Abstract
Research on genes and medications has advanced our understanding of the genetic basis of individual drug responses. The aim of pharmacogenomics is to develop strategies for individualizing therapy for patients, to optimize outcome through knowledge of human genome variability and its influence on drug response. Pharmacogenomics research is translational in nature and ranges from discovery of genotype-phenotype relationships to clinical trials which provide proof of clinical impact. Advances in pharmacogenomics offer significant potential for subsequent clinical application in individual patients; however, the translation of pharmacogenomics research findings into clinical practice has been slow. Key components to successful clinical implementation of pharmacogenomics will include consistent interpretation of pharmacogenomic test results, availability of clinical guidelines for prescribing based on test results, and knowledge-based decision support systems.
Keywords: pharmacogenomics, genetic testing, personalized medicine
Introduction
This is an exciting time in clinical medicine. Remarkable progress has been made in developing therapies for many common diseases, thanks in part to advances in knowledge about disease biology and pathogenesis. As more drug therapies are used to manage diseases, it is increasingly apparent that the majority of drugs or their dosages do not have a uniform effect across patients. A given therapy may be effective and cause serious adverse events in one subset of patients, while delivering no response in terms of toxicity or therapeutic effect in others. Mounting evidence demonstrates that an individual’s genetic makeup is a major factor in this differential outcome, accounting for an estimated 20%-95% of variability in drug disposition and effects.1
Pharmacogenomics is the study of genetic variation of drug-metabolizing enzymes, receptors, transporters and targets, and how these genetic variations interact to produce drug-related phenotypes, such as drug response or toxicity. Further, genetic markers can indicate novel drug targets or modifiers and serve to functionally classify patients’ disease and therefore influence the design of treatment.2 Although the terms pharmacogenetics and pharmacogenomics are often used interchangeably, “pharmacogenomics” is increasingly used to describe the study of drug response in relation to genome variations (either inherited, acquired or both). In this review, we use the word “pharmacogenomics” to apply to both singlegene and polygenic models. Advances in pharmacogenomics offer significant potential for subsequent clinical application to improve outcomes in individual patients.3 The intent of pharmacogenomics is to develop strategies to individualize therapy, with the goal of optimizing efficacy and safety through better understanding of human genetic variability and its influence on drug response. Here we discuss current and future initiatives in pharmacogenomics, the extent to which findings have already made an impact on patient care, and touch on some of the scientific challenges that lie ahead if we are to accelerate the translation of genomic science into safer and more effective drug therapy.
Current state of pharmacogenomics research
Research on genes and medications relevant to a wide range of diseases has advanced our understanding of the genetic basis of individual drug responses. Common genetic variations that have been studied include single nucleotide polymorphisms (SNPs), genomic insertions and deletions, and genetic copy number variations (CNVs). SNPs are the most frequent inherited sequence variations, constituting approximately 90% of all human genome variation and occurring every 100 to 300 base pairs. However, on average, CNVs account for larger regions of genome variation than do SNPs; normal individuals carry 4 Mb of CNVs (1 in every 800 bp).4,5 It is unclear whether SNPs or CNVs are more important in pharmacogenomics, but it is likely that both play a role, perhaps to varying degrees in different phenotypic outcomes and measures. Pharmacogenomics research has identified genes that contribute to individual patients’ drug sensitivity, resistance and toxicity; it has also identified the causes of variation in the expression and function of many of these genes among individuals, including the roles of microRNA, DNA methylation, copy number alterations, and single nucleotide differences, either inherited SNPs or somatically acquired single nucleotide variants (SNVs). Humans are estimated to have approximately 7 million SNPs whose minor allele frequency is greater than 5%.6-8 The importance of rare variants is being increasingly recognized in pharmacogenomics.9
SNPs that occur within the same region of DNA (usually < 50 kb apart) are typically inherited together as haplotypes. The human genome can be viewed as haplotype blocks in high linkage disequilibrium (LD; regions with a high level of concomitant inheritance), separated by regions of low LD and a low level of concomitant inheritance.8,10 Therefore, SNPs that are in strong LD and are associated with a disease or drug-response phenotype can identify the chromosomal position of a susceptibility gene or a functional SNP, although the SNPs themselves may not cause the phenotypic trait.11
Cancer pharmacogenomics presents additional challenges for research and for translation of findings because both germline and somatic mutations must be considered when selecting candidate genes. Tumor cells carry the same germline genetic polymorphisms as do normal cells (unless the tumors somatically acquire genomic deletions); however, increased genomic instability of malignant cells can produce a high frequency of additional mutations or can increase the copy number of inherited variant alleles.12 These genomic changes can include the acquisition of additional copies of chromosomes that carry genes encoding drug metabolizing enzymes or drug transporters, which can lead to alterations in the disposition of active drugs at the site of the tumor. Over the past several decades, a large amount of knowledge has been gained regarding the somatic molecular alterations that are involved in tumor development and progression in many types of cancers. This knowledge has led to the development of targeted cancer therapy, such as targeting the BCR/ABL1 fusion transcript in Philadelphia chromosome-positive chronic myeloid leukemia or acute lymphoblastic leukemia.
The long-term goal of pharmacogenomics is to translate findings regarding the genetic basis of drug responses into more effective and less toxic treatment for individual patients. The good news is that if we accurately define a person’s genome, we only need to do this once for each patient and then it can be used to guide drug therapy for a lifetime, making this a potentially very cost-effective diagnostic tool.
Candidate-gene approach and pharmacogenomics
Early progress in pharmacogenomics came from small candidate-gene studies of genes encoding drug-metabolizing enzymes. These studies focused on the effect of a SNP or a combination of SNPs in one or more candidate genes in a treatment context. One of the best-established genotype-phenotype relationships is that of the thiopurine methyltransferase (TPMT) gene and its effect on thiopurine therapy for acute lymphoblastic leukemia and for immune modulation.13,14 TPMT catalyzes the S-methylation of thiopurines, thus regulating the balance between cytotoxic thioguanine nucleotides (TGNs) and inactive metabolites in hematopoietic cells. Polymorphisms in the TPMT gene have been characterized.15 Clinical interest in TPMT pharmacogenomics is based on studies showing that TPMT genotype or phenotype can be used to identify patients at high risk of hematopoietic toxicity after thiopurine therapy.16
Another candidate gene is the hepatic cytochrome P450 gene CYP2D6, which catalyzes the metabolism of many drugs. One drug whose metabolism is strongly associated with CYP2D6 genotype or phenotype is the analgesic codeine, a prodrug which must be bioactivated to morphine, a strong opioid agonist, by CYP2D6; hence, the efficacy and safety of codeine have been shown to be influenced by CYP2D6 polymorphisms.17,18
Drug efficacy is not influenced solely by variation in drug-metabolism genes but also by polymorphisms in genes that encode drug receptors, transporters and drug targets. For example, a common promoter variant in the molecular target of warfarin (VKORC1) strongly influences the dose levels required by individual patients. VKORC1 encodes the vitamin K-epoxide reductase protein, the target enzyme of warfarin. Variants in VKORC1 are significantly associated with warfarin sensitivity and reduced dose requirements.19 Several transporters have likewise been shown to have pharmacogenomic relationships with drug pharmacokinetics or effects. For example, a synonymous SNP in the ABCB1 gene has been associated with the maximally achievable digoxin concentration.20 Similarly, polymorphisms in the transporter SLCO1B1 have been associated with several phenotypes, including an increased risk of simvastatin-induced myopathy,21 methotrexate-related gastrointestinal toxicity,22 and disposition of the cyclin-dependent kinase inhibitor flavopiridol.23
A disadvantage of the traditional candidate gene approach is its reliance on a priori knowledge about the biology of the disease or phenotype of interest. This can result in an “information bottleneck” wherein discovery is limited by the identification of candidate genes involved in genomic traits of specific interest.24 For this reason, broad approaches such as genome-wide association studies or whole genome sequencing may be better suited for pharmacogenomics studies of complex diseases for which the detailed molecular architecture behind their etiology is not well characterized, and for which the genomic causes are likely polygenic.
Genome-wide association studies and pharmacogenomics
Since the completion of the human genome project, genome-wide profiling platforms have become available for use as an unbiased approach to identify genes associated with phenotypes of interest. The possibilities for pharmacogenomics research have expanded greatly with data generated by the International HapMap Consortium,25,26 the 1000 Genomes Project Consortium,27 and with the availability of high-throughput sequencing technologies. The International HapMap Project has constructed a high-density haplotype map of the human genome that provides an important tool for studying complex phenotypes such as drug response in different racial or ethnic populations.26 Initiated in 2008, the 1000 Genomes Project is an international public-private consortium that aims to build a detailed map of human genetic variation, ultimately including data from the genomes of more than 2,600 people from 26 populations worldwide.
Genome-wide association studies (GWAS) are aimed at discovering new biological pathways and advancing our understanding of the genetic basis for complex phenotypic traits. This strategy uses genotyping arrays containing 100,000 to 2.3 million SNPs per array to screen the genome for genetic variants associated with a given phenotype. The design of GWAS studies is based on the fact that genetic variants are inherited as haplotypes,25 allowing assessment of SNPs that tag a haplotype and thereby obviating the need to genotype all variants present in the haplotype.28
GWAS approaches have been successfully applied to pharmacogenomics.28-30 Some studies have confirmed previous results, such as the association of a CYP2C19 variant with reduction of the antiplatelet effect of clopidogrel31 and the association of CYP2C9 and VKORC1 variants with warfarin dose requirements.32 Other studies have discovered new associations, such as the association of SNPs in the interleukin 15 gene with disposition of antileukemic drugs in acute leukemia.33 In addition, a GWAS for the phenotype of bisphosphonate-induced jaw osteonecrosis in patients with multiple myeloma identified SNPs in CYP2C8 that may increase the risk for osteonecrosis.34
GWAS have provided breakthroughs in the understanding of the genetic basis for complex drug response phenotypes. However, GWAS can be considered only the first step, as the SNPs found by these studies to be associated with specific phenotypes are not necessarily the causal variants, which must be identified by comprehensive resequencing and functional analyses.
Whole genome sequencing
Sequencing of the entire exome or the whole genome of a patient is an evolving approach to providing the most dense coverage of human genome variation for research and, ultimately, patient care. As the cost of whole exome sequencing (now ~ $1000 per whole exome) and whole genome sequencing (now approaching $3000 per genome) continues to decrease and their quality and rapidity continue to increase, they are increasingly viable strategies for assessing genomic variability. These strategies are particularly attractive for cancer genomics and pharmacogenomics, in which identification of acquired (somatic) genome variation is an evolving strategy for understanding individual patients’ carcinogenesis and how these patients may best be treated with available agents that target specific mutations or pathway perturbations. However, initial experiences in whole-genome sequencing of individual patients’ cancer cells have typically yielded many findings of potential biological relevance but far fewer that are deemed actionable.35 Further, the heterogeneity of most tumors poses a challenge; DNA extracted from a bulk tumor will likely reflect the dominant clone within the tumor, whereas a sub-clone, especially one residing in a metastatic site that is most refractory to treatment resulting in disease recurrence is less likely to be identified, as it constitutes only a small fraction of the whole.36 Until such time as whole genomes or whole exomes can be sequenced by using DNA from a single cell, this limitation will persist. Further, if interpretation of two genomes from a single patient (the germline and tumor-cell genomes) is a challenge, consider the daunting nature of the need to interpret three or more genomes for every patient. To gain a truly comprehensive view of genomic variations that alter disease risk and treatment response, it will be important to assess not only DNA sequence variations (SNPs, CNVs, structural variants, etc.) but also epigenetic modifications, metabolomics, proteomics, microRNAs, and other mechanisms that alter genome structure and function.37,38
Whole-genome or whole-exome sequencing offers a logical strategy for characterizing a person’s germline genome shortly after birth, before the onset of disease, and for depositing the sequence data into a secure repository; the parents, and ultimately the patient, can then authorize their physician, pharmacist, or other healthcare provider to utilize the data for disease prevention or treatment strategies. This scenario remains futuristic, but the great reduction in costs and improvement in technology have made it possible to incorporate a patient’s whole genome sequence into a preemptive clinical workup, as described by Ashley and colleagues.39
One of the immediate challenges of clinically implementing genome sequencing as a diagnostic tool is the need to perform the sequencing in Clinical Laboratory Improvement Amendments (CLIA)-approved laboratories, to meet the quality control standards used in other diagnostic tests. But this challenge is made more daunting by the fact that interpretation of human genome sequencing is not straightforward; it requires the correct use of bioinformatic tools to align genomes against a “reference,” the identification of variations (ruling out false positives and false negatives), and summarization of the data in a way that is useful for discovery or clinical purposes. Further, only a fraction of the genes responsible for most complex human diseases is currently known; therefore, genome sequencing of a large number of patients today is unlikely to yield a definitive diagnosis based on disease-associated variations identified in genes or pathways. Indeed, a clear diagnosis has been obtained in only 20%-40% of highly selected cohorts of patients (e.g., children with deafness) who have undergone whole-genome sequencing.40 However, this limitation will decline over time as science identifies more genes that drive disease risk and drug response.
Proper bioinformatic analysis of a whole genome is suggested to cost several times as much as sequencing a genome.41 Moreover, as DNA sequencing increasingly becomes a commodity, the greatest source of error may be in the interpretation of genome sequences rather than in the sequencing per se. Thus, CLIA certification of the interpretation, as well as of the DNA sequencing process, is important. Although the interpretation of most diagnostic laboratory tests is left to the judgment of the clinician, interpretation of millions of variants in each genome will be well beyond the capability of most clinicians. As alignment and interpretation become increasingly automated, certification of the analysis and interpretation of sequence data will become more manageable, but it will remain relatively imprecise until the genomic basis of diseases are more fully elucidated. Likewise, most pharmacogenomic traits, like most disease risk traits, are likely to be polygenic in nature, and considerable additional research is needed to adequately define these traits in a way that can be used to manage most medications. Therefore, the interpretation of whole-genome and whole-exome sequence data must evolve before robust guidelines can be developed for their clinical use. Consortiums such as the Clinical Pharmacogenomics Implementation Consortium (CPIC) of the National Institutes of Health’s Pharmacogenomics Research Network (PGRN) are now helping to establish and implement guidelines for monogenic traits (e.g., risk for clopidogrel-associated cardiovascular adverse events and CYP2C19 genotype; response to codeine and CYP2D6 genotype) and for some multi-gene traits (e.g., warfarin dosing requirements and CYP2C9/VKORC1 genotypes);.42-44 likewise, consensus guidelines for use of whole-genome data will be important in facilitating their translation into the clinic.
Research using publicly available genomic data
Pharmacogenomics investigators now use in silico methods to analyze publicly available genomic information to predict new uses for existing drugs.45 The data from the 1000 Genomes Project is now available as a public dataset “in the cloud.”46 Data for this project currently available include DNA sequencing results from approximately 1,700 people; data from the remaining 900 samples are expected to be available soon (www.1000genomes.org). Cloud access has been touted as a way to improve access to the data to decrease the cost and time needed for analyses; doing so will advance the goal of making the data from the 1000 Genomes Project as widely available as possible to accelerate discovery.
Another source of publicly available data is The National Center for Biotechnology Information Gene Expression Omnibus (GEO), a public repository that archives and provides access to microarray, sequencing, and other forms of high-throughput functional genomic data submitted by researchers. Two recent studies analyzed gene expression data accessed from the GEO to identify potential new therapeutic uses for approved drugs. The authors used two sets of gene expression data: microarray data associated with 100 diseases (from the GEO) and data from human cancer cell lines treated with 164 small molecules. The authors obtained a signature of genes that were significantly up- or down-regulated for each disease and compared each to drug-induced gene expression signatures to create drug-gene expression profiles. Several potential therapeutic agents were identified through this analysis for lung adenocarcinoma, including the histamine H2-receptor antagonist cimetidine. Follow-up preclinical studies validated the efficacy of cimetidine against lung adenocarcinoma.47 Likewise, several potential drugs were identified in silico as therapeutic matches for inflammatory bowel disease, including the antiepileptic drug topiramate. The efficacy of topiramate was subsequently validated in a preclinical rodent model of colitis.48 These reports provide proof of principle that analysis of public gene expression databases is a resourceful and affordable approach to discovering possible new uses for approved drugs. This approach may provide an efficient alternative to traditional drug discovery methods.
The potential is enormous for pharmacogenomics to yield a powerful set of methods with which to individualize therapy for patients. The question now is whether clinical medicine is ready to accept these methods as standard-of-care tools.
Translation
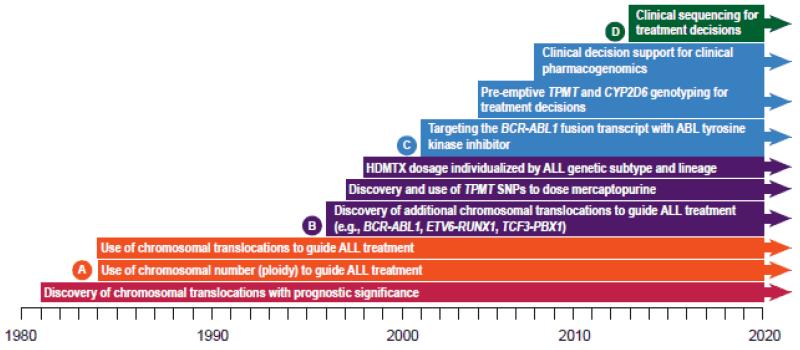
The continuum of pharmacogenomics research is complex, ranging from gene discovery to clinical trials. (Figure 1) Advances in pharmacogenomics offer significant potential to improve the clinical outcome of individual patients. However, the translation of pharmacogenomics research findings into clinical practice has been slow in most settings.
Figure 1.
The pathway of pharmacogenomics, from science to implementation.
Barriers to Translation of Pharmacogenomic Testing
Clinicians face many challenges to implementing pharmacogenomics in clinical practice, and the end result is that drugs are prescribed to patients whose relevant genotype is unknown. For example, codeine is still prescribed to patients whose CYP2D6 genotype is unknown, despite strong evidence that patients who are CYP2D6 poor metabolizers are not likely to experience analgesia, and ultrarapid metabolizers are at increased risk of toxicity. The challenges to widespread implementation are many, and include poor availability and scope of clinicians’ education in pharmacogenomics. Recent surveys of pharmacists and physicians in the U.S. reveal that many feel inadequately educated in pharmacogenomics.49-51 Reported deficiencies included knowledge about what tests are available, how to procure them, and how to interpret and apply the results to a patient’s care in the context of other clinical variables. 49,52 Interestingly, survey results have shown that clinicians who felt well-informed about the availability and applications of testing and had received pharmacogenomics instruction as part of their formal education were more often early adopters of pharmacogenomic testing.49
Cost is another barrier to the implementation of pharmacogenomics into routine clinical practice. In today’s economic environment, fiscal restraint is a top priority for both healthcare payers and healthcare providers. The results of a number of recent studies evaluating the clinical validity and utility of pharmacogenomic tests provide incentive for reimbursement of genetic testing and investment in implementation strategies.53-55 Pharmacy benefit managers such as Medco are partnering with health care organizations such as the Mayo Clinic to determine the benefit of modifying pharmacotherapy on the basis of genotyping. One such partnership found that CYP2C9 and VKORC1 genotyping of new warfarin recipients resulted in a 43% lower risk of hospitalization for bleeding or thromboembolism.56
One factor complicating cost-benefit analysis is the cost of genetic testing itself. The rapidity of probing a patient’s genome for variants continues to increase while the cost decreases as technology advances. Because the cost of genotyping is expected to decrease over time, it has been argued that any cost-benefit analysis should assume that the cost of genotyping is negligible.57 Previous benefit analyses, which used much higher cost figures, are likely to underestimate the cost-effectiveness of clinical pharmacogenomics. In the future, the cost of pharmacogenomics testing will shift from genotyping costs to the expense of personnel who interpret the results, produce reports for clinical use, and oversee the technology.
The increasing number of clinically relevant genes and their variants will soon far exceed the capacity of a clinician’s memory or ability to integrate them into clinical decision making. Fortunately, the difficulties of reporting, organizing, and interpreting complex pharmacogenomic test results will be reduced by the continuing adoption of electronic medical records. Further, in a system-based model of healthcare, expert collaboration, clinical guidelines, and knowledge-based decision support systems will be available to guide selection of therapy options. Decision support alerts allow point-of-care interventions and guide gene-based drug dosing months or years after a genetic test result is reported. Although the increased use of technology will enable integration of pharmacogenomics into clinical practice, the fragmentation of technology between health care systems will prevent the transfer of genetic test results between health care settings. Ultimately, the goal will be to overcome the fragmentation of health care databases so that genetic test results will follow patients from one healthcare setting to the next over the patient’s lifetime. As more complex, multigenic predictors of therapeutic response are identified, clinicians will increasingly rely on powerful decision-support tools to implement new testing protocols in clinical practice.
Even if genetic test results are conveyed to clinicians, consistent interpretation of the results remains an issue. The star-allele nomenclature was created to standardize genetic polymorphism annotation for the cytochrome P450 genes, wherein a discrete star allele represents either a single genetic variant or a haplotype (www.cypalleles.ki.se). However, translation to star-allele nomenclature itself can be problematic as new variants are continuously being described and published.58 For the many well-characterized pharmacogenes (e.g., CYP2D6, CYP2C19, CYP2C9, TPMT) with extensive population frequency linkage data that facilitate the use of star-allele nomenclature for genomic variants, assignment of a likely diplotype, or specific genetic variant combination, is possible.58 Because the function of the most commonly reported alleles has been described, a phenotype can be predicted for each patient and utilized for clinical recommendations. As the evidence supporting gene-based dosing has grown, there has been a move to develop specific, peer-reviewed, publicly available guidelines for the dosing or choice of drugs, based on specific pharmacogenopmic tests. Consortiums such as CPIC and the Pharmacogenomics Working Group of the Royal Dutch Association for the Advancement of Pharmacy (both highlighted on www.pharmgkb.org) are working to publish peer-reviewed guidelines that address practical issues in applying a patient’s pharmacogenomic test results to individualize therapy.52,59
Successful clinical pharmacogenomic test implementation
Even with the increasing availability of clinical guidelines for specific gene-drug pairs 42-44,60-62 that clearly indicate how prescribing should be modified based on test results, actual implementation of dose modifications remains a challenge. At St. Jude Children’s Research Hospital, we have gained experience in implementing pharmacogenomic tests in the clinic, first using single-gene tests63 and more recently, using array-based tests. This experience illustrates some of the steps needed for implementation and the importance of computational decision support tools.
Many health care practices begin implementing pharmacogenomics on a gene-by-gene basis; the decision to order a genetic test is individualized and based on the high likelihood that a “high-risk” drug (one substantially influenced by specific genetic variation) will be prescribed for any given patient or group of patients. An advantage is that the genetic test result is likely to be applied by the clinician, at least initially, because the prescribing decision was, by definition, linked to the ordering of the genetic tests. Pharmacogenomic test results may then be used as a covariate along with other patient characteristics to dose medications, as exemplified by warfarin-dosing algorithms that use both genetic and non-genetic factors to individualize warfarin doses. However, this “per gene” reactive testing has historically had disadvantages, in that it is expensive, and the turn-around time may be too slow to be useful for the initial prescribing decisions. In the context of reactive genetic testing, tests have usually not been ordered for patients who will eventually be prescribed a high-risk drug.
One example of a method to overcome these disadvantages has been introduced in the field of antiplatelet therapy. Recently, a novel “bedside” genetic test has been developed to identify carriers of the CYP2C19*2 allele with a buccal swab (Spartan RX CYP2C19, Spartan Biosciences, Ottawa, ON, Canada) for the purpose of identifying patients in whom clopidogrel should be avoided. The rapid nature of this technique is designed to ease the problem of turn-around time in allowing a pharmacogenomic approach to guiding antiplatelet treatment after percutaneous coronary intervention (PCI). A proof-of-concept study indicated that point-of-care genetic testing after PCI can be done effectively at the bedside,64 providing a valuable step toward improving the feasibility of individualized antiplatelet therapy.
A second approach to counteracting many of the disadvantages of reactive pharmacogenomic testing is the option of preemptive pharmacogenomic testing,65 made possible by the recent availability of inexpensive array-based pharmacogenomic testing platforms. Unlike individual pharmacogenomic testing for individual drugs, array-based preemptive testing can include a large number of relevant pharmacogenes that essentially cover most, if not all, “high-risk” drugs that may be prescribed to any individual, and is feasible for every patient entering the health care system because of its low cost. The test results would theoretically be available prior to any prescribing decision, consistent with the futuristic vision that patient genomes will be considered in every prescribing decision as an inherent patient characteristic,66 as are age and allergy status.
Our group has used single-gene tests, e.g. TPMT and CYP2D6, for several years to guide clinical prescribing decisions for thiopurines and codeine, respectively.13,63,67 More recently, we have implemented array-based genotyping for clinical purposes in a CLIA-approved laboratory, using the Affymetrix DMET Plus array, which tests for variants in 225 genes.68 Focusing on only two genes (TPMT and CYP2D6) to start, we have found that 46 of the first 220 patients (21%) had one or more “high-priority” actionable genotypes; thus, multigene arrays are likely to provide useful results for a substantial number of patients. For several reasons, we have elected to clinically implement an array-based pre-emptive genotyping approach via a research protocol (www.stjude.org/pg4kds) rather than as routine clinical care. One reason is that informed consent from patients is prudent, given that we will withhold some results: although arrays interrogate hundreds of genes, those for which there are no clear drug-related recommendations will be withheld from transfer to the medical record until such time as we have clear clinical recommendations for at least one drug for each genetic test. Consent is also required to inform patients that we may discover incidental findings during the course of the study that link a pharmacogene to disease risk, and to ascertain whether they wish to be notified of such incidental findings. In addition, the processes of prioritizing gene/drug pairs for clinical implementation, assessing their utilization, and ultimately communicating and acting on pharmacogenomic test results are complex and have been developed and tracked most easily in the context of a research protocol. All decisions on migration of gene/drug pair information into the medical record are approved by an oversight committee that reports to our institution’s Pharmacy and Therapeutics Committee, providing a mechanism for tracking integration of pharmacogenomic tests into the medical record.
Communication of pharmacogenomic test results is a critical step. Results must be communicated to clinicians and available statically in the medical record and at the point of care via clinical decision support. In our pharmacogenomics protocol, test results are translated into genotypes when possible, and each gene’s results are posted to the medical record with an accompanying written pharmacogenomic consultation. (Figure 2) The consultation contains information on the pharmacogenomic test result itself, phenotype assignment, genotype interpretation, basic dosing recommendations for our highest-use drugs, and a hyperlink to educational sites (Hicks JK, Crews KR, Hoffman JM, Kornegay NM, Wilkinson MR, Lorier R, et al., unpublished data). The phenotype assignment is categorized as “routine” or “high-priority” (high-risk); that is, phenotypes that should prompt alteration of dosing (such as poor or ultrarapid metabolizer of CYP2D6, and homozygous or heterozygous TPMT deficiency) are characterized as high-priority and added to the patient’s Problem List in the medical record. High-priority test results are also communicated to each attending physician via email, and they prompt the firing of alerts to prescribers and dispensers of high-risk drugs at the point of care.
Figure 2.
Steps involved in translating a genotype test result into therapy recommendations in a patient’s health record. Several well-characterized genes have variants which can be translated into likely star-allele (*) nomenclature and from there into diplotypes, or specific genetic variant combinations. Each diplotype is translated into a probable phenotype, which drives dosing recommendations for affected medications. The steps required vary based on whether the result is determined to be high-priority or routine, where high-priority refers to a result that has an actionable gene-based dosing recommendation. Dosing recommendations can be delivered to the clinician by static written pharmacogenomic consultations in the electronic medical record (EMR), and by decision-support based automated alerts which fire when an affected drug is ordered to a patient with an actionable pharmacogenomic test result. Patients are informed about each phenotype result and, if necessary, about recommended changes to therapy.
Decision-support alerts that link high-priority genotypes to high-risk drugs must be amenable to updates, as data linking additional drugs to known gene variants or linking additional gene variants to high-risk drugs are constantly emerging. The most practical site at which to pre-empt potentially dangerous prescribing/dispensing decisions is at the point of care. For example, after the CYP2D6 genetic test result has been placed in the medical record to guide codeine prescribing, the test should also guide the prescribing of tramadol and of tricyclic antidepressants, SSRIs, and many other agents. A system that can accommodate updates is necessary to allow the addition of high-risk drugs to a single pharmacogenomic test result.
A personalized letter describing the results of each gene test is written to each participant at the time the result (high-priority or routine) is entered into the medical record, and a copy of the letter is uploaded to the pharmacogenomics section of the medical record and sent to the attending physician. At present, this “hard copy” letter to the patient is the primary means by which non-St. Jude clinicians may be informed of the patient’s genetic test results and their implications, requiring patients to be their own advocates in communicating these results. The lack of universal access to critical information in patients’ medical records is of course an issue not only for pharmacogenomics but across the entire health care system. However, this communication problem in the context of pharmacogenomic test information, with its lifelong implications for prescribing for each patient, fully illustrates the inefficiency and suboptimal care inherent in fragmented health care, with its lack of free access to information in most care settings. It will be an important undertaking to implement a more comprehensive approach to health care records on a nationwide scale, which will improve the reporting and disseminating of pharmacogenomic test results. Obviously, the optimal system will allow inpatient, clinic, and outpatient prescribers and pharmacies access to pharmacogenomic test results, preferably at the time of prescribing and dispensing.
Influence of pharmacogenomics on drug development
The U.S. Food and Drug Administration (FDA) has required that the labels of several dozen medicines (e.g., abacavir, carbamazepine, cetuximab, imatinib mesylate, irinotecan, mercaptopurine, traztuzumab) be revised to include pharmacogenomic information. In the case of clopidogrel, new findings demonstrated that CYP2C19*2 carriers treated with clopidogrel were at higher risk of major adverse cardiovascular events, particularly stent thrombosis, than were non-carriers.69,70 This information was added to the prescribing information in 2010. A similar action based on evidence of a pharmacogenomic relationship has been taken for warfarin. In the case of the cancer drug panitumumab, new findings suggested that patients be tested for KRAS mutations before starting therapy to determine whether they may benefit from the drug,71 prompting the FDA to issue a new label warning in 2009. Changes in drug labels are likely to be ongoing as evidence for gene-drug associations continues to emerge for both approved drugs and new agents.
In recognition of the role that pharmacogenomics data play in evaluating drug safety and efficacy, in 2003 the FDA initiated a voluntary data exchange program (i.e., safe harbor agreement) through which companies may voluntarily submit genomic data with their new drug applications; many drug companies now do so. Notably, the FDA has recently approved two genomically-targeted cancer treatments, vemurafenib and crizotinib. In both cases, the FDA simultaneously approved a diagnostic test developed to identify patients eligible for treatment with the agent. Vemurafinib, a BRAF inhibitor, is approved for treatment of BRAFV600E mutation-positive metastatic melanoma. Crizotinib, an anaplastic lymphoma kinase (ALK) inhibitor, is approved for ALK-positive non-small cell lung cancer. For both drugs, the genomic markers of interest were known to be associated with disease pathology before either the drug or the associated diagnostic test was approved. Both drugs are intended for patient subpopulations with limited treatment options, and the FDA worked with the sponsors to grant accelerated approval to quickly bring these treatments to market. Both of the drug and diagnostic test combination products were approved on the basis of data from two single-arm studies; in these cases, sponsors were not required to enroll biomarker-negative patients, signaling a break from the FDA’s historical preference for at least one well-controlled, randomized study comparing an investigational anticancer agent to the standard of care. These approvals represent a proof of concept that the approval process can help speed the translation of a targeted therapy from science to practice. The growing frequency with which companion diagnostic tests accompany new agents will continue to spur the implementation of pharmacogenomics in an increasing number of therapeutic areas.
Future directions
Pharmacogenomics research is making steady progress toward understanding how genes influence drug responses and disease outcomes. To prepare for the inclusion of clinical pharmacogenomics tests as a medical standard of care, medical, pharmacy, and nursing professionals must significantly expand the availability and scope of pharmacogenomics education for new and practicing clinicians in all healthcare fields.
For pharmacogenomics, rigorous and systematic measurement of drug response phenotypes (e.g., drug toxicity, drug response) is often more difficult than is the measurement of genomic variability. Therefore, clinical trials should routinely include both informed consent to obtain samples of genomic DNA (and cancer cell DNA, when appropriate) and the collection of standardized prospective phenotype measurements for pharmacogenomics studies.72 Because a specific genotype may be important in determining the effects of a medication in one population or disease but not in another, gene-drug relationships must be extensively validated for each therapeutic indication and in different racial and ethnic groups. Ideally, clinical trial protocols will be designed so that subsequent genomic studies can be performed as new knowledge about candidate genes grows and as new platforms emerge. These approaches should facilitate future pharmacogenomic discoveries and their translation into practice.
It is now time to develop processes for translating pharmacogenomic findings to clinical care, so that clinicians can use existing and future data to personalize treatment and prospectively identify patients at high risk of treatment failure due to excessive toxicity or inferior efficacy. Key components to successful clinical implementation of pharmacogenomics will include consistent interpretation of pharmacogenomic test results, availability of clinical guidelines for prescribing based on test results, and knowledge-based decision support systems. Pharmacogenomic discoveries hold promise for optimizing treatment, for developing more effective therapies, and ultimately, for improving patient outcomes.
Acknowledgments
We thank Betsy Williford for assistance with the artwork. We thank Steven Paugh, J. Robert McCorkle, and Laura Ramsey for their critical review of the manuscript. This work is supported by the NIH/NIGMS Pharmacogenomics Research Network (U01 GM92666 and U01 HL65899), and by the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Conflict of interest:
W.E.E. is director and CEO of St. Jude Children’s Research Hospital. W.E.E., M.V.R., and St. Jude Children’s Research Hospital have received patent royalties from TPMT genotyping tests. The other authors declared no conflict of interest.
References
- 1.Kalow W, Tang BK, Endrenyi L. Hypothesis: comparisons of inter- and intra-individual variations can substitute for twin studies in drug research. Pharmacogenetics. 1998;8:283–289. doi: 10.1097/00008571-199808000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Cheok MH, Pottier N, Kager L, Evans WE. Pharmacogenetics in acute lymphoblastic leukemia. Semin Hematol. 2009;46:39–51. doi: 10.1053/j.seminhematol.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hopkins MM, et al. Putting pharmacogenetics into practice. Nat Biotechnol. 2006;24:403–410. doi: 10.1038/nbt0406-403. [DOI] [PubMed] [Google Scholar]
- 4.Eichler EE, et al. Completing the map of human genetic variation. Nature. 2007;447:161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherer SW, et al. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39:S7–15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson CS, et al. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat Genet. 2003;33:518–521. doi: 10.1038/ng1128. [DOI] [PubMed] [Google Scholar]
- 7.Hinds DA, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 8.Sachidanandam R, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 9.Ramsey LB, et al. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 2012;22:1–8. doi: 10.1101/gr.129668.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 11.Roses AD. Pharmacogenetics and the practice of medicine. Nature. 2000;405:857–865. doi: 10.1038/35015728. [DOI] [PubMed] [Google Scholar]
- 12.Cheng Q, et al. Karyotypic abnormalities create discordance of germline genotype and cancer cell phenotypes. Nat Genet. 2005;37:878–882. doi: 10.1038/ng1612. [DOI] [PubMed] [Google Scholar]
- 13.Relling MV, Pui CH, Cheng C, Evans WE. Thiopurine methyltransferase in acute lymphoblastic leukemia. Blood. 2006;107:843–844. doi: 10.1182/blood-2005-08-3379. [DOI] [PubMed] [Google Scholar]
- 14.Schwab M, et al. Azathioprine therapy and adverse drug reactions in patients with inflammatory bowel disease: impact of thiopurine S-methyltransferase polymorphism. Pharmacogenetics. 2002;12:429–436. doi: 10.1097/00008571-200208000-00003. [DOI] [PubMed] [Google Scholar]
- 15.Eichelbaum M, Ingelman-Sundberg M, Evans WE. Pharmacogenomics and individualized drug therapy. Annu Rev Med. 2006;57:119–137. doi: 10.1146/annurev.med.56.082103.104724. [DOI] [PubMed] [Google Scholar]
- 16.Lennard L, Lilleyman JS, Van Loon J, Weinshilboum RM. Genetic variation in response to 6-mercaptopurine for childhood acute lymphoblastic leukaemia. Lancet. 1990;336:225–229. doi: 10.1016/0140-6736(90)91745-v. [DOI] [PubMed] [Google Scholar]
- 17.Lotsch J, et al. Can extremely low or high morphine formation from codeine be predicted prior to therapy initiation? Pain. 2009;144:119–124. doi: 10.1016/j.pain.2009.03.023. [DOI] [PubMed] [Google Scholar]
- 18.Kirchheiner J, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharmacogenomics J. 2007;7:257–265. doi: 10.1038/sj.tpj.6500406. [DOI] [PubMed] [Google Scholar]
- 19.Rieder MJ, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmeyer S, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97:3473–3478. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Link E, et al. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 22.Trevino LR, et al. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J Clin Oncol. 2009;27:5972–5978. doi: 10.1200/JCO.2008.20.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni W, et al. Flavopiridol pharmacogenetics: clinical and functional evidence for the role of SLCO1B1/OATP1B1 in flavopiridol disposition. PLoS One. 2010;5:e13792. doi: 10.1371/journal.pone.0013792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu M, Zhao S. Candidate gene identification approach: progress and challenges. Int J Biol Sci. 2007;3:420–427. doi: 10.7150/ijbs.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The International HapMap Project Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 26.A haplotype map of the human genome Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.A map of human genome variation from population-scale sequencing Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.di Iulio J, Rotger M. Pharmacogenomics: what is next? Front Pharmacol. 2011;2:86. doi: 10.3389/fphar.2011.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daly AK. Genome-wide association studies in pharmacogenomics. Nat Rev Genet. 2010;11:241–246. doi: 10.1038/nrg2751. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364:1144–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shuldiner AR, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper GM, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang JJ, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarasquete ME, et al. Bisphosphonate-related osteonecrosis of the jaw is associated with polymorphisms of the cytochrome P450 CYP2C8 in multiple myeloma: a genome-wide single nucleotide polymorphism analysis. Blood. 2008;112:2709–2712. doi: 10.1182/blood-2008-04-147884. [DOI] [PubMed] [Google Scholar]
- 35.Roychowdhury S, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Downing JR, et al. The pediatric cancer genome project. Nat Genet. 2012;44:619–622. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suhre K, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashley EA, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayden EC. Sequencing set to alter clinical landscape. Nature. 2012;482:288. doi: 10.1038/482288a. [DOI] [PubMed] [Google Scholar]
- 41.Mardis ER. The $1,000 genome, the $100,000 analysis? Genome Med. 2010;2:84. doi: 10.1186/gm205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott SA, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for Cytochrome P450-2C19 (CYP2C19) Genotype and Clopidogrel Therapy. Clin Pharmacol Ther. 2011 doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crews KR, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin Pharmacol Ther. 2012;91:321–326. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson JA, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90:625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dudley JT, Pouliot Y, Chen R, Morgan AA, Butte AJ. Translational bioinformatics in the cloud: an affordable alternative. Genome Med. 2010;2:51. doi: 10.1186/gm172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.1000 Genomes Project data available on Amazon Cloud. 2012 < http://www.nih.gov/news/health/mar2012/nhgri-29.htm>. [Google Scholar]
- 47.Sirota M, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dudley JT, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3:96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanek EJ, et al. Adoption of Pharmacogenomic Testing by US Physicians: Results of a Nationwide Survey. Clin Pharmacol Ther. 2012 doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
- 50.Shields AE, Lerman C. Anticipating clinical integration of pharmacogenetic treatment strategies for addiction: are primary care physicians ready? Clin Pharmacol Ther. 2008;83:635–639. doi: 10.1038/clpt.2008.4. [DOI] [PubMed] [Google Scholar]
- 51.McCullough KB, et al. Assessment of the pharmacogenomics educational needs of pharmacists. Am J Pharm Educ. 2011;75:51. doi: 10.5688/ajpe75351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong WB, Carlson JJ, Thariani R, Veenstra DL. Cost effectiveness of pharmacogenomics: a critical and systematic review. Pharmacoeconomics. 2010;28:1001–1013. doi: 10.2165/11537410-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 54.van den Akker-van Marle ME GD, Detmar SB, Enzing CM, Hopkins MM, Gutierrez de Mesa E, Ibarreta D. Cost-effectiveness of pharmacogenomics in clinical practice: A case study of thiopurine methyltransferase genotyping in acute lymphoblastic leukemia in Europe. Pharmacogenomics. 2006;7:783–792. doi: 10.2217/14622416.7.5.783. [DOI] [PubMed] [Google Scholar]
- 55.Hughes AR, et al. Pharmacogenetics of hypersensitivity to abacavir: from PGx hypothesis to confirmation to clinical utility. Pharmacogenomics J. 2008;8:365–374. doi: 10.1038/tpj.2008.3. [DOI] [PubMed] [Google Scholar]
- 56.Epstein RS, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study) J Am Coll Cardiol. 2010;55:2804–2812. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 57.Altman RB. Pharmacogenomics: “noninferiority” is sufficient for initial implementation. Clin Pharmacol Ther. 2011;89:348–350. doi: 10.1038/clpt.2010.310. [DOI] [PubMed] [Google Scholar]
- 58.Robarge JD, Li L, Desta Z, Nguyen A, Flockhart DA. The star-allele nomenclature: retooling for translational genomics. Clin Pharmacol Ther. 2007;82:244–248. doi: 10.1038/sj.clpt.6100284. [DOI] [PubMed] [Google Scholar]
- 59.Swen JJ, et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther. 2011;89:662–673. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 60.Relling MV, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther. 2011;89:387–391. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin MA, et al. Clinical pharmacogenetics implementation consortium guidelines for hla-B genotype and abacavir dosing. Clin Pharmacol Ther. 2012;91:734–738. doi: 10.1038/clpt.2011.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilke RA, et al. The Clinical Pharmacogenomics Implementation Consortium: CPIC Guideline for SLCO1B1 and Simvastatin-Induced Myopathy. Clin Pharmacol Ther. 2012 doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crews KR, et al. Development and implementation of a pharmacist-managed clinical pharmacogenetics service. Am J Health Syst Pharm. 2011;68:143–150. doi: 10.2146/ajhp100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roberts JD, et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet. 2012 doi: 10.1016/S0140-6736(12)60161-5. [DOI] [PubMed] [Google Scholar]
- 65.McLeod HL, Isaacs KL. Preemptive pharmacogenetic testing: insufficient data equal unsatisfactory guidance. Ann Intern Med. 2011;154:842–844. doi: 10.7326/0003-4819-154-12-201106210-00016. [DOI] [PubMed] [Google Scholar]
- 66.Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med. 2010;363:301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 67.Stocco G, et al. Genetic polymorphism of inosine triphosphate pyrophosphatase is a determinant of mercaptopurine metabolism and toxicity during treatment for acute lymphoblastic leukemia. Clin Pharmacol Ther. 2009;85:164–172. doi: 10.1038/clpt.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fernandez CA, et al. Concordance of DMET Plus genotypes with orthogonal genotyping methods. Clin Pharmacol Ther. 2012 doi: 10.1038/clpt.2012.95. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mega JL, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sofi F, et al. Cytochrome P450 2C19*2 polymorphism and cardiovascular recurrences in patients taking clopidogrel: a meta-analysis. Pharmacogenomics J. 2011;11:199–206. doi: 10.1038/tpj.2010.21. [DOI] [PubMed] [Google Scholar]
- 71.Amado RG, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 72.Evans WE, McLeod HL. Pharmacogenomics--drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]