Abstract
One of the remaining challenges in Alzheimer's disease (AD) research is the establishment of biomarkers for early disease detection. As part of a prospective study spanning a period of five years, we have collected serial serum samples from cognitively normal, mild cognitively impaired (MCI), and mild AD participants, including same patient samples before and after cognitive decline. Using mass spectrometry we identified several promising leads for biomarker development, such as prosaposin, phospholipase D1, biliverdin reductase B, and S100 calcium binding protein A7. Selected candidate markers were verified using reverse phase protein microarray assays. Of 15 protein/protein abundance ratios that were significantly altered in sera from subjects with mild AD compared to Normal or MCI subjects, 14 were composed of ratios containing heme oxygenase-1, biliverdin reductase A, or biliverdin reductase B. Moreover, an increase in the protein abundance ratio of matrix metallopeptidase 9/biliverdin reductase differentiated stable MCI subjects from MCI subjects progressing into mild AD before the onset of cognitive decline. These findings strongly implicate the heme degradation pathway as a promising source of protein biomarkers for the early detection of AD.
Keywords: Alzheimer's disease, biomarker, BLVR, BVR, complement factor H, heme, heme oxyengase-1, phospholipase D1, prosaposin, S100A7, serum
INTRODUCTION
To date, the only methods to diagnose Alzheimer's disease (AD) are clinical diagnosis based on screening tests and postmortem brain histochemical staining for clinical hallmarks such as neurofibrillary tangles and amyloid-β deposits in the parenchyma and blood vessel walls. Although several therapies for AD are being tested in clinical trials, no biomarker exists to estimate the effectiveness of treatment. The ideal biomarker must facilitate the detection of early stage disease while there is still potential to therapeutically prevent neurodegeneration. A diagnostic marker for late stage disease may have no clinical therapeutic utility because late stage disease is largely irreversible. Mild cognitive impairment (MCI) has been recognized as a transitional stage between normal aging and AD [1] and may offer the critical treatment window before significant and irreversible neurodegeneration occurs. Thus a biomarker for MCI could provide significant clinical utility for early disease detection.
AD biomarkers based on imaging and body fluid analytes have been proposed, with in vivo magnetic resonance imaging providing the best results. For example, hippocampal atrophy is used to aid in the diagnosis of AD as well as predicting which MCI patients will progress into AD [2]. However, a considerable drawback is the significant fluctuation between individuals, which makes sequential measurements over a period of time necessary for correct interpretation of results. A source of potential protein biomarkers that has been studied extensively is cerebrospinal fluid (CSF), with levels of phospho-tau and Aβ in various abundance ratios providing the best results [3,4]. However, specificity and sensitivity vary between studies and the ability to differentiate between types of dementia is currently under debate [5].
Although farther removed from the brain, peripheral blood, serum, or plasma offer several advantages as potential biomarker sources. These fluids are much more accessible compared to CSF and therefore can be tested easily in a regular clinical setting. Furthermore, during the initial biomarker discovery phase, serum or plasma can be collected from patients at different stages of the disease, whereas antemortem CSF samples are significantly more difficult to obtain. Multiple alterations have been observed in AD blood, such as altered gene expression profiles in AD lymphocytes [6,7], increased serum copper [8], increased membrane fluidity, and an abnormal expression pattern of amyloid-β protein precursor isoforms in AD platelets [9]. Therefore it is not surprising that several groups are working on the identification of plasma biomarker candidates for AD [10, 11].
Serum proteome screening approaches are not limited by our current, incomplete understanding of the mechanisms involved in AD. However, they face the challenge that most of the protein mass in serum corresponds to a few highly abundant proteins such as albumin and immunoglobulins. Yet it is the low abundance, low molecular weight (LMW) proteome, which contains cleavage fragments and proteins small enough to passively enter the blood stream, which has been shown to contain disease associated biomarkers [12]. Whereas some studies of serum proteins have used two dimensional gel electrophoresis coupled with mass spec-trometry (MS) [13], we have developed a method that focuses on the detection of LMW proteins and protein fragments complexed with highly abundant serum proteins [14]. A similar technique has been independently applied by Lopez and colleagues to successfully identify unique mass fingerprints in AD serum [15]. We took this one step further, utilizing liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) to identify “free” and complexed LMW proteins and protein fragments in serial serum samples obtained from a community-based cohort of cognitively normal, MCI, and mild AD subjects.
MATERIALS AND METHODS
Subjects
Blood samples were collected from a community-based cohort of cognitively normal and MCI participants after obtaining informed consent. Collection protocols were approved by the Institutional Review Board of Loma Linda University. Subjects were recruited and followed clinically for a period of five years. Subject classification was based on extensive and repeated psychometric evaluation according to the criteria published by Petersen [1,16] and Reisberg [17]. A detailed description of the subject cohort and classification has been previously described by Kirsch and colleagues [18]. Histopathological confirmation of diagnosis was unavailable because no deaths occurred within the time-frame of this study.
In short, diagnosis was based on bi-yearly cognitive testing, including Logical Memory I and II, Wisconsin Card Sorting Test, Trail Making Test A and B, Boston Naming Test, Draw-A-Clock, Geriatric Depression Scale, Word Fluency (Phonemic and Semantic) as well as videotaped Global Clinical Dementia Rating (CDR) with informant. Cognitively normal subjects had a CDR of 0, a CDR memory component of 0, and a maximum sum of CDR boxes of 1 at baseline. MCI subjects had a CDR of 0.5 with confirmed memory complaint, abnormal memory according to age and education but no dementia, normal general cognitive function, and normal daily living activities. Progression to dementia (mild AD) was determined by a sum of CDR boxes of 3.5 or more, NINCDS-ADRDA criteria and clinical judgment. Patient demographic data is shown in Tables 1 and 2.
Table 1.
Demographic subject data for experiments that were analyzed by disease group (Normal, MCI, mild AD). Errors represent SD. Ages were compared using the Mann-Whitney non-parametric test. No significant difference in age between groups was found
| Experiment | Disposition | n (female/male) | Age [years] |
|---|---|---|---|
| Reverse Phase Protein Array: analysis by disease group | Normal | 10 (5/5) | 78.3 ± 2.5 |
| MCI | 10 (5/5) | 80.4 ± 3.8 | |
| mild AD | 10 (5/5) | 79.3 ± 3.4 |
Table 2.
Demographic subject data for experiments that were analyzed longitudinally before and after cognitive decline in the same subject. Cognitive Development: subjects marked as “decline” deteriorated cognitively over the time span indicated under Δ Time, while “stable” subjects did not change cognitively over the same time span (MCI => MCI). Sampling date: a = first serum collection, b = second serum collection. Δ Time: average time between first and second serum collection in the same subject. Errors represent SD. Differences in Age and Δ Time were evaluated using the Mann-Whitney non-parametric test. No significant differences between sample sets were found
| Experiment | Cognitive development | Sampling date | n (female/male) | Age [years] | Δ Time [years] |
|---|---|---|---|---|---|
| Mass Spectrometry: analysis by disease progression | decline | a | 3 (2/1) | 79.0 ± 2.7 | |
| b | 3 (2/1) | 80.0 ± 2.7 | 1 ± 0.0 | ||
| Reverse Phase Protein Array: analysis by disease progression | stable | a | 6 (4/2) | 80.5 ± 5.6 | |
| b | 6 (4/2) | 81.8 ± 5.7 | 1.5 ± 0.5 | ||
| decline | a | 6 (4/2) | 80.2 ± 7.2 | ||
| b | 6 (4/2) | 82.0 ± 6.9 | 1.8 ± 0.4 |
Serum sample collection
Consecutive blood samples were collected approximately every six months from a cohort of cognitively normal controls as well as MCI and mild AD subjects who we have followed clinically for the past five years. Blood collection tubes containing no anticoagulant were stored at 4°C overnight to allow the blood to clot. Subsequently, the serum was separated by centrifugation at 1800 g, 4°C for 10 min. Following separation, the serum was mixed by gently inverting it in a 15 mL Falcon tube and aliquots of 500 μL were stored at −80°C until analysis.
Low molecular weight fractionation
Serum samples were prepared in a loading solution as 25 μL serum, 75 μL 2× SDS Tris Glycine Sample buffer, 15 μL 1M DTT, 35 μL ddH20, and 3 μL Bromophenol Blue. A Mini Prep Cell Apparatus (Bio-Rad) was used according to manufacturer specifications to isolate low molecular weight proteins. A 4% stacking and 10% cylindrical gel were used for electrophoretic separation, followed by elution with a peristaltic pump into five 500 μL aliquots. Fractions containing proteins and peptides with molecular weights < 35 kDa were combined and concentrated with Microcon Ultracel YM-3 (Millipore) filter cartridges according to manufacturer specifications. A final volume of 40 μL was achieved by adding 1× Tris-Glycine SDS Running Buffer. For reverse phase protein microarray printing, samples were diluted 1:2 in a solution of 2× Tris-Glycine SDS Sample Buffer with 20% glycerol and 2.5% 2-mercaptoethanol. For mass spectrometry analysis, SDS was removed from the LMW fraction by tricholoroacetic acid (TCA) precipitation. Samples were incubated with an equal volume of 10% TCA (w/v) on ice for 1 h and then centrifuged at 15,000 g, 4°C for 30 min. The pellet containing the precipitated proteins/peptides was washed in cold acetone and dissolved in 8 M urea.
Mass spectrometry
LC-MS/MS analysis was performed using a Thermo hybrid LTQ-Orbitrap mass spectrometer. LMW serum fractions from three individual patients collected before and after significant cognitive decline (Fig. 1 and Table 1) were each reduced by reaction with 15 mM DTT, alkylated by 50 mM iodoacetamide, and followed by trypsin digestion.
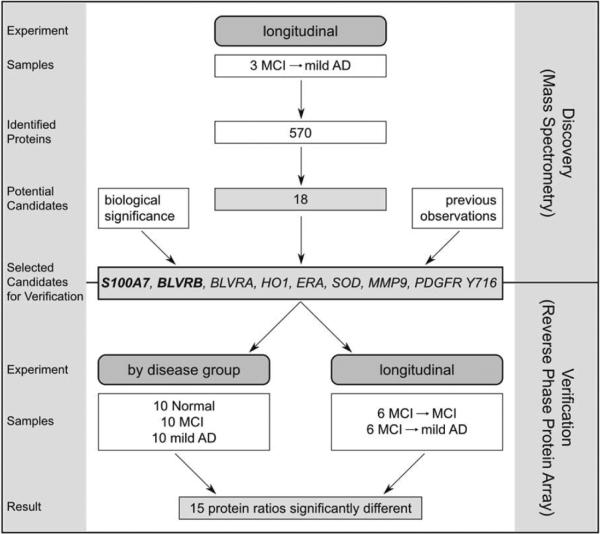
Fig. 1.
Flowchart of experimental setup. During the discovery phase of the project low molecular weight (LMW) serum samples from the same subjects before and after cognitive decline were analyzed using LC-MS/MS. During the validation phase relative abundances of selected biomarker candidates were measured in LMW serum fractions using reverse phase protein arrays.
Peptide and protein identification was performed using the SEQUEST algorithm to search the MS data against the human protein database available at NCBI. To obtain high confidence identifications the search results were filtered based on rank of match (RSp = 1), cross-correlation score for the peptide molecular ion charge state (XCorr > 1.9 (1+), 2.2 (2+) and 3.5 (3+)), difference between the first and second ranked match (Δ Cn > 0.1) and the probability of a random match (p < 0.01). An estimated “false discovery rate” of lower than 0.1% was obtained by searching a combined forward-reversed protein sequence database as described by Elias and colleagues [19]. Raw data were visualized, filtered and sorted using Scaffold (Proteome Software Inc.). Candidates for further analysis were selected by filtering according to the following criteria: protein identification by at least four spectra within a single disease group (before or after cognitive decline), ≥ 100% difference in number of MS-MS spectra between before and after cognitive decline and the direction of change in spectral count had to be the same in at least two subjects and could not be counter directional in the third subject. Percent difference was calculated as the difference between two variables divided by the average of both variables.
Analysis according to biological significance was performed using Ingenuity Pathway Analysis (Ingenuity Systems), DAVID bioinformatics database (National Institute of Allergy and Infectious Diseases, NIH), GeneCards (Weizmann Institute of Science and Xennex), GNF SymAtlas (Genomics Institute of the Novartis Research Foundation), and a custom software program developed in-house that allows batch searching of Medline through PubMed using automatically combined lists of proteins and specified search terms.
Reverse-phase protein arrays
To verify selected biomarker candidates, study samples were evaluated: (A) by disease group: serum LMW fractions from Normal (n = 10), MCI (n = 10), and mild AD (n = 10) subjects (Fig. 1 and Table 1); (B) by disease progression: LMW fractions of serum samples from six patients collected before and after significant cognitive decline, as well as six patients that remained stable with MCI during the same time span (Fig. 1 and Table 2).
Protein levels were measured using reverse phase protein microarrays (RPPA), a well established quantitative method with high precision and sensitivity that was first described in 2001 [20]. Precision, sensitivity, linearity and antibody validation for RPPAs have been previously reported by VanMeter et al. [21].
Samples were heated at 100°C for 5 min prior to printing onto FAST nitrocellulose slides (Whatman) with an Aushon 2470 arrayer equipped with 350 μm solid pins. Arrays were stored with dessicant at −20°C prior to immunostaining. Arrays were blocked (I-Block, Applied Biosystems) for 1 h and subsequently probed with primary antibodies, previously validated by Western blotting, to biliverdin reductase B (BLVRB, Abnova), biliverdin reductase A (BLVRA, Stressgen), Cu/Zn superoxide dismutase (SOD, Stressgen), estrogen receptor alpha (ERA, Cell Signaling Technology), heme oxygenase-1 (HO1, BIOMOL International, LP), matrix metallopeptidase-9 (MMP9, BIOMOL International, LP), platelet-derived growth factor receptor Tyr716 (PDGFR Tyr716, Upstate), and S100 calcium binding protein A7 (S100A7, Abnova). Immunostaining was performed on an automated slide stainer per manufacturer's instructions (Autostainer CSA kit, Dako). Secondary antibody was biotinylated goat anti-rabbit IgG H+L (1:5000) (Vector Labs, Burlingame, CA) or biotinylated rabbit anti-mouse IgG (1:10) (Dako). Subsequent protein detection was amplified via horseradish peroxidase mediated biotinyl tyramide with chromogenic detection (Diaminobenzidine) per manufacturer's instructions (Dako). Each antibody array was scanned on a flatbed scanner (UMAX PowerLook 1120), spot intensity analyzed, and a standardized, negative control subtracted, single data value was generated for each sample on the array (Image-Quant 5.2, Molecular Dynamics).
Statistical analysis
We used a Spearman's Rho non-parametric analysis of the non-normalized spot intensities to identify potential protein linkages (JMP 5.2, SAS). Correlations with a Spearman's Rho coefficient ≥ 0.85 and a p value ≤ 0.05 were considered for further analysis as protein abundance ratios. Differences in protein abundance ratios were determined using the Mann-Whitney nonparametric test (SPSS 16). A difference was considered significant if both arrangements of the ratio (a/b and b/a) resulted in a p value < 0.05.
RESULTS
Mass spectrometry
We used LC-MS/MS to identify candidate protein biomarkers in LMW serum fractions from three MCI subjects before and after cognitive decline to mild AD. Of the 570 proteins identified we selected 18 candidate biomarkers, of which 9 had increased and 9 had decreased spectral counts corresponding to subjects with cognitive decline (Table 3). In fact, 7 proteins were exclusively found either before or after cognitive decline. Several of the 18 candidates have been associated with AD, such as prosaposin, phospholipase D1, complement factor H, S100A7, and BLVRB (see Discussion for details).
Table 3.
LMW serum proteins with a different spectral count in three same subject samples before versus after cognitive decline. Qualifications: 1. Proteins were identified by at least four spectra within a single disease group, 2. Proteins demonstrated ≥ 100% difference in spectral count between before and after cognitive decline, 3. The direction of change in spectral count was the same in at least two subjects and could not be counter directional in the third subject. Percent difference was calculated as difference between before and after cognitive decline samples divided by average of before and after cognitive decline samples. Proteins in bold were exclusively found either before or after cognitive decline
| Regulation | Accession | Protein name | Spectra |
% difference after/before cognitive decline | |||||
|---|---|---|---|---|---|---|---|---|---|
| before Sample A | cognitive Sample B | decline Sample C | after Sample A | cognitive Sample B | decline Sample C | ||||
| ↑ | P78547 | Prosaposin | 0 | 0 | 0 | 1 | 2 | 1 | 200% |
| ↑ | P08493 | Matrix gla protein | 0 | 0 | 0 | 3 | 0 | 1 | 200% |
| ↑ | Q32LZ2 | Biliverdin reductase B | 0 | 0 | 0 | 1 | 0 | 3 | 200% |
| ↑ | Q86UQ9 | Citron | 0 | 0 | 0 | 2 | 2 | 0 | 200% |
| ↑ | 095740 | Serine (or cysteine) proteinase inhibitor, clade B, member 4 | 0 | 0 | 0 | 1 | 0 | 3 | 200% |
| ↑ | Q6EZF6 | Predicted: similar to neutrophil defensin 1 precursor | 0 | 2 | 1 | 0 | 4 | 18 | 152% |
| ↑ | P31151 | S100 calcium-binding protein A7 | 0 | 1 | 0 | 1 | 1 | 4 | 143% |
| ↑ | Q59EA4 | Phospholipase D1, phophatidylcholine-specific | 0 | 0 | 1 | 2 | 1 | 2 | 133% |
| ↑ | Q16782 | Serum amyloid A2 | 0 | 0 | 1 | 1 | 1 | 2 | 120% |
| ↓ | O00109 | Keratin 9 | 6 | 1 | 7 | 0 | 0 | 0 | 200% |
| ↓ | Q9P190 | Inter-alpha (globulin) inhibitor H4 | 2 | 4 | 4 | 0 | 0 | 0 | 200% |
| ↓ | Q02985 | Complement factor h isoform a precursor | 4 | 2 | 4 | 1 | 0 | 0 | 164% |
| ↓ | Q2KHQ6 | Apolipoprotein 11 isoform a precursor | 3 | 3 | 1 | 0 | 1 | 0 | 150% |
| ↓ | Q6GSJ0 | Keratin 1 | 36 | 5 | 7 | 3 | 4 | 3 | 131% |
| ↓ | 043608 | Roundabout, axon guidance receptor, homolog 2 | 2 | 2 | 0 | 0 | 1 | 0 | 120% |
| ↓ | Q9UGQ0 | Rsb-66 protein | 2 | 0 | 2 | 0 | 0 | 1 | 120% |
| ↓ | Q5SRP4 | Apolipoprotein m | 4 | 4 | 2 | 2 | 1 | 0 | 108% |
| ↓ | Q7L5M9 | Keratin 10 | 8 | 7 | 11 | 2 | 3 | 3 | 106% |
Reverse phase protein arrays
To verify the protein abundance of selected candidate biomarkers identified by mass spectrometry analysis, RPPAs were constructed using age- and gender-matched LMW serum samples from Normal, MCI, and mild AD participants. Furthermore, LMW serum samples from two groups of MCI participants were included: group (a) that progressed into mild AD, and group (b) who remained stable at MCI over the same time span of approximately 2 years. For each participant blood samples were collected at two distinct time points, thus providing before and after cognitive decline samples from the same patient. Two potential biomarker candidates were selected for verification based upon our mass spectrometry analysis: BLVRB and S100A7. To evaluate the expression of additional proteins involved in heme degradation we investigated HO1 and BLVRA. BLVRA and BLVRB both reduce biliverdin to bilirubin but their protein sequences and primary locations are very different. BLVRA is found primarily in adult human liver, while BLVRB is abundant in adult erythrocytes [22,23]. We further included SOD, MMP9, PDGFR Tyr716, and ERA based on their biological significance and previous observations in our laboratory.
No significant difference in abundance of any of the investigated proteins was found (data not shown). We subsequently used a Spearman's Rho non-parametric analysis to identify potential protein-protein linkages. We calculated a ratio for each antibody pair that met the cut-off criteria and compared these ratios in serum samples from Normal, MCI, and mild AD subjects, as well as before and after cognitive decline samples.
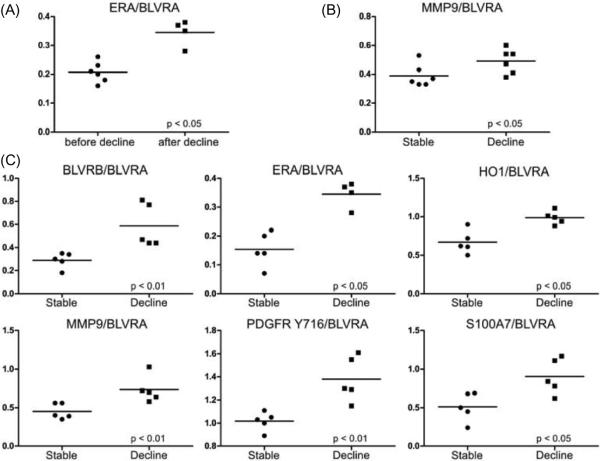
Comparing serum collection 1 (before cognitive decline) and collection 2 (after cognitive decline) in the MCI group progressing to mild AD revealed an increase in the ratio of ERA/BLVRA (Fig. 2A). This same ratio did not change in the stable MCI group over the same time span (data not shown). Furthermore, before progression to mild AD, cognitively declining MCI participants had an increased ratio of MMP9/BLVRA compared to patients with stable MCI (Fig. 2B). After cognitive decline six protein abundance ratios were elevated compared to the stable MCI group: BLVRB/BLVRA, ERA/BLVRA, HO1/BLVRA, MMP9/BLVRA, PDGFR Tyr716/BLVRA, and S100A7/BLVRA (Fig. 2C).
Fig. 2.
Ratio of staining intensities. Low molecular weight serum samples were analyzed using reverse phase protein arrays. (A) Same subject samples before and after significant cognitive decline. (B) Samples of stable MCI subjects (stable) versus cognitively declining MCI subjects (decline), before cognitive decline in the second group. (C) Samples of stable MCI subjects (stable) versus cognitively declining MCI subjects (decline), after cognitive decline in the second group (about 2 years later). Differences were evaluated using the Mann-Whitney non-parametric test and were considered significant if both arrangements of the protein abundance ratio (a/b and b/a) had a p <0.05.
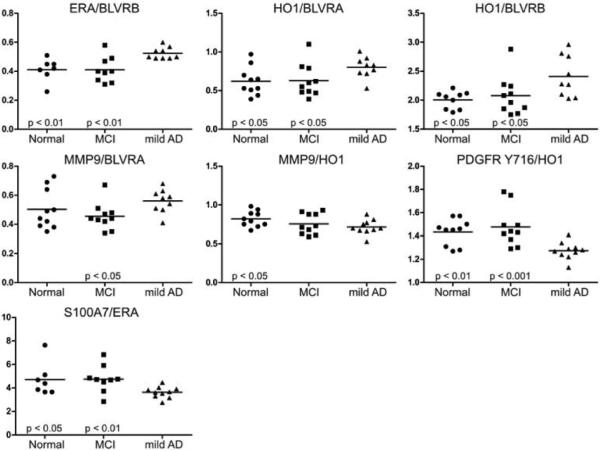
To further compare potential protein biomarkers based on disease groups, LMW serum samples from 10 age- and gender-matched Normal, MCI, and mild AD participants were printed on RPPAs and analyzed with the same antibodies. The protein ratios of ERA/BLVRB, HO1/BLVRA, and HO1/BLVRB were elevated in mild AD serum versus Normal and MCI (Fig. 3). A similar ratio increase was significant only in serum from patients with mild AD versus MCI for MMP9/BLVRA. PDGFR Tyr716/HO1 and S100A7/ERA ratios were reduced in the serum of patients with mild AD compared to Normal subjects, as well as sera from patients with MCI. The reduction of MMP9/HO1 was significant only in mild AD versus Normal subjects.
Fig. 3.
Ratio of staining intensities. Low molecular weight serum samples were analyzed using reverse phase protein arrays. Samples were analyzed by disease group. Differences were evaluated using the Mann-Whitney non-parametric test and were considered significant if both arrangements of the protein abundance ratio (a/b and b/a) had a p < 0.05.
DISCUSSION
One of the remaining challenges in AD treatment and/or prevention is early disease diagnosis. To implement successful therapy, it is necessary to begin treatment before irreversible neurodegeneration has progressed. However, AD is currently diagnosed by neuropathologic examination of postmortem brain tissue or neuropsychological evaluation, which relies on symptoms triggered by severe neurodegeneration. In an attempt to identify potential serum protein biomarkers of AD, we performed mass spectrometry and reverse phase protein microarray analysis of low molecular weight serum proteins from a community-based cohort of Normal, MCI, and mild AD participants. These subjects were followed with extensive psychometric evaluation bi-annually over a period of five years. Using stringent and generally accepted criteria [16,17] for subject classification as well as subject inclusion and exclusion, we secured a very well-characterized set of samples. Moreover, due to the prospective nature of this study and the increased progression rate of MCI to AD (12% per year) compared to age-matched cognitively normal controls (1–2% per year) [1], we were able to collect serum samples from study participants before and after the onset of dementia. Thus, although the sample set evaluated during the course of this study was comparatively small, it proved to be very valuable for biomarker discovery.
Using LC-MS/MS, we identified 570 proteins in LMW fractions (< 35 kDa) of serum obtained from three subjects before and after cognitive decline. Eighteen of these proteins were selected as potential biomarkers, several of which have been correlated with AD. Prosaposin, the precursor protein for saposins A-D, is involved in lysosomal degradation of sphingolipids [24], which are thought to affect γ-secretase activity and amyloid-β protein precursor cleavage [25]. We identified prosaposin only in subject samples after cognitive decline. This correlates well with the recent report that prosaposin expression is increased in zebrafish following interference with splicing of presenilin 1 and 2 genes [26]. Phospholipase D1 catalyzes the hydrolysis of phosphatidylcholine,and it was shown that binding to presenilin 1 decreases amyloid-β production [27]. We found phospholipase D1 to be increased with cognitive decline, which agrees with reports that phospholipase D1 is upregulated in AD brain mitochondria [28]. Complement factor H, an inhibitor for the alternative pathway of complement [29], is present in amyloid-β plaques [30] and has been shown to be increased in AD plasma [13]. In contrast, we observed a decrease in complement factor H with cognitive decline. However, we have focused on the LMW fraction of the serum proteome, which excludes full-length complement factor H due to its size. Although the abundance of protein fragments is linked to the abundance of the corresponding full-length protein, factors such as increased/decreased cleavage or secretion can influence this balance. S100A7 is an inflammatory response protein that promotes non-amyloidogenic cleavage of amyloid-β protein precursor by enhancing the activity of α-secretase [31]. We found S100A7 to be increased with cognitive decline, which is in agreement with the recent finding of Qin and colleagues that S100A7 is elevated in AD CSF [31]. BLVRB reduces biliverdin, a degradation product of heme, to bilirubin. We identified BLVRB only in subject sera after cognitive decline. Although BLVRB itself has not been implicated in AD, another component of the heme degradation pathway, HO1, is upregulated in AD brain tissue and is associated with neurofibrillary tangles and senile plaque neurites [32]. In fact, upregulation of HO1 appears to be an early occurrence in AD brain and correlates with decreased cognitive function [33].
To verify the abundance of selected biomarker candidates identified during the MS-based discovery phase of the project, we measured relative protein levels using RPPA technology. Of the candidates identified, we first chose to verify the serum level of BLVRB and S100A7. Based on our previous observations and biological relevance in AD, we also included MMP9, PDGFR Y716, SOD, and ERA in the analysis. Furthermore, to extend our evaluation to the entire heme degradation pathway, we included antibodies against BLVRA and HO1. While BLVRB is found abundantly in adult erythrocytes, BLVRA is the primary biliverdin reductase located in the human adult liver [22]. In fact, BLVRB shares very little sequence identity with BLVRA, but rather was found to be identical with flavin reductase [23].
Of the 15 protein ratios observed to be significantly altered in sera from patients with mild AD compared to Normal or MCI, 14 contained HO1, BLVRA, or BLVRB. This strongly implicates the heme degradation pathway as a potential biomarker target for AD. Previous evidence indicates that heme biosynthesis and catabolism are altered in AD. The expression of aminolevulinate synthase and porphobilinogen deaminase, two of the rate limiting enzymes for heme biosynthesis, is downregulated in AD brain [34]. In contrast, the level of ferrochelatase, which inserts iron into protoporphyrin IX during the final step of heme biosynthesis, is increased [35]. Heme complexes with amyloid-β, which is believed to cause free heme deficiency [36]. This, in turn, could be involved in the mitochondrial dysfunction observed in AD [37]. Alterations in heme degradation include the early upregulation of HO1 and the increased level of bilirubin in AD CSF [33,38]. In contrast, due to the inhibitory action of α1-antitrypsin, reduced levels of HO1 and bilirubin have been reported in plasma of patients with AD [?]. This is consistent with our observations, in which all 12 protein ratios with BLVRA or BLVRB as the denominator resulted in ratio increases, thereby indicating a reduction in either BLVRA or BLVRB. Comparing subjects after cognitive decline with those that remained stable at MCI, we found an increase in the ratio of BLVRB/BLVRA and HO1/BLVRA. This points to a skewed ratio of heme degradation enzymes. Furthermore, we discovered an increased ratio of MMP9/BLVRA, which distinguished cognitively stable subjects from those with progressing cognitive decline.
To our knowledge, the expression and activity patterns of BLVRA and BLVRB in AD brain have not been investigated. We believe that this protein pair may be a key component of several observations typical of AD pathology. BLVRA is not only essential for heme catabolism but also is a soluble dual specificity kinase that is critically involved in cell signaling [42,43]. It functions as a kinase in the insulin receptor/MAPK pathway and a transcription factor for ATF-2/CREB, influencing apoptosis, glucose transport, and protein synthesis. BLVRA is an ERK activator in insulin-like growth factor (IGF) 1 signaling, a pathway that regulates maximum lifespan [44,45]. Insulin signaling is perturbed in AD and the expression of several key components, such as IGF1 and IGF1 receptor, is downregulated [46]. IGF1 receptor signaling also modulates the ratio of the neurotrophin receptors p75NTR and TrkA, which are known to influence beta cleavage of amyloid-β protein precursor [45]. Interestingly, another protein that is known to regulate p75NTR and TrkA expression is estrogen [47]. Although the beneficial effect of estrogen on the risk for AD is well known [48], the involved mechanisms are still unclear. In our study the only significant difference found between the same patient samples before and after cognitive decline was an increase in the ratio of ERA/BLVRA. We demonstrated that this is not due to an increase in age because this ratio did not change in a similar group that remained stable at MCI over the same time span. It remains to be determined if both proteins are mechanistically related or if their correlation relies on respective downstream effects.
In conclusion, we identified 11 protein abundance ratios that distinguished mild AD sera from either MCI or Normal, indicating their potential for monitoring the progression from cognitively normal or MCI status to mild AD. Additionally, one protein ratio distinguished MCI subjects who would progress into mild AD from those who remained stable at MCI, before the onset of cognitive decline. As over 90% of these protein ratios contained at least one enzyme involved in heme catabolism and because heme metabolism has a well-established biological correlation with AD, these data justify further studies evaluating these biomarker candidates in blinded validation study sets of MCI patients and matched Normal subjects. The results also provide motivation for further targeted proteomic studies of the heme protein biochemistry that could yield more detailed insights into this AD-associated sub-proteome. Ultimately, it will be necessary to combine a panel of such biomarkers, containing a variety of proteins with different functions to reduce dependence on one particular pathway and provide the necessary sensitivity and specificity to correctly diagnose AD.
ACKNOWLEDGMENTS
The authors would like to thank the study participants and their families for providing serum samples and investing the time for clinical and neuropsychological tests. This work was supported by the National Institute on Aging, NIH, AG20948.
Footnotes
Authors' disclosures available online (http://www.jalz.com/disclosures/view.php?id=179).
REFERENCES
- [1].Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256:183–194. doi: 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]
- [2].Schott JM, Kennedy J, Fox NC. New developments in mild cognitive impairment and Alzheimer's disease. Curr Opin Neurol. 2006;19:552–558. doi: 10.1097/01.wco.0000247611.44106.76. [DOI] [PubMed] [Google Scholar]
- [3].Clark CM, Xie S, Chittams J, Ewbank D, Peskind E, Galasko D, Morris JC, McKeel DW, Farlow M, Weitlauf SL, Quinn J, Kaye J, Knopman D, Arai H, Doody RS, DeCarli C, Leight S, Lee VM, Trojanowski JQ. Cerebrospinal fluid tau and beta-amyloid: how well do these biomarkers reflect autopsy-confirmed dementia diagnoses? Arch Neurol. 2003;60:1696–1702. doi: 10.1001/archneur.60.12.1696. [DOI] [PubMed] [Google Scholar]
- [4].Andreasen N, Sjögren M, Blennow K. CSF markers for Alzheimer's disease: total tau, phospho-tau and Abeta42. World J Biol Psychiatry. 2003;4:147–155. doi: 10.1080/15622970310029912. [DOI] [PubMed] [Google Scholar]
- [5].Nagy Z. The dysregulation of the cell cycle and the diagnosis of Alzheimer's disease. Biochim Biophys Acta. 2007;1772:402–408. doi: 10.1016/j.bbadis.2006.11.001. [DOI] [PubMed] [Google Scholar]
- [6].Palotas A, Kalman J, Palotas M, Juhasz A, Janka Z, Penke B. Beta-amyloid-induced increase in the resting intra-cellular calcium concentration gives support to tell Alzheimer lymphocytes from control ones. Brain Res Bull. 2002;58:203–205. doi: 10.1016/s0361-9230(02)00773-6. [DOI] [PubMed] [Google Scholar]
- [7].Kalman J, Kitajka K, Pakaski M, Zvara A, Juhasz A, Vincze G, Janka Z, Puskas LG. Gene expression pro?le analysis of lymphocytes from Alzheimer's patients. Psychiatr Genet. 2005;15:1–6. doi: 10.1097/00041444-200503000-00001. [DOI] [PubMed] [Google Scholar]
- [8].Squitti R, Pasqualetti P, Dal Forno G, Moffa F, Cassetta E, Lupoi D, Vernieri F, Rossi L, Baldassini M, Rossini PM. Excess of serum copper not related to ceruloplasmin in Alzheimer disease. Neurology. 2005;64:1040–1046. doi: 10.1212/01.WNL.0000154531.79362.23. [DOI] [PubMed] [Google Scholar]
- [9].Kozubski W, Swiderek M, Kloszewska I, Gwozdzinski K, Watala C. Blood platelet membrane ?uidity and the exposition of membrane protein receptors in Alzheimer disease (AD) patientsflpreliminary Study. Alzheimer Dis Assoc Disord. 2002;16:52–54. doi: 10.1097/00002093-200201000-00009. [DOI] [PubMed] [Google Scholar]
- [10].Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K, Friedman LF, Galasko DR, Jutel M, Karydas A, Kaye JA, Leszek J, Miller BL, Minthon L, Quinn JF, Rabinovici GD, Robinson WH, Sabbagh MN, So YT, Sparks DL, Tabaton M, Tinklenberg J, Yesavage JA, Tibshirani R, Wyss-Coray T. Classi?cation and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- [11].Song F, Poljak A, Smythe GA, Sachdev P. Plasma biomarkers for mild cognitive impairment and Alzheimer's disease. Brain Res Rev. 2009;61:69–80. doi: 10.1016/j.brainresrev.2009.05.003. [DOI] [PubMed] [Google Scholar]
- [12].Liotta LA, Ferrari M, Petricoin E. Clinical proteomics: written in blood. Nature. 2003;425:905. doi: 10.1038/425905a. [DOI] [PubMed] [Google Scholar]
- [13].Hye A, Lynham S, Thambisetty M, Causevic M, Campbell J, Byers HL, Hooper C, R?sd?k F, Tabrizi SJ, Banner S, Shaw CE, Foy C, Poppe M, Archer N, Hamilton G, Powell J, Brown RG, Sham P, Ward M, Lovestone S. Proteome-based plasma biomarkers for Alzheimer's disease. Brain. 2006;129:3042–3050. doi: 10.1093/brain/awl279. [DOI] [PubMed] [Google Scholar]
- [14].Lowenthal MS, Mehta AI, Frogale K, Bandle RW, Araujo RP, Hood BL, Veenstra TD, Conrads TP, Goldsmith P, Fishman D, Petricoin EF, Liotta LA. Analysis of albumin-associated peptides and proteins from ovarian cancer patients. Clin Chem. 2005;51:1933–1945. doi: 10.1373/clinchem.2005.052944. [DOI] [PubMed] [Google Scholar]
- [15].Lopez MF, Mikulskis A, Kuzdzal S, Bennett DA, Kelly J, Golenko E, DiCesare J, Denoyer E, Patton WF, Ediger R, Sapp L, Ziegert T, Lynch C, Kramer S, Whiteley GR, Wall MR, Mannion DP, Cioppa GD, Rakitan JS, Wolfe GM. High-resolution serum proteomic pro?ling of Alzheimer disease samples reveals disease-speci?c, carrier-protein-bound mass signatures. Clin Chem. 2005;51:1946–1954. doi: 10.1373/clinchem.2005.053090. [DOI] [PubMed] [Google Scholar]
- [16].Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch. Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- [17].Reisberg B. Global measures: utility in defining and measuring treatment response in dementia. Int Psychogeriatr. 2007;19:421–456. doi: 10.1017/S1041610207005261. [DOI] [PubMed] [Google Scholar]
- [18].Kirsch W, McAuley G, Holshouser B, Petersen F, Ayaz M, Vinters HV, Dickson C, Haacke EM, Britt W, III, Larsen J, Kim I, Mueller C, Schrag M, Kido D. Serial Susceptibility Weighted MRI Measures Brain Iron and Microbleeds in Dementia. J Alzheimers Dis. 2009;17:599–609. doi: 10.3233/JAD-2009-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Meth. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- [20].Paweletz CP, Charboneau L, Bichsel VE, Simone NL, Chen T, Gillespie JW, Emmert-Buck MR, Roth MJ, Petricoin EF, III, Liotta LA. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- [21].VanMeter A, Signore M, Pierobon M, Espina V, Liotta LA, Petricoin EF. Reverse-phase protein microarrays: application to biomarker discovery and translational medicine. Expert Rev Mol Diagn. 2007;7:625–633. doi: 10.1586/14737159.7.5.625. [DOI] [PubMed] [Google Scholar]
- [22].Pereira PJ, Macedo-Ribeiro S, Párraga A, Pérez-Luque R, Cunningham O, Darcy K, Mantle TJ, Coll M. Structure of human biliverdin IXbeta reductase, an early fetal bilirubin IXbeta producing enzyme. Nat Struct Biol. 2001;8:215–20. doi: 10.1038/84948. [DOI] [PubMed] [Google Scholar]
- [23].Shalloe F, Elliott G, Ennis O, Mantle TJ. Evidence that biliverdin-IX beta reductase and flavin reductase are identical. Biochem J. 1996;316(Pt 2):385–387. doi: 10.1042/bj3160385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vaccaro AM, Salvioli R, Tatti M, Ciaffoni F. Saposins and their interaction with lipids. Neurochem Res. 1999;24:307–314. doi: 10.1023/a:1022530508763. [DOI] [PubMed] [Google Scholar]
- [25].Grimm MOW, Grimm HS, Pätzold AJ, Zinser EG, Halonen R, Duering M, Tschäpe JA, De Strooper B, Müller U, Shen J, Hartmann T. Regulation of cholesterol and sphingomyelin metabolism by amyloid-beta and presenilin. Nat. Cell Biol. 2005;7:1118–1123. doi: 10.1038/ncb1313. [DOI] [PubMed] [Google Scholar]
- [26].Newman M, Tucker B, Nornes S, Ward A, Lardelli M. Altering presenilin gene activity in zebrafish embryos causes changes in expression of genes with potential involvement in Alzheimer's disease pathogenesis. J Alzheimers Dis. 2009;16:133–147. doi: 10.3233/JAD-2009-0945. [DOI] [PubMed] [Google Scholar]
- [27].Cai D, Netzer WJ, Zhong M, Lin Y, Du G, Frohman M, Foster DA, Sisodia SS, Xu H, Gorelick FS, Greengard P. Presenilin-1 uses phospholipase D1 as a negative regulator of beta-amyloid formation. Proc Natl Acad Sci U S A. 2006;103:1941–1946. doi: 10.1073/pnas.0510708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jin J, Kim N, Lee Y, Kim Y, Choi E, Kozlowski PB, Park MH, Kim H, Min DS. Phospholipase D1 is up-regulated in the mitochondrial fraction from the brains of Alzheimer's disease patients. Neurosci Lett. 2006;407:263–267. doi: 10.1016/j.neulet.2006.08.062. [DOI] [PubMed] [Google Scholar]
- [29].Zipfel PF, Skerka C. FHL-1/reconectin: a human complement and immune regulator with cell-adhesive function. Immunol Today. 1999;20:135–140. doi: 10.1016/s0167-5699(98)01432-7. [DOI] [PubMed] [Google Scholar]
- [30].Strohmeyer R, Shen Y, Rogers J. Detection of complement alternative pathway mRNA and proteins in the Alzheimer's disease brain. Brain Res Mol Brain Res. 2000;81:7–18. doi: 10.1016/s0169-328x(00)00149-2. [DOI] [PubMed] [Google Scholar]
- [31].Qin W, Ho L, Wang J, Peskind E, Pasinetti GM. S100A7, a novel Alzheimer's disease biomarker with non-amyloidogenic alpha-secretase activity acts via selective promotion of ADAM-10. PLoS ONE. 2009;4:e4183. doi: 10.1371/journal.pone.0004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Smith MA, Kutty RK, Richey PL, Yan SD, Stern D, Chader GJ, Wiggert B, Petersen RB, Perry G. Heme oxygenase-1 is associated with the neuro?brillary pathology of Alzheimer's disease. Am J Pathol. 1994;145:42–47. [PMC free article] [PubMed] [Google Scholar]
- [33].Schipper HM, Bennett DA, Liberman A, Bienias JL, Schneider JA, Kelly J, Arvanitakis Z. Glial heme oxygenase-1 expression in Alzheimer disease and mild cognitive impairment. Neurobiol Aging. 2006;27:252–261. doi: 10.1016/j.neurobiolaging.2005.01.016. [DOI] [PubMed] [Google Scholar]
- [34].Dwyer BE, Smith MA, Richardson SL, Perry G, Zhu X. Down-regulation of aminolevulinate synthase, the rate-limiting enzyme for heme biosynthesis in Alzheimer's disease. Neurosci Lett. 2009;460:180–184. doi: 10.1016/j.neulet.2009.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Atamna H, Frey WH. A role for heme in Alzheimer's disease: Heme binds amyloid beta and has altered metabolism. Proc Natl Acad Sci U S A. 2004;101:11153–11158. doi: 10.1073/pnas.0404349101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Atamna H, Boyle K. Amyloid-beta peptide binds with heme to form a peroxidase: relationship to the cytopathologies of Alzheimer's disease. Proc Natl Acad Sci U S A. 2006;103:3381–3386. doi: 10.1073/pnas.0600134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Atamna H, Frey WH. Mechanisms of mitochondrial dysfunction and energy deficiency in Alzheimer's disease. Mitochondrion. 2007;7:297–310. doi: 10.1016/j.mito.2007.06.001. [DOI] [PubMed] [Google Scholar]
- [38].Kimpara T, Takeda A, Yamaguchi T, Arai H, Okita N, Takase S, Sasaki H, Itoyama Y. Increased bilirubins and their derivatives in cerebrospinal fluid in Alzheimer's disease. Neurobiol Aging. 2000;21:551–554. doi: 10.1016/s0197-4580(00)00128-7. [DOI] [PubMed] [Google Scholar]
- [39].Kim T, Pae C, Yoon S, Jang W, Lee NJ, Kim J, Lee S, Lee C, Paik I, Lee C. Decreased plasma antioxidants in patients with Alzheimer's disease. Int J Geriatr Psychiatry. 2006;21:344–348. doi: 10.1002/gps.1469. [DOI] [PubMed] [Google Scholar]
- [40].Schipper HM, Chertkow H, Mehindate K, Frankel D, Melmed C, Bergman H. Evaluation of heme oxygenase-1 as a systemic biological marker of sporadic AD. Neurology. 2000;54:1297–1304. doi: 10.1212/wnl.54.6.1297. [DOI] [PubMed] [Google Scholar]
- [41].Maes OC, Kravitz S, Mawal Y, Su H, Liberman A, Mehindate K, Berlin D, Sahlas DJ, Chertkow HM, Bergman H, Melmed C, Schipper HM. Characterization of alpha1-antitrypsin as a heme oxygenase-1 suppressor in Alzheimer plasma. Neurobiol Dis. 2006;24:89–100. doi: 10.1016/j.nbd.2006.06.009. [DOI] [PubMed] [Google Scholar]
- [42].Maines MD. New insights into biliverdin reductase functions: linking heme metabolism to cell signaling. Physiology (Bethesda) 2005;20:382–389. doi: 10.1152/physiol.00029.2005. [DOI] [PubMed] [Google Scholar]
- [43].Lerner-Marmarosh N, Miralem T, Gibbs PEM, Maines MD. Human biliverdin reductase is an ERK activator; hBVR is an ERK nuclear transporter and is required for MAPK signaling. Proc Natl Acad Sci U S A. 2008;105:6870–6875. doi: 10.1073/pnas.0800750105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Longo VD, Finch CE. Evolutionary medicine: from dwarf model systems to healthy centenarians? Science. 2003;299:1342–1346. doi: 10.1126/science.1077991. [DOI] [PubMed] [Google Scholar]
- [45].Puglielli L. Aging of the brain, neurotrophin signaling, and Alzheimer's disease: is IGF1-R the common culprit? Neurobiol Aging. 2008;29:795–811. doi: 10.1016/j.neurobiolaging.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Steen E, Terry BM, Rivera EJ, Cannon JL, Neely TR, Tavares R, Xu XJ, Wands JR, de la Monte SM. Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer's disease–is this type 3 diabetes? J Alzheimers Dis. 2005;7:63–80. doi: 10.3233/jad-2005-7107. [DOI] [PubMed] [Google Scholar]
- [47].Hasan W, Smith HJ, Ting AY, Smith PG. Estrogen alters trkA and p75 neurotrophin receptor expression within sympathetic neurons. J Neurobiol. 2005;65:192–204. doi: 10.1002/neu.20183. [DOI] [PubMed] [Google Scholar]
- [48].Tang MX, Jacobs D, Stern Y, Marder K, Schofield P, Gurland B, Andrews H, Mayeux R. Effect of oestrogen during menopause on risk and age at onset of Alzheimer's disease. Lancet. 1996;348:429–432. doi: 10.1016/S0140-6736(96)03356-9. [DOI] [PubMed] [Google Scholar]