Introduction
Advances in pharmacogenetic testing will expand the number of clinically important pharmacogenetic variants. Communication and interpretation of these test results are critical steps in implementation of pharmacogenetics into the clinic. Computational tools that integrate directly into the electronic medical record (EMR) are needed to translate the growing number of genetic variants into interpretive consults to facilitate gene-based drug prescribing. Herein, we describe processes for incorporating pharmacogenetic tests and interpretations into the EMR for clinical practice.
In pharmacogenetics, there are several well-characterized genes whose variants can be amalgamated into likely star-allele (*) nomenclature and therefore probable diplotypes for each patient.(1) For many of the pharmacogenes, it is possible to assign a phenotype based on the diplotype and to make clinical recommendations for the use of affected medications.(2–4) Such recommendations can be delivered to the clinician in at least two ways: as pharmacogenetic consults (clinical test interpretations that are diplotype-specific and present statically in the EMR), and as decision-support-based automated alerts which fire only when an affected drug is ordered or dispensed to a patient with an actionable pharmacogenetic test result.
A pharmacogenetic consult provides a diplotype-specific interpretation of each test result, conveys the assigned phenotype, and provides general recommendations for gene-based prescribing. Comprehensively documenting the interpretation of the pharmacogenetic test result, such as annotating the extent of functional consequence for each genetic variant, is pitted against the need to succinctly and clearly translate the genetic variants into meaningful results for clinicians. A pharmacogenetic consult that is concise, easily interpretable, and quickly retrievable from the EMR provides an effective means for communicating test results to clinicians. Because pharmacogenetic tests have life-long implications, a mechanism is needed to alert clinicians of gene-based drug prescribing recommendations at the time a drug is ordered, which may be months or years after the test results and consults are documented in the EMR. Coupling high-risk diplotypes to high-risk drugs using clinical decision support will alert clinicians of a priority pharmacogenetic test result at the point of care, i.e., when a high-risk drug is about to be prescribed or dispensed.
High-throughput genotyping interrogates genetic variants in hundreds or even thousands of genes. Providing consults for many of the putative pharmacogenes is hindered by a lack of consensus on how to clinically interpret the data. Our approach has been to demonstrate proof-of-principle for clinical pharmacogenetics by generating consults for select genes that are ready for implementation, such as CYP2D6, CYP2C19, CYP2C9, VKORC1, and TPMT.(2–5) As new information emerges, additional drugs will be associated with a particular pharmacogenetic test, and the interpretation of previous tests may change. Therefore, both pharmacogenetic consults and decision support must be amenable to updates.
For these reasons, we have developed a modifiable system for construction of interpretative pharmacogenetic consults, and decision support alerts. Gene-specific pharmacogenetic consult templates are divided into modular sections, with each modular section having multiple versions. The consult for each unique diplotype is constructed by assigning one version of each modular section, which when linked together constitute a logical interpretation of the pharmacogenetic test result. Once diplotype-specific consults have been constructed, they are stored and available for integration into the EMR when a corresponding test result is posted. The automated consult for each test result can be released into the EMR unaltered, or with additional patient-specific comments. Individual modular sections of each consult can be updated as new data emerge, without the need to rewrite the entire template. High-risk diplotype-drug pairs coupled to decision support alerts can be updated as new drugs are linked to a pharmacogenetic test.
To illustrate our clinical implementation strategy, we provide examples of pilot data from our research protocol, St. Jude PG4KDS, the purpose of which is to selectively migrate array-based genotypes for clinically relevant genes into each patient’s EMR preemptively. DNA is genotyped in a CLIA-certified laboratory at the Medical College of Wisconsin using the DMET Plus array (Affymetrix, Inc., Santa Clara, CA), which probes for 1,936 genomic variants in 225 genes, and is supplemented with a CYP2D6 copy number assay. For each patient, the results for all genes are parsed into 225 individual files (one per gene), stored in a database, and selectively integrated into the EMR (Cerner Millennium v2010.02, PowerChart application, Cerner Corporation, Kansas City, MO) (Supplemental Figure S1). The initial 2 genes chosen for migration into the EMR were CYP2D6 and TPMT. Genomic variants are detected by 32 CYP2D6 probe sets and 8 TPMT probe sets, which are translated into 29 possible CYP2D6 alleles and 9 possible TPMT alleles using the DMET Console Software (v1.2, Affymetrix, Inc., Santa Clara, CA). A total of 46 unique CYP2D6 and 8 unique TPMT diplotypes have been observed in the first 200 patients studied (Figure 1A and 1B, Supplemental Tables S1 and S2).
Figure 1.
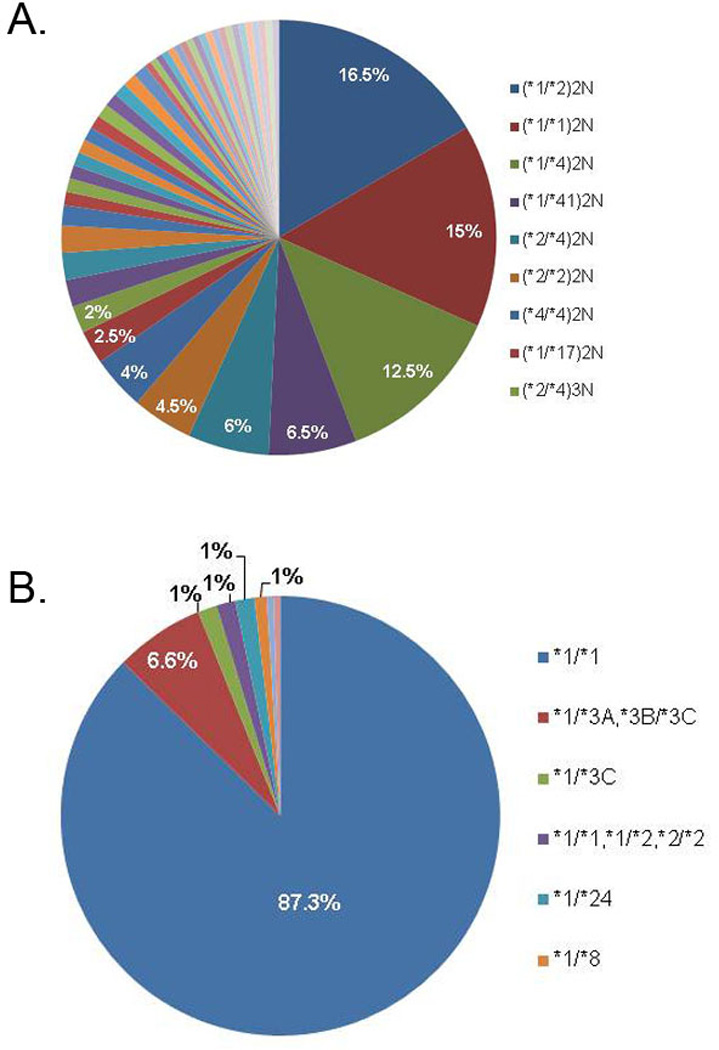
(A) 46 unique CYP2D6 diplotypes and (B) 8 unique TPMT diplotypes have been observed in the first 200 patients enrolled on PG4KDS. The most common diplotypes along with frequency are highlighted. For a complete list of all observed diplotypes and frequencies see Supplemental Tables S1 and S2. CYP2D6 copy number is determined by 3 independent sets of primers.
“Look-up” tables, or translation tables, (Supplemental Tables S3 and S4) for CYP2D6 and TPMT relate each unique diplotype result to phenotype, as well as to clinical priority status, appropriate versions of each modular section for creation of a pharmacogenetic consult (Supplemental Tables S5 and S6), problem list entry (if applicable), and decision support alerts. The clinical priority status is categorized as either routine or high-priority (high-risk), where high-priority refers to a result that has an actionable gene-based dosing recommendation. The look-up tables are stored in a standard database management system.
Phenotypes are assigned to each unique CYP2D6 or TPMT diplotype based on assessments of functional allele activity from prior studies, generally as summarized in Clinical Pharmacogenetics Implementation Consortium Guidelines.(2, 3, 5) CYP2D6 diplotypes are assigned to four phenotype categories: extensive metabolizer, intermediate metabolizer, poor metabolizer, and ultrarapid metabolizer, with the latter two considered as high-priority phenotypes (Supplemental Table S3). TPMT diplotypes are assigned to three phenotype categories: normal (high) TPMT activity, intermediate TPMT activity, and low or absent TPMT activity, with the latter two phenotypes considered high-priority phenotypes (Supplemental Table S4). Less commonly, phenotype ambiguity is possible due to copy number variation that cannot be clearly assigned to one particular allele or the inability to assign a definite diplotype based upon “no calls” from a single probe set. In some instances, communication of the ambiguous phenotype assignment may be informative, such as a possible ultrarapid CYP2D6 metabolizer [e.g., (*2/*10)3N, when the exact allele that is duplicated is unknown], when a high-risk drug such as codeine is prescribed. A possible high-risk diplotype might lead a clinician to choose alternative agents that are not substrates of CYP2D6.
A total of 187 diplotype-specific CYP2D6 pharmacogenetic consults and 31 diplotype-specific TPMT pharmacogenetic consults have been created for observed and likely allele combinations (examples, Figure 2A and 2B). The CYP2D6 consults are built from 5 modular sections, with each section having multiple versions: phenotype (6 versions), diplotype interpretation (32 versions), dosing recommendations (6 versions), activity score (11 versions), and an educational link. TPMT consults are built from 4 modular sections: phenotype (5 versions), diplotype interpretation (5 versions), dosing recommendations (5 versions), and an educational link.
Figure 2.
An example of a (A) CYP2D6 and a (B) TPMT pharmacogenetic consult template. Each consult template is divided into modular sections, and specific versions of each modular section are linked to a unique diplotype. For a complete list of versions for each modular section see Supplemental Tables S5 and S6.
After approval by an appropriately trained clinician, the diplotype results and pharmacogenetic consults are accessible in the EMR via a customized Pharmacogenetics tab located in the flow sheet (Supplemental Figure S2). Aggregating pharmacogenetic test results into a single section of the EMR on a per-patient basis, rather than on a chronological basis, allows for a summative view of all test results regardless of the date of the pharmacogenetic test. In the first 200 patients studied, a total of 400 pharmacogenetic consults have been entered into patients’ EMRs. Thirty-seven of the test results and corresponding consults are of high-priority status and are highlighted (using font changes) to differentiate them from routine consults. Additionally, high-priority phenotypes are automatically placed into the problem list of the EMR, facilitating custom-built, drug-specific decision support alerts linked to problem list entries (Supplemental Figure S3).
This modular approach to creating pharmacogenetic consults has several advantages. As new data emerge, modifications only need to be made once in the database to alter all affected consults. As an example, if dosing recommendations change due to new information, all consults can be automatically updated by changing the wording of the applicable modular section version (on dosing) in only one place. Codeine is prescribed to approximately 20% of our patient population; therefore gene-based prescribing is specifically referenced for codeine in the CYP2D6 pharmacogenetic consult. To highlight drugs likely to be prescribed at other institutions, the dosing recommendation modular section could be altered to include the most pertinent medications. The modular approach also allows for efficient integration of preemptive pharmacogenetics into clinical practice. Based on the platform utilized for genotyping, all possible allele combinations can be predicted a priori, permitting the creation of hundreds of pharmacogenetic consult templates before any results are ever uploaded into the EMR.
Well-characterized pharmacogenes have the advantage that extensive population frequency and linkage data allows the assignment of star-alleles to genomic variants.(1) The function of most of these alleles has been described, thus allowing simplification of genomic variants to likely diplotypes and assignment of a predicted phenotype. For most other genes, such as G6PD, genomic variants and linkage disequilibrium have not been sufficiently studied to permit designation of star-allele nomenclature. Therefore, translating genomic variants into clinical recommendations and reportable results into the EMR will be much more difficult. Thus, the need for computational systems to summarize genetic variants into clinically useful and informative results, and interpretive consults with actionable dosing recommendations will become even greater. The system we describe can easily expand to accommodate additional genes and drugs, and such scalable systems are critical to successfully integrate pharmacogenetics into clinical practice.
Supplementary Material
Acknowledgements
The authors thank the clinical staff, research nurses, patients, and their parents for participation, and especially Pam McGill, David Jenkins, and Charles Hurmiz.
Support:
Supported by NCI grants CA 36401, and CA 21765 and the NIH/NIGMS Pharmacogenomics Research Network (U01 GM92666), and by the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Conflict of Interest/Disclosure:
M.V.R: receives a portion of the income St. Jude receives from licensing patent rights related to TPMT. W.E.E.: receives a portion of the income St. Jude receives from licensing patent rights related to TPMT. All other authors have no financial disclosures.
Authorship Contributions:
Contribution: J.K.H., K.R.C., J.M.H., and M.V.R. interpreted the data and drafted the manuscript. R.L., A.S., U.B. genotyped DNA samples using the Affymetrix DMET Plus array, performed the CYP2D6 copy number assay, and created the gene-specific file reports. W.Y. and C.S. developed DMET Tracker. N.M.K. and M.R.W. developed a database allowing for automated incorporation of pharmacogenetic consults into the EMR. D.K.B, J.M.H., J.K.H., K.R.C., C.H, S.J.C., and M.V.R. developed decision support rules. All authors contributed to the writing of the manuscript.
References
- 1.Robarge JD, Li L, Desta Z, Nguyen A, Flockhart DA. The star-allele nomenclature: retooling for translational genomics. Clin Pharmacol Ther. 2007;82:244–248. doi: 10.1038/sj.clpt.6100284. [DOI] [PubMed] [Google Scholar]
- 2.Crews KR, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for Codeine Therapy in the Context of Cytochrome P450 2D6 (CYP2D6) Genotype. Clin Pharmacol Ther. 2012;91:321–326. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Relling MV, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther. 2011;89:387–391. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott SA, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther. 2011;90:328–332. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83:234–242. doi: 10.1038/sj.clpt.6100406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



