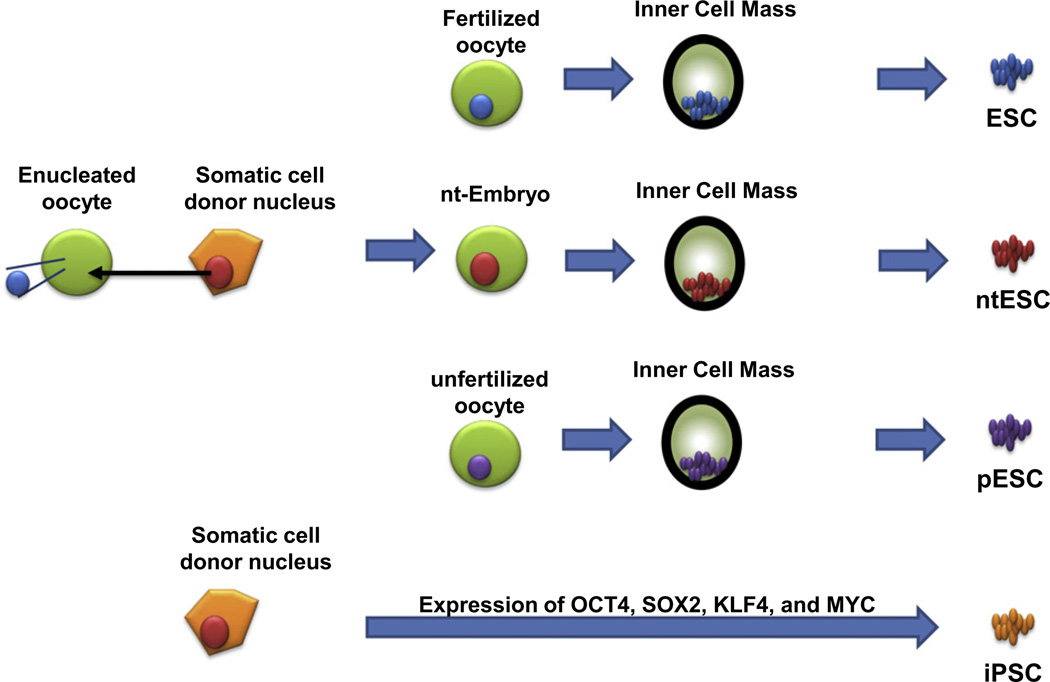
Embryonic stem cells (ESC) are present in the inner cell mass (ICM) of a blastocyst-stage embryo. Compared with other somatic cells, ESC have two unique properties—self-renewal and pluripotency. With these two properties, ESC represent an unlimited source of tissue (self-renewal), and can generate any type of tissue (pluripotency). However, the use of ESC for tissue replacement or transplantation not only raises significant ethical concerns, but also requires the creation of a large stem cell bank in order to produce tissues that could be immunologically matched with any recipient patient. Generation of ESC by somatic cell nuclear transfer to an enucleated oocyte (ntESC) has been proposed as an alternative source of transplantable cells and tissues, primarily because this technique may provide genetically identical and immunologically compatible stem cells for individual somatic cell donors (i.e., patient recipients; Fig. 1) [1]. Unfortunately, this method is both cumbersome and inefficient.
Fig. 1.
Derivation of the various pluripotent stem cells: ESC from a fertilized oocyte, ntESC from a nuclear transferred embryo, pESC from an unfertilized oocyte, and iPSC from the direct reversion of somatic cells by ectopic expression of four cellular reprogramming factors. Of note, whereas derivation of ESC, ntESC, and pESC utilizes all reprogramming factors present in the ooplasm, derivation of iPSC involves only the minimal four cellular reprogramming factors present in ESC.
Yet another technique has been proposed for producing histocompatible stem cells suitable for tissue replacement therapy: the generation of parthenogenetic ESC (pESC) (Fig. 1). This procedure does not require destruction of viable embryos, as pESC are derived from unfertilized oocytes that have the potential to develop into only a limited number of tissues [2,3]. In addition, pESC appear to have fewer epigenetic abnormalities than ntESC because the nuclear transfer technique requires reversion of the epigenetic status of a terminally differentiated adult somatic cell nucleus to an embryonic stage, whereas pESC are already at an embryonic stage. However, pESC have a haploid duplicated homozygous genome [4,5] and large duplicated regions of homozygosity, which, theoretically, might trigger natural killer cell-mediated immune rejection if these cells are used for tissue replacement therapy [6]. Clinically, partial major histocompatibility complex (MHC) antigen matching enhances allograft survival, but fully MHC-matched tissues are the most favorable for transplant [7]. Therefore, an alternative method for generating pESC that are fully matched to the oocyte donor/patient recipient has been developed, in which a subset of pESC that have undergone crossing-over events in their leukocyte antigen region during meiosis are selected [8,9]. Nonetheless, pESC are generated from donor eggs, which means that this approach can only be applied to female patients.
Generation of ESC, ntESC, and pESC require the use of oocytes, and the limited availability of human oocytes poses a significant obstacle to producing customized ESC (selected from a large ESC bank) or producing ntESC and pESC suitable for therapeutic use. Therefore, alternative methods continue to be developed in order to generate other types of stem cells. In these efforts, ESC continue to represent the “gold standard” in terms of pluripotency, and are used as a critical control against which the degree of pluripotency of new types of derived stem cells is measured (Fig. 1).
In 2006, the generation of induced pluripotent stem cells (iPSC) by ectopic expression of four reprogramming factors present in ESC (OCT4, SOX2, KLF4, and MYC) revolutionized the stem cell field [10]. iPSC not only represented a valuable scientific tool to study the mechanisms of cellular reprogramming, but also a potential source of patient-specific, histocompatible adult stem cells. Importantly, the generation of iPSC does not require the use of oocytes, as the recipient patient would donate his or her own cells to generate stem cells for autotransplantation. Therefore, iPSC opened up the possibilities for therapeutic application of pluripotent stem cells by removing the ethical and histocompatibility limitations of ESC, ntESC, and pESC (Fig. 1).
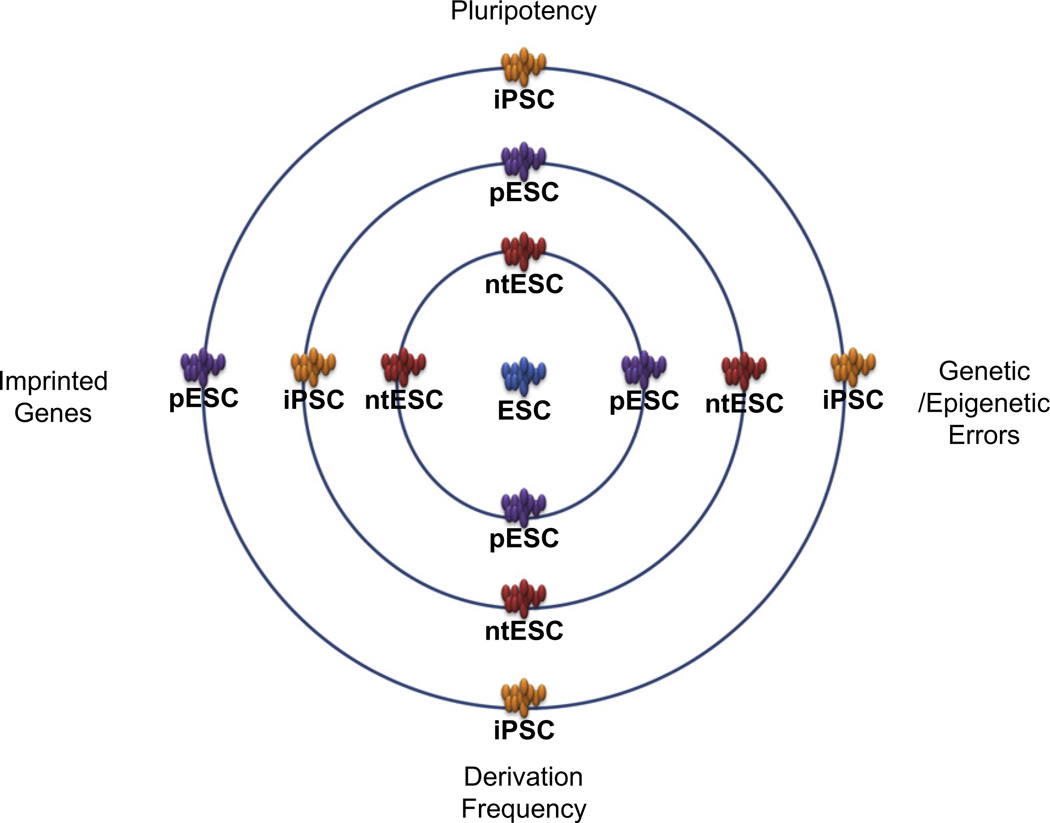
Yet while iPSC have a great degree of similarity to ESC, several reports have also shown significant differences in DNA methylation, gene expression, aberrant reprogramming, and differentiation potential of iPSC compared with other types of pluripotent stem cells (Fig. 2). It is now clear that further research is needed to assess the dynamic changes that occur during iPSC cellular reprogramming and to evaluate the characteristics of the resulting pluripotent stem cells—in terms of differentiation potential, functionality, and safety—if future therapeutic applications of iPSC are to be realized.
Fig. 2.
Cellular reprogramming and characteristics of the various pluripotent stem cells. Relative similarity of the various pluripotent stem cell types to the “gold standard” ESC (center) in terms of pluripotency, genetic/epigenetic errors, imprinted gene status, and derivation frequency. Improving the quality of iPSC will involve moving iPSC closer to ESC in each of these areas.
2. Epigenetic and functional differences between ESC and iPSC
Early studies comparing ESC and iPSC at the genome level found a high degree of similarity in terms of overall gene expression profile and chromatin modification [11–13], with statistical analyses suggesting that gene expression and chromatin modification were identical in ESC and iPSC. The most stringent test of pluripotency, tetraploid embryo complementation, has been achieved with a few mouse iPSC clones, using either iPSC generated from embryonic fibroblasts or via serial selection of specific imprinted gene expression [14,15]. However, while iPSC are highly similar to ESC in terms of cell surface marker expression, morphology, teratoma formation, tissue contribution in chimeric mice, and pluripotent gene expression, functional analyses of the cells found that iPSC have reduced differentiation potential toward neural and cardiovascular lineages [16,17], and show signs of premature aging [16,17]. Because ESC are isolated from early-stage embryos and iPSC are generated from post-natal tissues, ESC and iPSC show differential effects of cellular aging on gene expression and epigenetic alterations.
Because of the observed functional differences between ESC and iPSC, the findings of comparative genome-wide analyses have been challenged on a number of fronts. First, there is the question of sensitivity. The minimal unit for detecting differential chromatin modification extends across a relatively large region of the chromosome, whereas molecular analyses (e.g., single gene and microRNA expression) require as few as a hundred base pair regions, and DNA methylation analysis is based on a single nucleotide modification. Therefore, the sensitivity of molecular analyses is much higher than that of chromatin modification by the nature of the analysis unit used. Second, overall gene expression profile analysis of samples from undifferentiated stages of pluripotent stem cells is problematic. Because pluripotent stem cells co-regulate all pluripotency-related genes within a network [18], undifferentiated stem cells highly express pluripotency genes and suppress genes that are specific to differentiated cells. Therefore, the gene expression patterns in samples from undifferentiated pluripotent stem cells does not represent the expression patterns in differentiated pluripotent stem cells and the resulting tissue. Yet the evaluation of pluripotency depends on the functional differentiation potential of stem cells. For this reason, gene expression profile analysis of undifferentiated pluripotent stem cells is limited in its ability to predict the function of the same genes in stem cell-derived differentiated tissues.
The final concern is whether numerical statistics can be applied to this biological system. While ESC and iPSC are statistically identical in terms of overall gene expression and chromatin modification patterns, it is clear that these two types of stem cells are not identical in terms of their functional biology. We know that a number of single gene mutations and epigenetic alterations in a single chromosomal region can significantly impair the function of undifferentiated stem cells and stem cell-derived tissues. As an example, significant functional differences were observed in iPSC that contained a single region of epigenetic alterations in an imprinted gene [19] and development-related genes [20,21]. Genome-wide analyses of two stem cell types cannot capture these discrete genetic and epigenetic differences, and so the cells will appear to be statistically identical. Therefore, it is essential that we examine these subtle differences between ESC and iPSC—at the level of single gene expression [22], microRNA expression [23], and DNA methylation [20,21]—to define the specific molecular profiles of and the functional differences between the differentiated cells and tissues that are derived from ESC and iPSC pluripotent stem cell types.
3. Clonal variability
In the comparative analysis of ESC and ntESC, ntESC showed much higher clonal variability [24,25]. This epigenetic instability is due to a systematic instability of reprogramming after the somatic cell nuclear transfer procedure. It has been hypothesized that ntESC generation requires a specific stoichiometry of reprogramming factors from the ooplasm during the ntESC derivation procedure. As a result, ntESC are often obtained from several hundred nuclear transferred embryos. iPSC show an even greater clonal variability than ntESC; this is primarily because ESC and ntESC derivation involves a stringent cell selection step in vivo during early embryo development that in turn increases the stringency of cell selection in in vitro culture that greatly reduces stem cell variability. By contrast, iPSC generation involves in vitro culture of somatic cells after introduction of four reprogramming factors. These factors include the well-known oncogene, Myc, which supports rapid and uncontrolled growth of the cells on the tissue culture plate. iPSC colonies are then selected based on morphological criteria. As a result, the stringency of iPSC selection is much lower than that of ESC and ntESC. To overcome the clonal variability of iPSC, reporter genes have been expressed under the control of promoters of pluripotency-related genes such as OCT4 and NANOG; however, pluripotent stem cell marker-based selection depends on the expression of only one pluripotent reporter gene, and this approach is not likely to be applied to human iPSC. Clonal variability can be reduced by more faithful reprogramming of iPSC, but how to achieve this is a crucial question for future research that must be answered before clinical application of iPSC to human tissue replacement therapy.
4. Tissue-specific epigenetic memory
Reprogramming of somatic cells to produce iPSC is incomplete, and leaves residual epigenetic markers—DNA methylation, chromatin modification, and transcriptional regulation—in the resulting iPSC genome. Recent genome-wide DNA methylation studies [19,20] showed that reprogrammed iPSC contain the DNA methylation signature of the tissue of origin. Compared with the gold standard of pluripotency, ESC, ntESC exhibited a much smaller tissue-specific epigenetic signature than did iPSC generated from different somatic cell origins (blood, fibroblast, and brain) [20]. We concluded that this was due to an incomplete reset of the tissue-specific epigenetic signature to the default embryonic stages during iPSC reprogramming. In functional analyses, the residual tissue-specific epigenetic DNA methylation signature affected the differentiation potential and function of the cells and tissues derived from iPSC, favoring differentiation to the tissue of origin. Neither ntESC nor ESC showed this preferential tissue differentiation potential. While these findings indicated that epigenetic memory in iPSC directs tissue differentiation, the study also showed that this epigenetic memory interferes with current tissue differentiation protocols. Nevertheless, identifying the unique epigenetic signatures in iPSC-derived from various somatic cell origins will inform efforts to enhance tissue differentiation from pluripotent stem cells toward certain tissue lineages.
Although the differentiation potential of iPSC has been demonstrated, producing blood, pancreatic beta cells, and cardiac muscle [21,26,27], tissue-specific epigenetic memory can interfere with iPSC differentiation. Tissue-specific epigenetic memory originates during tissue development in the embryo, at certain stages of somatic cell differentiation and de-differentiation under tightly regulated gene expression. During differentiation of iPSC, the residual epigenetic memory can alter these highly controlled patterns of gene expression, resulting in lower differentiation efficiency and abnormal function of the resulting cells and tissues. With the ultimate goal of using iPSC in therapeutic applications, it is crucial that the epigenetic memory in iPSC be minimized. In terms of epigenetic signature, ntESC more closely resemble ESC and are thus a better source of pluripotent stem cells than iPSC (Fig. 2).
5. Dissection of the epigenetic changes during iPSC reprogramming
Recent studies of human iPSC compared with ESC and somatic cells suggest that high-resolution DNA methylation analysis may be a better approach to dissecting the epigenetic changes that occur during iPSC reprogramming [21,28]. In this approach, differentially methylated regions (DMRs) are identified in iPSC and compared with those in ESC and somatic cells, allowing the identification of the source of the DMRs during reprogramming. Surprisingly, only half of the DMRs were defined as related to incomplete reprogramming and decreased pluripotency, whereas the other half of the DMRs were identified as an epigenetic errors that were not found in either somatic cells or ESC. These methylation errors were transmitted to the iPSC-derived differentiated tissues [28].
In examining the relationship between cellular reprogramming and pluripotency, we can consider that the generation of ESC after fertilization involves cellular reprogramming of the sperm and oocyte. During early embryo development, the sperm- and oocyte-specific epigenetic memory must be erased to the default stage, and the epigenetic signature of the ESC must be established to complete cellular reprogramming. The derivation of ntESC mimics the cellular reprogramming of the ESC, similarly erasing the epigenetic memory in the somatic cell nucleus to the default stage, and re-establishing the ntESC epigenetic signature. In both cases, fewer epigenetic errors are incorporated into the resulting ntESC and ESC compared with iPSC (Fig. 2). This indicates that, to reach a pluripotent state, iPSC reprogramming may proceed down a different pathway than in either ESC or ntESC, or iPSC may be missing important components that protect against abnormal epigenetic alterations that occur during reprogramming. How to reduce the number of epigenetic errors that arise during iPSC reprogramming and increase the quality of iPSC remains to be investigated.
6. Alteration of imprinted genes
Imprinting is a well-conserved epigenetic signature in various organisms, which evolutionarily discriminate between male- and female-specific gene expression established during sperm and oocyte development. The combination of the two imprinted gene expression patterns upon fertilization determines the balance of gene expression during embryo development. Abnormal regulation of imprinting often causes altered gene expression. Only the presence of an additional copy of the gene from either the paternal or maternal chromosome is sufficient to induce altered phenotypes, such as a developmental defect, tissue differentiation, or oncogenicity. Imprinted genes are erased during early germ cell development, and are re-established upon full development of the sperm and oocyte. As described above, epigenetic reprogramming to revert somatic cells to an embryonic stage must induce genome-wide epigenetic modifications that erase the tissue-specific epigenetic signature to a default stage, and then re-establish the appropriate tissue-specific signatures during differentiation of the pluripotent stem cells. During early-stage reprogramming, either by fertilization (reprogramming of the sperm and oocyte to create a zygotic embryo) or by nuclear transfer of a somatic cell nucleus to an enucleated oocyte (reprogramming of the somatic nucleus by factors in the ooplasm) [29], imprinted genes are protected from genome-wide epigenetic changes. Because pESC originate from unfertilized oocytes, these stem cells contain only female-specific imprinting; these oocyte-specific imprinted genes are protected during the pESC derivation procedure. By contrast, iPSC reprogramming is less discriminate, and imprinted regions are also altered, which frequently damages their epigenetic signature. Aberrant silencing of imprinted genes during iPSC reprogramming may lead to altered tissue development. In one study, iPSC were selected that did not contain abnormal imprinted genes and were used to generate normal differentiated tissue as well as full-term mice by tetraploid complementation (Fig. 3) [15,30].
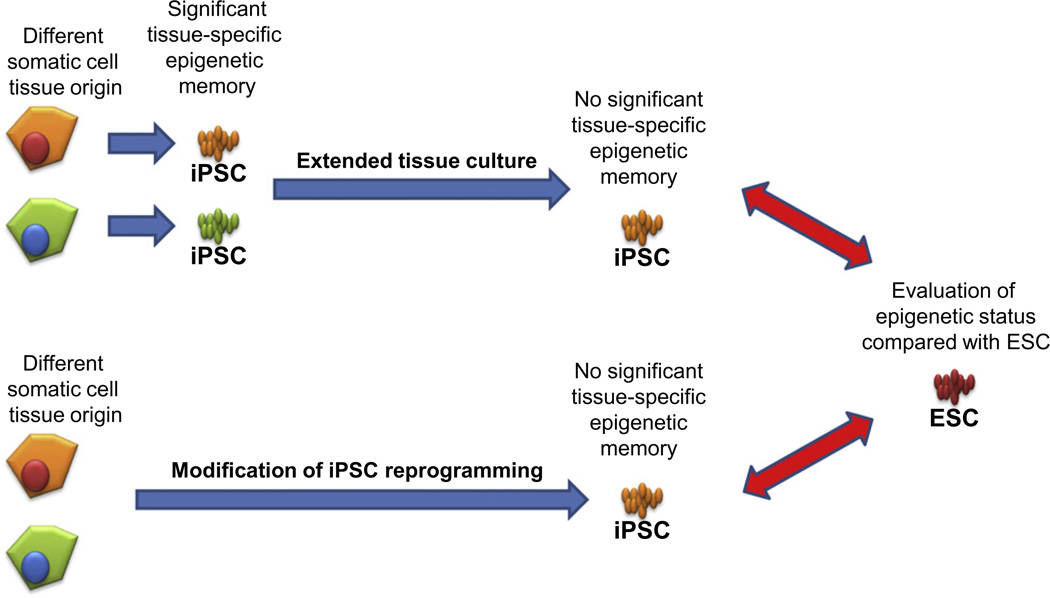
Fig. 3.
Approaches to erasing tissue-specific epigenetic memory and evaluating the resulting pluripotent stem cells. Extended tissue culture after iPSC reprogramming (top) and modification of the iPSC reprogramming procedure (bottom) have been suggested to increase the quality of the resulting iPSC. In both cases, the resulting cells need to be compared not only with iPSC from different tissues but also with ESC as the gold standard of pluripotency. Genetic and epigenetic stability and alteration rates will be different during extended tissue culture and with variations in iPSC reprogramming; therefore, rigorous verification procedures are required to evaluate the potential therapeutic utility of the resulting cells.
7. Approaches to correcting the iPSC epigenetic signature
Among the various pluripotent stem cells, ntESC are closer to the gold standard of ESC than any other pluripotent stem cell type, in terms of both molecular signature and function (Fig. 2) [20]. Although iPSC hold significant potential for therapeutic use and provide an alternative human stem cell research material, aberrant epigenetic alterations affect the differentiation potential and function of the resulting cells and tissues. Thus, a major interest in the pluripotent stem cell field is how to correct the iPSC epigenetic signature to produce high-quality pluripotent stem cells that are more similar to ESC in terms of differentiation potential, function, and, for clinical applications, safety. The functional phenotypes that we observe are limited, and there are no standard protocols to test all aspects of pluripotency, oncogenicity, and tissue function of iPSC. Therefore, epigenetic signatures determined by molecular analysis are often used as surrogate indicators of iPSC quality.
Epigenetic memory in iPSC can be erased by extended tissue culture [19], although it is not yet clear whether the extended culture of iPSC also improves their pluripotency, moving them closer to ESC (Fig. 3). However, use of earlier passages of pluripotent stem cells is favored in therapeutic applications to avoid genetic and epigenetic alterations that arise during extended tissue culture. Alternatively, application of a chromatin modifying drug (trichostatin A) [31] and a DNA demethylation agent (5-aza-cytidine) [32] can erase the epigenetic memory if iPSC, and can restore the ability to differentiate to various tissue lineages, different from the tissue of origin. Again, this approach is not ideal for therapeutic applications because these drug treatments do not improve the pluripotency of iPSC (i.e., move them toward ESC), and most likely damage other regions of the DNA that should not be modified, such as those containing imprinted genes.
Therefore, fundamental studies are needed to determine how to erase tissue-specific epigenetic memory in iPSC in order to generate better quality pluripotent stem cells that more closely resemble ESC. The first step will be to characterize the genetic and epigenetic differences between human ESC and iPSC. To achieve this, tight control of sampling procedures and a large numbers of samples from each stem cell type will be required, as it is especially difficult to control for the genetic backgrounds of human ESC and iPSC. Indeed, the detection of epigenetic differences between ESC and iPSC in humans has been limited by the collection of donor cells under genetically and epigenetically well-controlled conditions. Nevertheless, tissue-specific epigenetic memory in human iPSC has been verified by examining the differentiation potential of iPSC to their various tissues of origin, such as skin, blood, and pancreas [21,26]. The tissue-specific molecular signatures of human iPSC show the same DNA methylation patterns as those seen in mouse iPSC.
Parallel to efforts in erasing tissue-specific epigenetic memory in iPSC, it is also important to investigate ways to alter current iPSC reprogramming strategies to overcome the limitations posed by residual epigenetic modifications (Fig. 3). Because ntESC do not contain tissue-specific epigenetic memory, and are therefore more similar to ESC, we believe that the ooplasm contains additional factors needed to completely erase the tissue-specific epigenetic memory. There is an ongoing search for these additional factors, as well as research into the events that occur during ntESC reprogramming. One of the most interesting studies in this area asked whether the ooplasm is able to reverse the incomplete reprogramming status of iPSC after iPSC nuclear transfer to and enucleated oocyte [27]. Interestingly, iPSC nuclear transfer was unable to recover the poorly reprogrammed iPSC and produce faithfully reprogrammed pluripotent stem cells, which was demonstrated by generating iPSC-derived mice. Gene expression analysis also confirmed that the iPSC-ntESC clustered away from other pluripotent stem cells, with significant dissimilarity from ESC, despite the fact that both stem cell types utilize ooplasm reprogramming factors. These results indicated that the epigenetic alterations in iPSC are preserved after the nuclear transfer procedure, and a large portion of the aberrant epigenetic signatures in iPSC are permanent.
Further analysis is also needed of iPSC generated using different transgene introduction methods (e.g., RNA, adenovirus, and piggy-Bac virus), and containing various combinations of additional reprogramming factors to understand the effect on epigenetic alterations, tissue differentiation, safety, and tissue function in various therapeutic models.
8. Genomic integrity of iPSC
Cellular reprogramming leads to changes in epigenetic markers that then alter cellular properties and function. However, epigenetic alteration cannot be considered independent of genetic changes. Epigenetic modification induces differential gene expression patterns, including genes related to genomic integrity, which are frequently linked with cancer progression [33]. The genomic integrity of iPSC was defined by genome-wide DNA sequencing of 22 independent iPSC-derived from the various sources [34]. iPSC were found to contain a higher rate of DNA mutation. While half of the mutations existed at a low level in the donor somatic cells, the other half were de novo mutations that arose during the iPSC reprogramming procedure. Interestingly, the majority of the iPSC-specific mutations were related to oncogenicity, which raised serious concerns regarding the potential clinical application of iPSC. The results of this study implied that iPSC reprogramming enriched for an oncogenic genetic signature that is not seen during ESC derivation. Significantly higher copy number variations (CNVs) were also observed in iPSC compared with ESC and fibroblasts [35]. CNVs remained high during iPSC reprogramming and then gradually decreased during extended tissue culture. It has been hypothesized that progressive negative selection occurs against higher CNVs in the cultured cells. However, it is still not clear how CNVs are increased during iPSC reprogramming; this may be caused by either a selective advantage or active accumulation during the iPSC reprogramming process.
9. Key developmental regulators for pluripotency
As described above, the goal of iPSC research is to improve the faithful reprogramming of iPSC, mimicking as closely as possible the events that occur in the generation of ESC in the developing embryo, to produce high-quality iPSC that can be used for clinical applications. The dramatic developmental transitions that accompany early embryonic development and gastrulation are induced and regulated by environmental stimuli that include hormones, morphogens, and extracellular matrix proteins. Cell differentiation and the adoption of stable, irreversible cell fates involve the concerted action of transcription factors followed by consolidation of transcriptional programs through stable epigenetic modifications. The octamer-binding transcription factor 4 (OCT4), a central player in the early embryo, is zygotically expressed at the 4- to 8-cell stage and its continued expression defines the inner cell mass (ICM) of the blastocyst [36]. A zone of trophoblastic specification in the outer “rind” or “crust” cells of the compacting morula is established by the down-regulation of OCT4 [37]. In this cell fate choice within the mammalian embryo, OCT4 acts as a negative regulator of differentiation into trophectoderm [38], and a critical regulator of the pluripotent capacity of the ICM [37,39,40]. OCT4-null embryos fail to form the ICM. Instead, all cells default to trophectoderm [37]. A simple lack of OCT4 expression may be insufficient to elicit a default towards trophectoderm and may additionally require the expression of transcription factors like CDX2 and EOMES. Both CDX2 and EOMES expression is restricted to the trophectoderm. CDX2 [41] and EOMES [42] knockout embryos fail to implant, further indicating that both genes are required in trophectodermal development. Transient expression of OCT4 is observed in the hypoblast just prior to the specification of parietal and visceral endoderm (which don’t express OCT4), suggesting that ephemeral up-regulation of OCT4 further commits hypoblast differentiation [43]. OCT4 regulates several genes during early embryonic development, including SOX2, NANOG [44], FGF4, REX1, OPN, hCG, UTF1, creatine kinase B, Makorin 1, Importin, Histone H2A.Z, Ribosomal protein S7 [45], and INF [46]. OCT4 is also expressed in undifferentiated embryonic carcinoma (EC) cells, embryonic germ (EG) cells, and ESC. Continuous expression of OCT4 appears necessary but not sufficient to maintain pluripotency in embryonic cells [38].
In the blastocyst, expression of fibroblast growth factor 4 (FGF4) defines the ICM/epiblast, while expression of the FGF receptor 2 (FGFR2) defines trophectoderm [47]. FGF4 appears to serve as a paracrine signal for trophectoderm proliferation or differentiation [48]. The transcription factor SOX2 shows a similar expression pattern to that of OCT4 during early embryonic development, with the exception that it is continuously expressed in trophectoderm [49]. SOX2 is important in lineage specification, as SOX2 and OCT4 together regulate FGF4 expression within the epiblast, which is essential for proliferation of the trophectoderm [49]. SOX2 expression is also required for the formation of the ICM and epiblast [49]. SOX2 and OCT4 are co-expressed in the morula, ICM, epiblast, and germ cells, implying collaborative function in the maintenance of pluripotency [50]. SOX2 mutants demonstrate reduced cellular proliferation, leaving only trophoblast giant cells and extraembryonic ectoderm. Such mutants do not prevent formation of the blastocyst cavity, although the ICM is missing. Thus, in the murine SOX2 knockout model, ESC cannot be derived and the mice are not viable past early embryonic development. Epiblast defects in the Sox2 mutant can be rescued by the injection of wild-type ESC [49].
NANOG is a critical transcriptional factor underlying pluripotency in both ESC and EG cells [51]. Immunocytochemical analysis demonstrates that the NANOG protein is present in the nuclei of cells in morula-stage embryos, the ICM, and the epiblast, but not in primordial germ cells (PGC), the intraembryonic mesoderm, and extraembryonic endoderm. NANOG deficiency triggers the differentiation of ESC to the extraembryonic endoderm lineage, suggesting that this DNA-binding protein acts in part by transcriptionally repressing key regulators of this alternative tissue fate. NANOG-null embryos are unable to support the formation of the epiblast and also fail to generate ESC, producing only endodermal derivatives [51]. NANOG cannot be detected in OCT4-positive PGCs, supporting the hypothesis that regulation of NANOG expression may come from mechanisms independent of OCT4 or by the concerted action of OCT4 and other factors. OCT4 and NANOG have both been implicated in the maintenance of pluripotency. Overexpression of NANOG promotes the expansion of ESC independent of leukemia inhibitory factor (LIF), STAT3, and OCT4 pathways, suggesting that NANOG is important for self-renewal in ESC and cells of the early embryo [52]. The expression patterns of NANOG and OCT4 are not identical. NANOG turns over rapidly [51]. Thus, it is only proliferating ESC that are double-positive for OCT4 and NANOG. Recently, it was shown that a culture regimen including bone morphogenic proteins (BMPs) and LIF under serum-free and feeder-free conditions supports pluripotency of ESC, while over-expressing NANOG eliminates the need for BMPs [53]. Therefore, NANOG maintains the pluripotency of ESC independent of the LIF-STAT3, BMP, and OCT4 pathways.
10. The essential iPSC reprogramming factors
Reprogramming of somatic cells into iPSC relies on the ectopic expression of four transcription factors (OCT4, SOX2, KLF4, and MYC), which are highly expressed in ESC [10]. Two additional factors (NANOG and LIN28) have been identified that enhance iPSC reprogramming; these factors are mechanistically associated with the four essential reprogramming factors [54]. OCT4, SOX2, KLF4, and MYC are predicted to have unique functions. OCT4 and SOX2 induce the pluripotent gene regulatory pathway, and enhance expression of NANOG. KLF4 is known to interact with pluripotency network proteins including OCT4 and SOX2, and also inhibits cell death. MYC is dispensable for reprogramming but is important to regulate chromatin structure to facilitate cellular reprogramming [55]. MYC is mechanistically associated with LIN28, which is a downstream target that enhances MYC expression. LIN28 is also involved in microRNA regulation by inhibition of LET7 family proteins, which are important for tissue differentiation [56]. Depending on the donor tissue types and the reprogramming method used, additional reprogramming factors are not required for iPSC reprogramming [57]. However, how the requirement for the cellular reprogramming is controlled and how the cellular regulatory pathways are established during cellular reprogramming need to be further defined. Although the individual function of each reprogramming factor in somatic cells has been studied, the synergistic interactions of all four factors and their contribution to the overall reprogramming process still need to be understood.
Although these reprogramming factors are expressed in somatic cells using an inducible promoter, reprogramming efficiency is still below 0.1–1% [58]. This indicates that reprogramming is not a unidirectional process in all cells, and variable epigenetic differences select for subpopulation of cells to generate iPSC. Due to the low reprogramming efficiency, we cannot isolate cells at intermediate stages of iPSC reprogramming, and a detailed analysis of the epigenetic changes that occur during cellular reprogramming has been challenging. Developing methods to select intermediate forms of reprogrammed iPSC will allow us to study the epigenetic changes that occur throughout the iPSC reprogramming. In addition, these intermediate forms will help us discover factors or new approaches that can overcome the genetic and epigenetic abnormalities of iPSC, and new ways to increase reprogramming specificity such that only pluripotency-related genomic regions are altered. Together, this research will move us closer to generating iPSC of equivalent quality to ESC, and realizing the opportunities for using iPSC to safely and reliably generate patient-specific, histocompatible transplantable tissues and cells.
11. Future perspectives: investigating other pluripotent stem cells beyond iPSC
iPSC have the potential to provide enormous benefit, not only for future therapeutic applications but also as research tools to clarify the mechanism of cellular reprogramming of terminally differentiated aged tissue to the embryonic stage. A number of reports have demonstrated that iPSC are highly similar to the pluripotency gold standard ESC, and could substitute for multiple types of the pluripotent stem cells. More careful analysis has shown that iPSC are not identical to ESC in terms of their molecular signature, differentiation potential, and function. To improve the quality of iPSC, basic studies on the regulatory mechanisms present in the developing embryo is needed to identify the key factors involved in the generation of pluripotent ESC. As such, research in ESC biology has become even more valuable since the introduction of iPSC reprogramming methods, and these cells continue to serve as the gold standard of the pluripotency as we move forward in the development of other stem cell lines. Yet we must also continue to study the mechanisms governing reprogramming after nuclear transfer in ntESC, in order to understand what other factors may improve iPSC quality and pluripotency for use in regenerative medicine. While iPSC mimic the cellular reprogramming procedure in ntESC by activating the few essential factors necessary to reverse differentiation of a somatic cell, the reprogramming factors present in the enucleated oocyte more efficiently and faithfully reprogram the somatic cell nucleus [20], in both molecular signature and differentiation potential. We are therefore obligated to study the differences in reprogramming that occur during the nuclear transfer procedure and during the generation of iPSC to discover those factors that will promote faithful iPSC reprogramming. Another challenge in the development of iPSC is that these cells often show alterations in imprinting [15]. This imprinting damage is also observed in ntESC [24]. Studies with pESC and androgenetic ESC (aESC) derived from a uniparental origin are needed in order to investigate the epigenetic signature of the imprinted genes and how they impact the pluripotency and function of the stem cells. In conclusion, although there are many reasons—ethical, immunological, and technical—for using iPSC rather than other pluripotent stem cells, a great deal of research on all stem cell types is still needed in order to achieve high-quality iPSC that are suitable for clinical application in tissue replacement therapy. It is therefore essential that the public and policymakers recognize the importance of understanding the mechanistic relationships among the various pluripotent stem cells and that supporting research on only one stem cell type will not only obstruct the scientific community’s efforts, but also impede progress in regenerative medicine that will improve human health and well-being.
Acknowledgements
K.K. is supported by a Gerstner Young Investigator Award and NIH (4R00HL093212-03).
References
- 1.Vogelstein B, Alberts B, Shine K. Genetics. Please don’t call it cloning! Science. 2002;295:1237. doi: 10.1126/science.1070247. [DOI] [PubMed] [Google Scholar]
- 2.Cibelli JB, et al. Parthenogenetic stem cells in non-human primates. Science. 2002;295:819. doi: 10.1126/science.1065637. [DOI] [PubMed] [Google Scholar]
- 3.Lin H, Lei J, Wininger D, Nguyen MT, Khanna R, Hartmann C, Yan WL, Huang SC. Multilineage potential of homozygous stem cells derived from metaphase II oocytes. Stem Cells. 2003;21:152–161. doi: 10.1634/stemcells.21-2-152. [DOI] [PubMed] [Google Scholar]
- 4.Robertson EJ, Evans MJ, Kaufman MH. X-Chromosome instability in pluripotential stem cell lines derived from parthenogenetic embryos. J. Embryol. Exp. Morphol. 1983;74:297–309. [PubMed] [Google Scholar]
- 5.Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 6.Hoglund P, et al. Host MHC class I gene control of NK-cell specificity in the mouse. Immunol. Rev. 1997;155:11–28. doi: 10.1111/j.1600-065x.1997.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 7.Opelz G, Wujciak T, Dohler B, Scherer S, Mytilineos J. HLA compatibility and organ transplant survival. Collaborative transplant study. Rev. Immunogenet. 1999;1:334–342. [PubMed] [Google Scholar]
- 8.Kim K, et al. Histocompatible embryonic stem cells by parthenogenesis. Science. 2007;315:482–486. doi: 10.1126/science.1133542. Epub 2006 Dec 14. [DOI] [PubMed] [Google Scholar]
- 9.Kim K, et al. Recombination signatures distinguish embryonic stem cells derived by parthenogenesis and somatic cell nuclear transfer. Cell Stem Cell. 2007;1:346–352. doi: 10.1016/j.stem.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. Epub 2006 Aug 10. [DOI] [PubMed] [Google Scholar]
- 11.Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat. Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 13.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 14.Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- 15.Stadtfeld M, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narsinh KH, et al. Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J. Clin. Invest. 2011;121:1217–1221. doi: 10.1172/JCI44635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Q, et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28:704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polo JM, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim K, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat. Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin MH, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM, 3rd, Biniszkiewicz D, Yanagimachi R, Jaenisch R. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 25.Humpherys D, et al. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12889–12894. doi: 10.1073/pnas.192433399. Epub 2002 Sep 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Jiang J, et al. Different developmental potential of pluripotent stem cells generated by different reprogramming strategies. J. Mol. Cell Biol. 2011;3:197–199. doi: 10.1093/jmcb/mjr012. [DOI] [PubMed] [Google Scholar]
- 28.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brambrink T, Hochedlinger K, Bell G, Jaenisch R. ES cells derived from cloned and fertilized blastocysts are transcriptionally and functionally indistinguishable. Proc. Natl. Acad. Sci. U.S.A. 2006;103:933–938. doi: 10.1073/pnas.0510485103. Epub 2006 Jan 17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L, et al. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J. Biol. Chem. 2010;285:19483–19490. doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toth KF, Knoch TA, Wachsmuth M, Frank-Stohr M, Stohr M, Bacher CP, Muller G, Rippe K. Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J. Cell Sci. 2004;117:4277–4287. doi: 10.1242/jcs.01293. [DOI] [PubMed] [Google Scholar]
- 32.Schneider-Stock R, et al. 5-Aza-cytidine is a potent inhibitor of DNA methyltransferase 3a and induces apoptosis in HCT-116 colon cancer cells via Gadd45- and p53-dependent mechanisms. J. Pharmacol. Exp. Ther. 2005;312:525–536. doi: 10.1124/jpet.104.074195. [DOI] [PubMed] [Google Scholar]
- 33.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gore A, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hussein SM, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 36.Bortvin A, Eggan K, Skaletsky H, Akutsu H, Berry DL, Yanagimachi R, Page DC, Jaenisch R. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development. 2003;130:1673–1680. doi: 10.1242/dev.00366. [DOI] [PubMed] [Google Scholar]
- 37.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 38.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 39.Plachta N, Bollenbach T, Pease S, Fraser SE, Pantazis P. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat. Cell Biol. 2011;13:117–123. doi: 10.1038/ncb2154. [DOI] [PubMed] [Google Scholar]
- 40.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chawengsaksophak K, James R, Hammond VE, Kontgen F, Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–87. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- 42.Russ AP, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- 43.Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- 44.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 45.Du Z, Cong H, Yao Z. Identification of putative downstream genes of Oct-4 by suppression-subtractive hybridization. Biochem. Biophys. Res. Commun. 2001;282:701–706. doi: 10.1006/bbrc.2001.4636. [DOI] [PubMed] [Google Scholar]
- 46.Ezashi T, Ghosh D, Roberts RM. Repression of Ets-2-induced transactivation of the tau interferon promoter by Oct-4. Mol. Cell. Biol. 2001;21:7883–7891. doi: 10.1128/MCB.21.23.7883-7891.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niswander L, Martin GR. Fgf-4 expression during gastrulation, myogenesis, limb and tooth development in the mouse. Development. 1992;114:755–768. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- 48.Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 49.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Gene Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pesce M, Scholer HR. Oct-4: control of totipotency and germline determination. Mol. Reprod. Dev. 2000;55:452–457. doi: 10.1002/(SICI)1098-2795(200004)55:4<452::AID-MRD14>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 51.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 52.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 53.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 54.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. Epub 2007 Nov 20. [DOI] [PubMed] [Google Scholar]
- 55.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 56.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim JB, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 58.Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat. Methods. 2009;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]