Abstract
TRIM28 is critical for the silencing of endogenous retroviruses (ERVs) in embryonic stem (ES) cells. Here, we reveal that an essential impact of this process is the protection of cellular gene expression in early embryos from perturbation by cis-acting activators contained within these retroelements. In TRIM28-depleted ES cells, repressive chromatin marks at ERVs are replaced by histone modifications typical of active enhancers, stimulating transcription of nearby cellular genes, notably those harboring bivalent promoters. Correspondingly, ERV-derived sequences can repress or enhance expression from an adjacent promoter in transgenic embryos depending on their TRIM28 sensitivity in ES cells. TRIM28-mediated control of ERVs is therefore crucial not just to prevent retrotransposition, but more broadly to safeguard the transcriptional dynamics of early embryos.
TRIM28 (tripartite motif-containing protein 28, also known as KAP1, KRAB-associated protein 1, or TIF1β) is a co-repressor that is highly expressed in embryonic stem (ES) cells and is crucial to early mouse development, because homozygous Trim28 knock-out (KO) embryos arrest shortly after implantation and fail to gastrulate (Cammas et al. 2000). TRIM28 is tethered to DNA by sequence-specific Krüppel-associated box zinc finger proteins (KRAB-ZFPs) (Friedman et al. 1996; Emerson and Thomas 2009; Thomas and Schneider 2011) and induces local heterochromatin formation through the histone methyltransferase SETDB1 (or ESET), responsible for trimethylating histone 3 at lysine 9 (Schultz et al. 2002; Ivanov et al. 2007; Frietze et al. 2010), the NuRD (nucleosome remodeling and deacetylation) complex (Schultz et al. 2001), which contains the histone deacetylases HDAC1 and HDAC2 (for review, see McDonel et al. 2009), and heterochromatin protein 1 (HP-1) (Lechner et al. 2000; Sripathy et al. 2006). TRIM28 is required for proper oocyte-to-embryo transition (Messerschmidt et al. 2012), for the maintenance of imprinting marks immediately after fertilization (Li et al. 2008; Quenneville et al. 2011; Zuo et al. 2012), and for the self-renewal of ES cells, which rapidly die or undergo differentiation upon its removal (Wolf and Goff 2007; Fazzio et al. 2008; Hu et al. 2009; Rowe et al. 2010; Seki et al. 2010). However, which specific genes are controlled by TRIM28 during this early embryonic period remains largely unknown.
In contrast, it has now been firmly established that TRIM28, in part through SETDB1, is responsible for maintaining endogenous retroviruses (ERVs) in a silent state in ES cells and early embryos (Matsui et al. 2010; Rowe et al. 2010). TRIM28-mediated repression acts on multiple subsets of ERVs including intracisternal A-type particles (IAPs) and early transposon (Etn)/MusD elements, as well as on MERVL and ERVK families (for review, see Rowe and Trono 2011), and also partakes in blocking the replication of murine leukemia virus (MLV) in murine embryonic cells (Wolf and Goff 2007, 2009). Preventing the genomic spread of these retroelements may intuitively appear as the primary role of this process, yet the vast majority of ERVs carry mutations that inactivate their retrotransposition potential. Accordingly, it is noteworthy that the long terminal repeats (LTRs) of ERVs harbor binding sites for numerous transcription factors, as expected from the needs of their own replication. Furthermore, rare ERV-contained sequences have been found to function as cis-acting regulatory elements during mouse, human, and chick development through their recruitment of proteins such as POU5F1 (also called OCT4), GATA4, and CTCF (Bourque et al. 2008; Kunarso et al. 2010; Mey et al. 2012; Schmidt et al. 2012). ERVs and cellular genes can additionally be coordinately controlled in ES cells (Karimi et al. 2011; Macfarlan et al. 2011, 2012). Based on this premise, we asked here whether a component of the TRIM28-mediated maintenance of ES cell homeostasis might be the control of cryptic ERV-associated transcriptional activators. Our results indicate that ERVs are, indeed, transcriptional landmines, the TRIM28-mediated control of which is essential to preserve the transcriptional dynamics of ES cells. Regulation of retrotransposons by a TRIM28 pathway is thus critical not just to prevent retrotransposition, but more broadly to safeguard the timely activation of genes during early development.
Results
Transcriptional deregulation in Trim28 knock-out ES cells
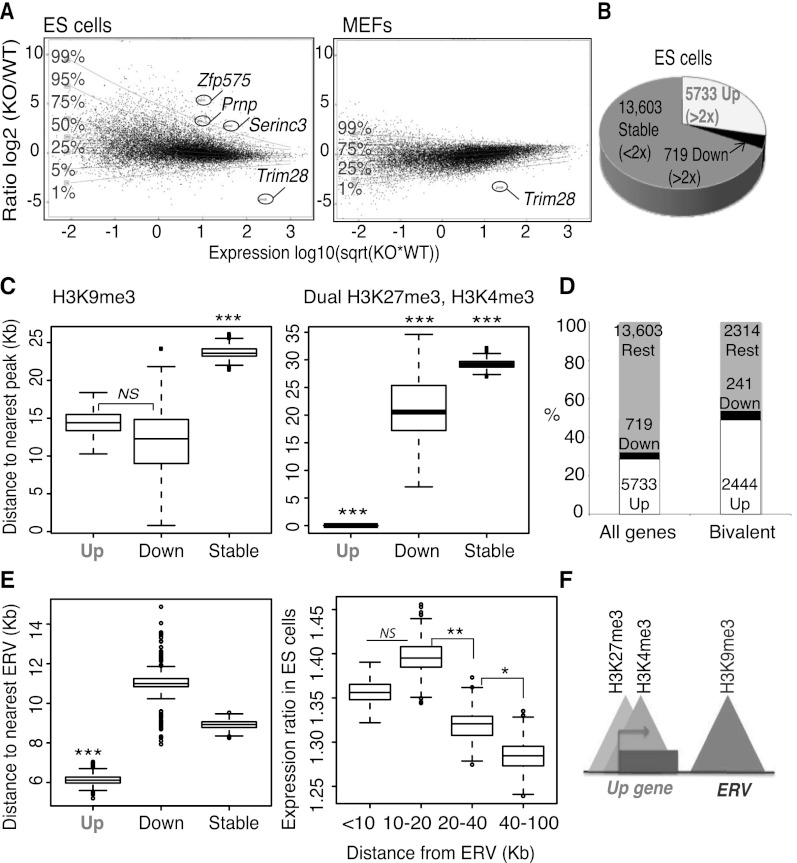
Using a previously described tamoxifen-inducible Cre/lox system (Rowe et al. 2010), we first compared mRNA-sequencing (mRNA-seq) data from control and Trim28-deleted murine ES cells (Fig. 1A,B). Transcripts from ∼20,000 genes were detected in control cells. Four days after Cre induction, based on a twofold cutoff and a significant difference of P ≤ 0.05, around 5700 of them were up-regulated (29%, including 1850 transcripts that were more than fivefold up-regulated), while around 720 were down-regulated (4%) and 13,600 unchanged (67%). From now on, we refer to these gene groups as “Up,” “Down,” and “Stable,” respectively. In contrast, in mouse embryonic fibroblasts (MEFs), transcriptional deregulation was only modest upon Trim28 deletion (Fig. 1A). This correlates the difference between the dramatic phenotype of Trim28-deleted ES cells, which die or differentiate after a few days and overexpress ERVs, and MEFs, which can be stably maintained and do not up-regulate ERVs (Rowe et al. 2010). Of note, genes affected by Trim28 deletion (both Up and Down) in ES cells were lowly expressed at baseline compared with genes unaffected by removal of this regulator (according to a Wilcoxon rank-sum test that was used to calculate significance here and for all boxplots) (Supplemental Fig. S1A). We decided to focus on up-regulated genes since they represented the larger category and Gene Ontology analysis indicated these genes to be involved in developmental pathways (see Supplemental Fig. S1B; Supplemental Table 1), including through expression at the embryonic two-cell stage as recently described (Macfarlan et al. 2012).
Figure 1.
Trim28 deletion in ES cells leads to up-regulation of genes close to ERVs, including many bivalent genes. (A) mRNA-seq in Trim28 wild-type (WT) and knock-out (KO) embryonic stem (ES) cells (left panel) or Trim28 WT and KO MEFs (right panel). Transcripts (assembly mm9) are plotted in black with the ratio on the y-axis and expression level on the x-axis. (Sqrt) Square root. (Horizontal lines) Levels of gene deregulation (e.g., only 1% of genes lie above the 99% line). The genes Zfp575, Prnp, and Serinc3 (referred to later) are highlighted, as well as Trim28. (B) Data from ES cells in A were used to group transcripts depending on whether they were greater than twofold up-regulated (Up), greater than twofold down-regulated (Down), or less than twofold affected (Stable). Up and Down genes were significantly changed based on a DESeq test (Anders and Huber 2010) (adjusted P-values ≤0.05). (C) The distance to the nearest peak (of either H3K9me3 on the left panel, 19,128 peaks, or dual H3K27me3, H3K4me3 peaks on the right panel, 12,390 peaks) from Up, Down, and Stable gene groups. (Left P-values) Up versus Down, not significant (NS), P = 0.48; Up versus Stable, P = 7.7 × 10−10; Down versus Stable, P = 0.0010. (Right P-values) Up versus Down, P = 9.9 × 10−11; Up versus Stable, P ≤ 2.2 × 10−16; Down versus Stable, P = 4.1 × 10−4. (D) Bivalent genes (as defined above by the presence of dual H3K27me3, H3K4me3 peaks) are enriched for up-regulated genes compared with all genes. (E) ERV locations (N = 82,382) were downloaded from the UCSC Genome Browser to include the categories ERV, ERV1, ERVK, and ERVL as defined by Repbase with a size cutoff of 500-bp minimum and used to plot the distance to the nearest ERV from Up, Down, and Stable gene groups (left). A Mann-Whitney Wilcoxon test was used to calculate significance: Up genes were significantly closer than the other two gene groups; (***) P ≤ 0.001. (Right) All genes were divided into groups based on their distance to the nearest ERV and their ratio between Trim28 WT and KO ES cells plotted on the y-axis. (P-values) The groups 10–20 versus 20–40 and 20–40 versus 40–100 are different: P = 0.0048 and P = 0.01, respectively. (F) Model showing that Up genes are close to H3K9me3 marks and ERVs and are often bivalent.
Chromatin state at genes affected by Trim28 deletion
Surprisingly, confrontation of these results with TRIM28 ChIP-seq data performed in the same cells revealed that <1% of up-regulated gene promoters were direct targets of TRIM28 (Supplemental Table 2). This suggested that Up genes could be indirectly affected by Trim28 deletion and/or were normally subjected to TRIM28-controlled nearby cis-acting influences. We thus compared the chromatin status of Up, Down, and Stable genes more broadly using available ChIP-seq data (Mikkelsen et al. 2007). We focused on H3K4me3, a Trithorax group– or TrxG-deposited mark typically associated with active transcription, H3K9me3, frequently a signature of TRIM28/SETDB1 recruitment (Matsui et al. 2010; Rowe et al. 2010), and H3K27me3, another repressive histone modification induced by the Polycomb repressive complex 2 (PRC2) (Bernstein et al. 2006; Gan et al. 2007; Guenther and Young 2010). As previously observed (Mikkelsen et al. 2007), H3K4me3 and H3K27me3 were significantly enriched at gene promoters, while H3K9me3 was generally depleted from these functional domains (Supplemental Fig. S1C). Genes deregulated upon TRIM28 depletion, whether up or down, were significantly closer to H3K9me3-enriched regions than unaffected genes (Fig. 1C, left). More revealingly, Up genes almost completely coincided with H3K27me3 peaks (Supplemental Fig. S1D). In ES cells, the H3K27me3 repressive mark is found together with its activating counterpart H3K4me3 at so-called bivalent promoters, which are rapidly induced upon differentiation (Bernstein et al. 2006). We thus compared the relative distribution of these two marks over the three gene groups. Genes unaffected by TRIM28 removal were the closest to H3K4me3-alone peaks and the farthest away from H3K27me3-alone peaks (Supplemental Fig. S1E), consistent with their average higher levels of expression than Up or Down genes. In contrast and most strikingly, Up genes almost completely overlapped bivalent H3K4me3/H3K27me3 peaks (Fig. 1C, right), indicating that the promoters of many of the genes induced upon Trim28 deletion are poised for transcription. Reciprocally, up-regulated genes (2444) were enriched among bivalent genes (4999) (Mikkelsen et al. 2007), compared with all genes (Fig. 1D, Fisher's exact test: P-value ≤ 1 × 10−16).
Genes up-regulated upon Trim28 deletion are located close to ERVs
Since few gene promoters were direct targets of TRIM28 (see above), we hypothesized that up-regulation of many genes could reflect the deregulation of TRIM28-controlled cis-acting elements situated in their nearby vicinity. In that respect, TRIM28, together with H3K9me3, is found enriched at ERV sequences in ES cells but not MEFs (Matsui et al. 2010; Rowe et al. 2010). Because ERVs are known to contain transcription-regulating sequences, we asked whether they were spatially associated with genes induced upon Trim28 deletion. Indeed, matching the genomic locations of ERVs (82,382 sites) with the three gene groups differentially affected by TRIM28 removal revealed that Up genes were on average significantly closer to these elements than Down or Stable genes (Fig. 1E, left). We also verified that it is not the case that all bivalent genes are enriched in ERVs but rather that bivalent Up genes (2444) are on average closer to ERVs than bivalent stable genes (2314, P = 0.001470) (Supplemental Fig. S2A). Interestingly, Up genes also clustered with long interspersed nuclear elements (LINE1s) but lay further from short interspersed nuclear elements (SINEs) than Down and Stable genes (Supplemental Fig. S2B–D), consistent with the previous observation that LINEs but not SINEs are modestly up-regulated in Trim28-deleted ES cells (Rowe et al. 2010). Reciprocally, the closer genes were to an ERV or particularly to an ERV of the subclass IAPs, the higher their average up-regulation upon TRIM28 removal, with genes also affected (although to a lesser extent) at distances of 100 kb (Fig. 1E, right; data not shown). Of note, this phenomenon of nearby cis-acting regulation is consistent with the previously documented modulation of the Agouti gene by an IAP located some 100 kb away, leading to variable coat colors in mice (Duhl et al. 1994; Michaud et al. 1994). In sum, these data indicate that many Up genes harbor bivalent promoters and lie close to H3K9me3 and ERVs (Fig. 1F).
Trim28 deletion triggers a switch from repressive to active chromatin marks at ERVs
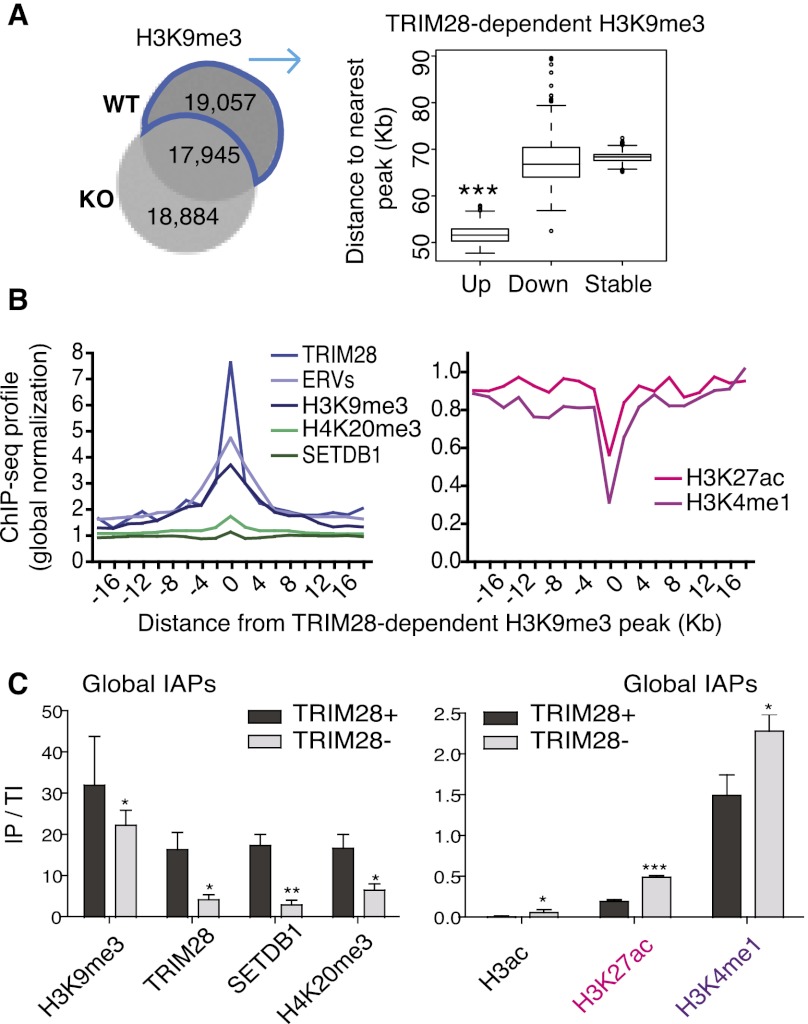
Mapping the genomic location of specific TRIM28-regulated ERVs based on a TRIM28 ChIP-seq is problematic because of the sharpness of the corresponding peaks, which only rarely extend beyond the borders of these multicopy elements. We thus turned to a comparison of H3K9me3 peaks in wild-type and Trim28-deleted ES cells, since this histone modification can spread a few kilobases into the junction of ERV proviruses with their flanking regions (Karimi et al. 2011; Rebollo et al. 2011). We found around 19,000 H3K9me3 peaks, that is, about half of those detected in control ES cells, to be TRIM28 dependent as indicated by their absence in knock-out cells (Fig. 2A, left). In agreement with their noted proximity to ERVs (see Fig. 1E), Up genes lay closer to TRIM28-dependent H3K9me3 peaks than Down and Stable genes (Fig. 2A, right). Likewise, in an element-centric analysis, we used the TRIM28-dependent H3K9me3 peaks to determine the nearest gene, generating a list significantly enriched for up-regulated genes (giving 2220 Up genes, Fisher's exact test, P ≤ 2.2 × 10−16) (Supplemental Fig. S3A; Supplemental Table 3), in line with the gene-centric analysis above. Of note, upon further examination of the high number of H3K9me3 peaks “newly present” in Trim28 knock-out cells, we found them to be in the same locations as the WT peaks but just slightly displaced and smaller in height and diameter rather than representing new peaks (Fig. 2A). These peaks thus most likely represent remnants of TRIM28-specific peaks, which is not surprising considering that our analyses were performed only 4 d after inducing Trim28 excision to avoid lethality.
Figure 2.
Trim28 deletion triggers a switch from repressive to active chromatin marks at ERVs. (A) Venn diagram of H3K9me3 ChIP-seq peaks in WT versus KO ES cells (left). 19,057 peaks are present in WT but lost in KO cells and so are defined as TRIM28-dependent peaks, which cluster closer to Up genes than Down (P = 0.001418) and Stable (P ≤ 2.2 × 10−16) genes (right). (B) TRIM28-dependent H3K9me3 peaks (see above) were assessed for correlation with ChIP-seq data sets. Positive correlations are shown on the left graph and anti-correlations on the right. All data displayed after global normalization of ChIP-seq counts. (C) ChIP results for repressive (left panel) and active (right panel) marks present at global IAPs (using IAP 5′-UTR primers). Bars show the mean and SD of three to four ChIPs per antibody with immunoprecipitate values normalized to total inputs (IP/TI) relative to Gapdh. Negative controls of no antibody were used in all experiments giving no enrichments, while the Pou5f1 enhancer served as a positive control with high enrichments for both H3K27ac and H3K4me1 of 1.1 and 7.5, respectively. Results were also reproduced in an independent ES cell line (Rex1). Paired t-tests were used to compare WT and TRIM28-depleted samples for each antibody: H3K9me3, P = 0.014; TRIM28, P = 0.027; SETDB1, P = 0.0036; H4K20me3, P = 0.0308; H3ac, P = 0.0337; H3K27ac, P ≤ 0.0001; H3K4me1, P = 0.011.
Interestingly, we observed that the TRIM28-dependent H3K9me3 peaks not only correlated with repressive histone marks, TRIM28, SETDB1 peaks (the latter data set obtained from Bilodeau et al. 2009), and with ERVs, but anti-correlated with H3K4me1 and H3K27ac, marks typically found together on active enhancers (Creyghton et al. 2010; Rada-Iglesias et al. 2010; Shen et al. 2012), while displaying no particular association with H3K4me3 or H3K27me3 (Fig. 2B; data not shown). In line with this, Up genes themselves also lay far from enhancer marks (Supplemental Fig. S3B). We therefore hypothesized that ERVs may gain these marks upon Trim28 deletion, thereby enhancing expression of neighboring genes. To test this idea, we focused on IAPs since we identified a motif highly represented in our H3K9me3 ChIP-seq peaks (in 64% of peaks) normally present in IAP consensus sequences (Supplemental Fig. S3C,D). Supporting this model, ChIP-qPCR with primers designed to amplify the majority of IAPs revealed that, indeed, in Trim28 knock-out ES cells, these elements not only lost TRIM28, SETDB1, and the repressive marks H3K9me3 and H4K20me3, but also gained active marks, including H3K27ac and H3K4me1 (Fig. 2C). This observation fits with the recent detection of H3K9me3 at poised enhancers (Zentner et al. 2011), and indicates that loss of this mark upon TRIM28 depletion may be sufficient to activate such regulatory elements, notably those located within IAPs and likely other ERVs. The derepression of cryptic enhancers within ERVs thus appears to be one prominent mechanism in the transcriptional deregulation triggered by Trim28 deletion in ES cells.
Activation of specific ERV-based enhancers upon loss of TRIM28 leads to activation of nearby genes
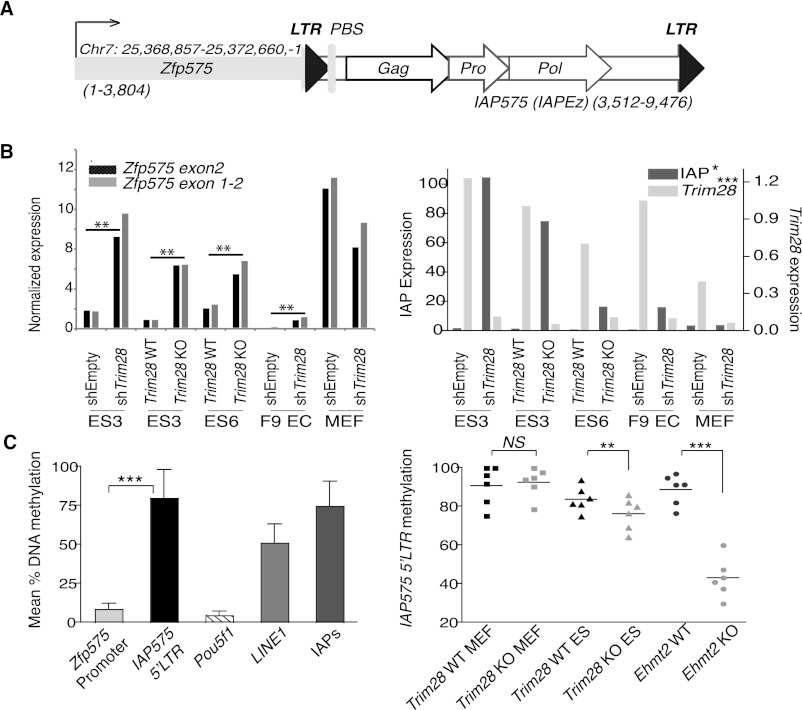
To explore the molecular mechanism of this process further, we examined transcription and chromatin state at specific ERV–Up gene pairs. We first focused on an element that was 90% identical to IAP sequences previously found to be TRIM28 regulated (Rowe et al. 2010) and named this ERV IAP575 because of its position 3′ to the bivalent gene Zfp575 (Mikkelsen et al. 2007; Bilodeau et al. 2009) in the sense orientation (Fig. 3A). Zfp575 was markedly up-regulated in TRIM28-depleted ES cells but not MEFs, consistent with our mRNA-seq data, paralleling the modulation of IAPs in these targets (Figs. 3B, 1A). Similar to its Pou5f1 counterpart, the Zfp575 promoter was unmethylated in ES cells. In contrast, the IAP575 LTR displayed high rates of CpG methylation, as did the IAP family as a whole, and to a lesser extent LINEs (Fig. 3C, left). The failure of DNA methylation to extend from the IAP575 LTR to the promoter of the adjacent Zfp575 gene fits with recent observations that (1) DNA methylation only spreads a few kilobases from TRIM28 binding sites (Quenneville et al. 2012; Rowe et al. 2013), and (2) ERV methylation rarely affects flanking regions (Rebollo et al. 2011). Interestingly, while methylation of the IAP575 LTR was unaltered by Trim28 deletion in MEFs, it significantly decreased in their ES cell counterparts, albeit not as dramatically as in ES cells deleted for Ehmt2 (G9a), a histone methyltransferase involved in the maintenance of DNA methylation (Fig. 3C, right; Dong et al. 2008; Tachibana et al. 2008). Perhaps explaining this latter difference, TRIM28 loss is lethal after a few days in ES cells (Rowe et al. 2010), while EHMT2-depleted cells can be stably maintained for many passages, allowing for extensive loss of cytosine methylation through multiple rounds of DNA replication. However, since this only modest decrease in DNA methylation was observed in parallel to the striking up-regulation of genes, it is possible that it contributes to this phenotype.
Figure 3.
Expression and cytosine methylation at the Zfp575 gene and adjacent IAP. (A) Map (drawn to scale) of the Zfp575 gene that just overlaps a full-length IAP (named IAP575 and of the IAPEz type) with both gene and IAP in the same orientation (sizes of each are stated). (LTR) Long terminal repeat; (PBS) primer binding site; (Gag) group-specific antigen; (Pro) protease; (Pol) polymerase. (B) TRIM28 knock-out and knockdown (comparing control, shEmpty and KD, shTRIM28) cell lines were assessed for their expression of Zfp575 (left panel) using two different primer sets, or TRIM28 or IAPs as controls (right panel). Unpaired t-tests were used to compare controls with TRIM28-depleted samples for all ES and EC cell lines: Zfp575, P = 0.0015; IAP, P = 0.0344; TRIM28, P = 0.0008. Since Zfp575 is normally expressed specifically in brain, we also verified it to be expressed in primary neurospheres and brain (data not shown). (C) Quantitative pyrosequencing was used to measure DNA methylation levels at the Zfp575 promoter versus the flanking 5′-LTR IAP575 promoter (left panel). Control primers were specific for the Pou5f1 promoter or global LINE1s or global IAP LTRs (IAPs). Bars represent means over multiple CpG positions with error bars showing the SD across all CpGs. (Right panel) Samples were compared (across six CpG positions) for their methylation levels at the IAP575 promoter. Primordial germ cells were also used to show that IAP575 is demethylated in germ cells to a level not much lower than in Trim28-deleted ES cells (e.g., to an average of 69% instead of 76%) (data not shown). Two-tailed paired t-tests display all significant differences: Trim28 WT versus KO ES, P = 0.0088; Ehmt2 WT versus KO, P = 0.0001.
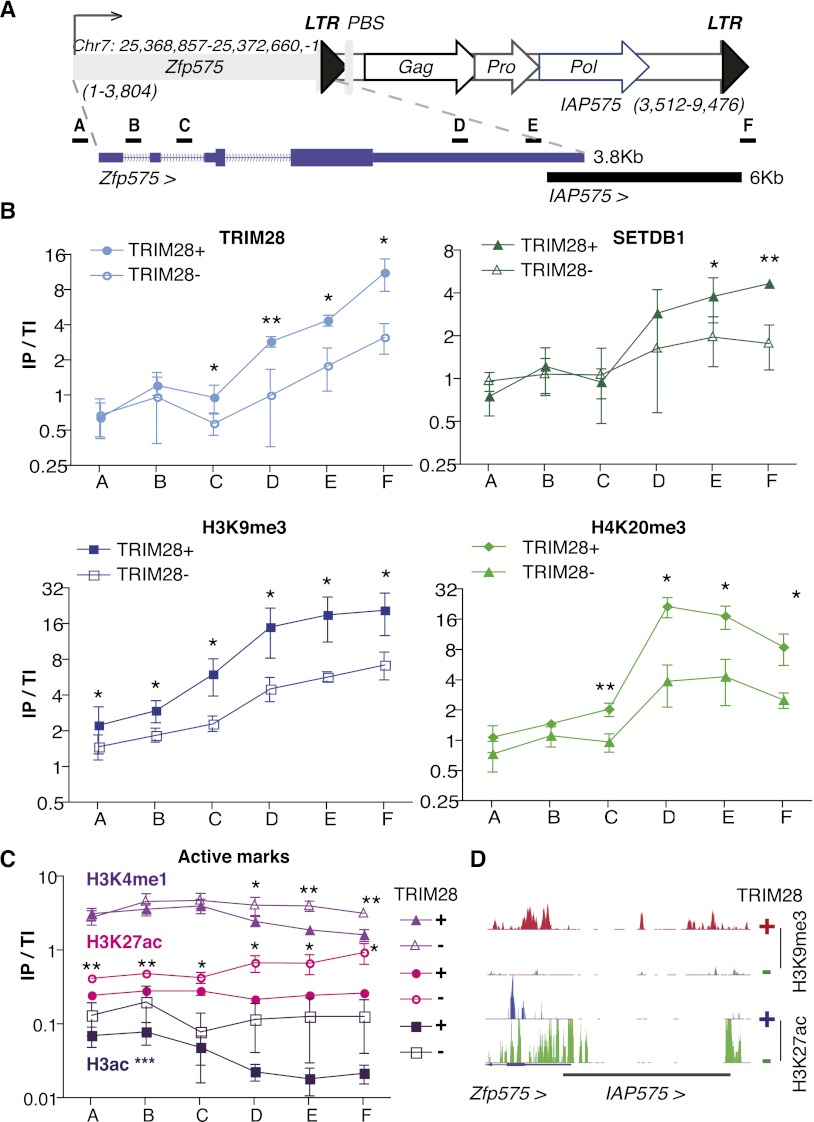
We then mapped histone marks across the Zfp575/IAP575 locus (Fig. 4). TRIM28, SETDB1, H3K9me3, and H4K20me3 were markedly enriched at IAP575, yet did not spread back to the zfp575 promoter. Upon Trim28 deletion, these repressive histone modifications collectively decreased, to be replaced by the active marks H3K4me1, H3K27ac, and H3Ac over the whole locus, albeit in the most pronounced fashion over its IAP575 part (Fig. 4B–D). We then further validated the up-regulation of several other ERV–Up gene pairs and verified that at these loci, TRIM28-dependent H3K9me3 is substituted by the active mark H3K27ac, as documented by ChIP-seq (Supplemental Figs. S4–S6), in support of our model.
Figure 4.
Zfp575 is regulated by a gain of active chromatin marks at its adjacent IAP575. (A) Map of Zfp575 and its adjacent IAP575 (for details, see Fig. 3A) with an enlargement shown underneath to show where primer pairs for ChIP are located. (B) ChIP results of repressive marks. (IP/TI) Immunoprecipitate values were normalized to their respective total inputs and to Gapdh. Bars represent the mean and SD of three to four ChIPs per antibody, and experiments were also reproduced in another ES cell line (Rex1) (data not shown). In each experiment, controls of no antibody were included giving no enrichments. Differences between WT and TRIM28-depleted samples were assessed for each primer set using paired t-tests with all significant differences given; (*) P ≤ 0.05, (**) P ≤ 0.01. (C) ChIPs this time on active marks were performed as described in B with data representing three to four ChIPs per antibody. Additionally, here the Pou5f1 enhancer was used as a positive control (data not shown) showing high enrichment for both H3K4me1 and H3K27ac but not for TRIM28 or H3K9me3. For H3K4me1 and H3K27ac, all significant differences are shown for each primer set, while for H3ac, WT samples were significantly different from TRIM28-depleted ones, not for individual points but over all primer sets; (***) P ≤ 0.001. (D) ChIP-seq maps of H3K9me3 and H3K27ac in TRIM28 WT and depleted ES cells (set to the same vertical scale) at the Zfp575-IAP575 locus. Note that reads within ERVs, especially conserved ones (in black), are usually missing due to the inability to map reads within highly repeated sequences. However, reads are present at the borders of these elements.
ERV sequences that escape TRIM28-mediated repression can act as activators during embryogenesis
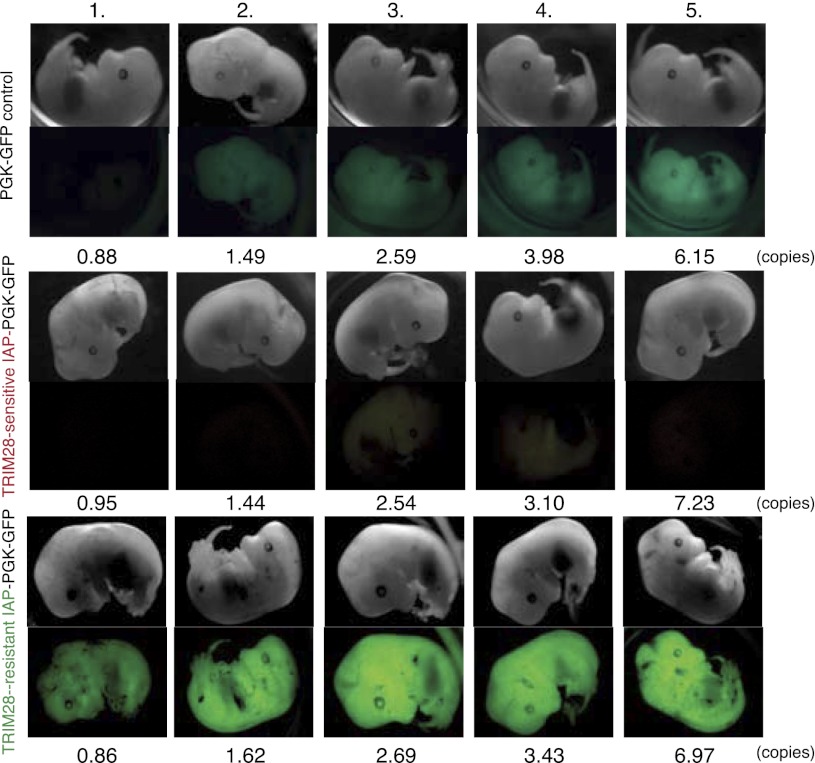
These results indicate that some ERVs carry intrinsic enhancer sequences that are silenced at the ES cell stage via TRIM28-induced repression. To probe this model further, we tested previously identified TRIM28-sensitive and TRIM28-resistant IAP sequences (Rowe et al. 2010) for their ability to modulate a nearby cellular promoter during embryonic development. To this end, we placed these elements in the antisense direction upstream of a phosphoglycerate kinase (PGK) promoter because at baseline this promoter drives only weak expression of GFP in embryos. We then used these lentiviral vectors for transgenesis via transduction of fertilized murine oocytes. Examination of the resulting embryos at E13 revealed that, while a TRIM28-sensitive IAP-derived sequence (IAP4) was able to limit expression from the PGK promoter contained in the lentiviral provirus, its TRIM28-resistant counterpart (IAP1, ∼87% identical) (see Rowe et al. 2010), in contrast, enhanced GFP expression (Fig. 5). Thus, TRIM28 susceptibility can condition the cis-acting transcriptional impact of specific ERV sequences in vivo during embryonic development.
Figure 5.
ERV sequences that escape TRIM28-mediated repression can act as activators during embryogenesis. Lentiviral transgenesis was performed with an empty PGK-GFP vector (PGK-GFP control, upper panels), or with the same vector including either an IAP4 (TRIM28-sensitive IAP-PGK-GFP, middle panels) or an IAP1 (TRIM28-resistant IAP-PGK-GFP, lower panels) sequence cloned antisense upstream of the PGK promoter. At E13, embryos were scored for GFP expression and vector copy numbers. For the PGK-GFP control, 13/29 embryos were green. For the TRIM28-sensitive IAP-PGK-GFP, 4/19 embryos were green (all with copy numbers above 16), and 4/19 pale green (including numbers 3 and 4 in this figure). For the TRIM28-resistant IAP-PGK-GFP, 12/17 embryos were green (including one with a copy number above 10), and 2/17 pale green (with copy numbers of 0.95 and 0.89). Embryos with similar copy numbers per vector group are shown in each column with increasing copy numbers by row. Vectors were injected twice with similar results. In one experiment, MEFs were derived from embryos to verify that microscopy differences were reproduced by flow cytometry (data not shown).
Discussion
The present work unveils a fundamental aspect of transcriptional regulation during the early embryogenesis of higher vertebrates. At the heart of this system lies, on one side, retroelements that have colonized eukaryotic genomes from the earliest times, and on the other side, the tetrapod-specific KRAB-ZFP gene family (Urrutia 2003; Huntley et al. 2006; Emerson and Thomas 2009; Wolf and Goff 2009; Thomas and Schneider 2011), which acts as the targeting machinery for TRIM28. We previously demonstrated that TRIM28 is responsible for the silencing of ERVs in ES cells and early embryos (Rowe et al. 2010). Here, we reveal that an important role of this process is to protect the transcriptional dynamics of early embryos from perturbation by cis-acting activators contained in these mobile elements.
For this, we deleted Trim28 in ES cells and monitored chromatin signatures at deregulated genes and ERVs. We found that half of the ∼5700 transcriptional units up-regulated upon Trim28 deletion in ES cells bore, at baseline, the bivalent histone marks H3K4me3 and H3K27me3 characteristic of genes poised for transcription (Bernstein et al. 2006). Moreover, we noted that, remarkably, these genes were on average located closer to ERVs than genes down-regulated or unaffected following TRIM28 removal. We then further observed that, while in wild-type ES cells, ERVs bound TRIM28 and SETDB1 and accordingly were enriched in H3K9me3 and H4K20me3, they lost these repressive marks upon Trim28 deletion and instead acquired chromatin modifications typically associated with active enhancers such as H3K4me1 and H3K27ac, a phenomenon that was documented both at global IAPs and at the level of specific ERV-up-regulated gene loci. Finally, we could demonstrate that ERV-derived sequences could either repress or activate an adjacent cellular promoter in transgenic mouse embryos, depending on whether they were recognized or not by a TRIM28-containing complex in ES cells.
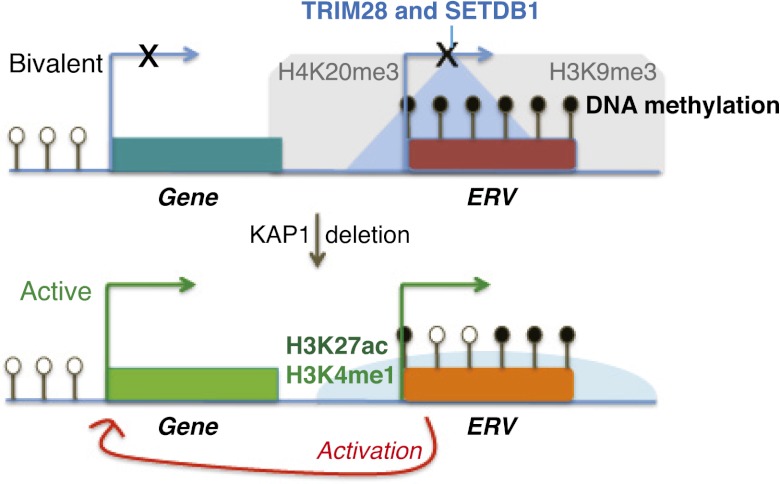
The model emerging from our study (Fig. 6) is one whereby, in ES cells, the recruitment of TRIM28 and its partners, including SETDB1, at ERV-contained enhancers leads to the maintenance of H3K9me3, H4K20me3, and DNA methylation, which prevents the untimely activation of nearby genes, in particular, those harboring bivalent promoters. Indeed, DNA methylation is known to anti-correlate with active marks (Okitsu and Hsieh 2007; Ooi et al. 2007; Weber et al. 2007; Stadler et al. 2011), and SETDB1 has previously been shown to maintain H3K9 trimethylation and, secondarily, the Suv420H1/2-mediated mark H4K20me3 at ERVs (Matsui et al. 2010). Inactivation of this machinery leads not only to the loss of silent histone marks and to a mild decrease in cytosine methylation but also to the acquisition of active enhancer marks at these loci, which tilts nearby genes, notably those poised for transcription, toward expression. Noteworthy, the NuRD complex, also recruited by TRIM28, is known to mediate deacetylation of H3K27 through its HDAC1 and HDAC2 subunits (Reynolds et al. 2011), which would explain the genome-wide anti-correlation observed between H3K27ac and TRIM28 target sites at baseline. Likewise, LSD1, which shares at least some targets with TRIM28 and NuRD (Macfarlan et al. 2011, 2012), is able to demethylate and therefore decommission the active mark H3K4me1 (Whyte et al. 2012). Accordingly, disruption of either SETDB1 or LSD1 leads to effects on cellular transcripts (Bilodeau et al. 2009; Yuan et al. 2009; Karimi et al. 2011; Macfarlan et al. 2011, 2012). In the case of SETDB1 deletion, this includes the induction of chimeric transcripts initiating from derepressed ERVs, which we also see evidence for here, since some of the same transcripts are induced (Karimi et al. 2011; this study). Here we demonstrate that in the absence of TRIM28, retrotransposon-based enhancers become active.
Figure 6.
Summary model: Substitution of TRIM28-dependent repressive chromatin by the active marks H3K4me1 and H3K27ac at specific ERV-Up gene pairs parallels activation of gene expression.
The heterogeneity of the TRIM28-recruiting ERV loci uncovered here, with sequences intrinsic to IAP, MERVL, and ERVK families, suggests that a large number of different KRAB-ZFPs engage in directing TRIM28 to ERVs in ES cells. Additionally, TRIM28 can also interact with KRAB-O proteins that lack zinc fingers but bridge DNA through other factors such as SRY (Peng et al. 2009). Remarkably, TRIM28 and some KRAB-ZFPs are also detected in adult tissues, albeit along exquisitely cell- and stage-specific fashions, where they have become coopted to influence tissue-specific gene regulation (Jakobsson et al. 2008; Bojkowska et al. 2012; Chikuma et al. 2012; Krebs et al. 2012; Santoni de Sio et al. 2012a,b). Whether some ERV-derived enhancers serve as docking sites for this repressor system in these adult tissues warrants exploration. There is evidence that some ERV sequences function as authentic regulators, including enhancers, in certain cells, not only during development but also in adult tissues (Pi et al. 2004; Bourque et al. 2008; Kunarso et al. 2010; Teng et al. 2011; Mey et al. 2012; Schmidt et al. 2012). Our data indicate that these rare coopted elements represent only exceptions within a large group, most members of which are repressed through TRIM28. This may explain why most KRAB-ZFP genes are expressed in both mouse and human ES cells, while at least in this latter species, most if not all endogenous retroviruses have accumulated mutations that would anyway preclude their retrotransposition. The need to preserve the transcription dynamics of ES cells, rather than to protect the genome from further spread of these elements, is likely what constitutes the strongest selective pressure on the KRAB/TRIM28 system in higher species.
Methods
Lentiviral vectors
For in vivo experiments, the transfer vector pRRLSIN.cPPT.PGK-GFP.WPRE (available from Addgene) was used with either IAP1 or IAP4 sequences (Rowe et al. 2010) included upstream of the PGK (phosphoglycerate kinase-1) promoter in the antisense orientation (Rowe et al. 2013). For TRIM28 knockdown experiments, shRNA lentiviral plasmids (against mouse Trim28 or the empty vector control) were ordered from Sigma-Aldrich (pLKO.1-puro). All vectors were produced by transient transfection of 293T cells with the transfer vector, packaging, and VSVG envelope plasmids (Barde et al. 2010) and titrated on 3T3 fibroblasts.
Cell culture
ES cells were cultured in standard conditions as described (Rowe et al. 2013). The ES cell lines used were two Trim28loxP/loxP lines called ES3 and ES6 and their derived Trim28-conditional knock-out cell lines that are transduced with a tamoxifen (4-0HT)–inducible Cre vector (Rowe et al. 2010). For analysis of expression and chromatin marks, knock-out cells were collected 4 d after treatment with 4-0HT (used overnight at 1 μM, Sigma-Aldrich: H7904) due to the lethality of Trim28 knock-out for longer time periods. Rex1GFP ES cells (Wray et al. 2011) were additionally used where stated (kind gift from A.G. Smith, University of Cambridge, UK) or Ehmt2 parental or stable knock-out ES cells (Dong et al. 2008; Tachibana et al. 2008) (a kind gift from Yoichi Shinkai, RIKEN Institute, Japan). TRIM28-knockdown was induced with shRNA vectors (see above), and cells selected with puromycin 2 d post-transduction and collected 4 d post-puromycin selection, a time point giving similar expression changes to 4 d post-knock-out. Knockdown efficiency was verified by qRT-PCR. TRIM28loxP/loxP 4-0HT-inducible MEFs were used to delete Trim28, while TRIM28 knockdowns were also performed in MEFs and F9 EC cells where stated.
Flow cytometry
Vector titers and GFP repression were measured by FACS, as well as the differentiation status of ES cells as monitored by staining with an SSEA-1 PE- conjugated antibody or isotype control (BD Pharmingen: 560142 and 555584).
RNA extraction and quantification
Total RNA was extracted with TRIzol (Invitrogen: 15596-018), purified using a PureLink RNA kit (Ambion: 12183018A), treated with DNase (Ambion: AM1907) and 500 ng reverse-transcribed using random primers and SuperScript II (Invitrogen: 18064-022). Primers (see Supplemental Table 4) were designed for an Applied Biosystems 7900HT machine using Primer Express (Applied Biosystems) and used for SYBR Green qPCR. Primer specificity was confirmed by dissociation curves and samples were normalized to Gapdh, although Actin gave similar results.
mRNA sequencing
Total RNA (10 μg) from TRIM28 WT and KO ES cells and MEFs was subject to mRNA selection, fragmentation, cDNA synthesis, and library preparation for Illumina high-throughput sequencing, after checking RNA quality on a Bioanalyzer. Single read sequencing was performed on a Genome Analyzer IIx machine with 40 cycles generating ∼33 million reads per sample. Additionally, mRNA sequencing was performed on Trim28 control (shEmpty) and knockdown (shTRIM28) Rex1 ES cells with 50 cycles on an Illumina HiSeq 2000 machine generating around 200 million reads per sample and confirming our knock-out ES cell results.
Chromatin immunoprecipitation (ChIP)
ES cell samples were washed twice (in PBS + 2% FCS), counted to normalize by cell number, cross-linked (10 min rotation in 1% formaldehyde), quenched with glycine (at 125 mM on ice), washed three times (PBS), and pelleted at 107 cells per Eppendorf. Pellets were lysed, resuspended in 1 mL of sonication buffer on ice (10 mM Tris at pH 8, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% NaDOC, 0.25% NLS, and protease inhibitors), transferred to glass 12 × 24-mm tubes (Covaris: 520056), and sonicated (Covaris settings: 20% duty cycle, intensity 5, 200 cyles/ burst, 30 min). Sonication was then assessed by reverse cross-linking overnight in the presence of proteinase K and RNase, followed by DNA extraction and quantification on a Bioanalyzer (Agilent 2100 machine). Fragment sizes were equivalent between wild-type and knock-out samples, which were done in parallel (with mean fragment sizes of ∼200 bp for Experiment 1 and ∼400 bp for Experiments 2 and 3). Samples were also checked for the absence of single-stranded DNA by Exonuclease I treatment. Immunoprecipitations were performed in duplicates or triplicates with Dynabeads (100.03D) using 1 × 106 to 2 × 106 cells, 80 μL of pre-blocked beads, and 5 μg of antibody (or no antibody as a control) per sample in IP buffer (167 mM NaCl, 16.7 mM Tris at pH 8.1, 1.2 mM EDTA, 0.5 mM EGTA, 1.1% Triton X-100, and protease inhibitors) overnight. After washing and reverse cross-linking (also overnight) and DNA extraction, results were quantified by SYBR Green qPCR (for primers, see Supplemental Table 4). The antibodies used were TRIM28 (Tronolab, rabbit polyclonal SY 3267-68, 30–50 μL per sample), H3K9me3 (Abcam: ab8898), SETDB1 (Santa Cruz, 50 μL per sample), H4K20me3 (Millipore: 07-463), H3ac (Millipore: 06-599), H3K27ac (Abcam: ab4729), and H3K4me1 (Abcam: ab8895).
ChIP sequencing
Total input (TI) and corresponding immunoprecipitated (IP) ChIP libraries were prepared using 10 ng of material with gel selection of 200-bp- to 300-bp-sized fragments. Libraries were ligated with Illumina adaptors and paired-end sequenced (or single-end for H3K27ac) on an Illumina HiSeq 2000 machine with 50–100 cycles and two samples multiplexed in one lane, generating ∼100 million sequences per sample. TI samples gave background enrichment patterns distinct from IPs.
Quantitative bisulfite pyrosequencing
Genomic DNA was converted (200 ng/sample) and used for PCR and pyrosequencing as previously described (Rowe et al. 2013). We thank A. Reymond (CIG, UNIL, Lausanne) for kind use of the pyrosequencer. Results were analyzed using Pyro Q-CpG Software.
Lentiviral transgenesis
Lentiviral vectors for transgenesis were prepared using Episerf medium (Invitrogen: 10732022), the particle concentration obtained by p24 ELISA (PerkinElmer: NEK050B001KT), and the infectious titer determined on HCT116 cells by GFP flow cytometry. Ratios for the three vectors were between 1/319 and 1/428 of infectious to physical particles with titers between 2 and 2.4 × 109 infectious units/mL. Transgenesis was performed by perivitelline injection of vectors into fertilized oocytes that were transferred to foster mothers (strain B6D2F1/J) and then recovered at embryonic day 13 (E13). Photographs were taken using the same saturation, gain, and exposure settings and image settings for all embryos.
Bioinformatics analyses and statistics
mRNA-seq analysis
Reads were mapped to the mouse genome mm9 using the short read aligner program Bowtie (Langmead et al. 2009) with reads (three mismatches allowed) excluded that mapped more than five times. The SAMtools and bedtools suites (Li et al. 2009; Quinlan and Hall 2010) were used to generate files to be visualized on the UCSC Genome Browser (http://genome.ucsc.edu/) (Kent et al. 2002).
MA plots
MA plots were generated from rpkm values (number of reads normalized by gene length and total reads) using the maplot Python package (https://github.com/delafont/maplot).
Boxplots
Boxplots showing bootstrapped values (generated using R: http://www.R-project.org) were used in gene-centric analyses to determine if up-regulated (Up) genes were closer to the indicated histone marks/ERVs compared with two control gene groups (down-regulated, “Down” or unaffected, “Stable” genes). Statistical significance was calculated using the Wilcoxon rank-sum test.
H3K9me3 ChIP-seq analysis
Paired-end reads were mapped to the mouse genome (three mismatches allowed) mm9 using the short read aligner program Bowtie (Langmead et al. 2009). Several analyses were performed, showing the same global results where reads were either excluded if mapping more than one time, five times, or 20 times to the genome. Peaks were called from the data where reads were mapped with a cutoff of 20 to allow more coverage of repeats, although individual peaks of interest were validated using the analysis where a cutoff of one was used (in this case, only exact matches were allowed). Enriched regions were defined using the ChIP-Part analysis module from the ChIP-seq analysis suite (http://ccg.vital-it.ch/chipseq/). H3K27ac ChIP-seq data were confirmed to correlate (by 53%) with previous H3K27ac ChIP-seq in ES cells (Creyghton et al. 2010) and verified to be normally present at active genes and gained at specific ERV loci (see Supplemental Figs. S5, S6). TRIM28 ChIP-seq peaks were defined using MACS (default threshold P-value <1 × 10−5) and normalized to the total input generating 3099 peaks. Direct binding sites to promoters of up-regulated genes were identified using a cutoff of ±2 kb from the TSS giving 49 genes, 13 of which were excluded due to the binding being through an ERV.
Public ChIP-seq data
Raw or already mapped reads were downloaded from publicly available ChIP-seq data (GEO IDs: GSE12241, GSE18371, and GSE24165) and peaks called using MACS. ChIP-correlation analyses were performed with bed files, using the online tool ChIP-Cor (http://ccg.vital-it.ch/chipseq/chip_cor.php). Histograms were analyzed using raw counts and count densities, and those showing a correlation were displayed after global normalization, where ChIP-seq counts are normalized by the total number of counts and the window width to allow visualization of multiple data sets on the same plot.
Motif identification
The MotifRegressor and motifsComparator softwares were used to identify DNA sequence binding motifs (Conlon et al. 2003; Carat et al. 2010).
Other statistical analyses
GraphPad Prism version 4.00 (http://www.graphpad.com) was used for other statistical analyses, where control and knock-out groups were compared with paired or unpaired t-tests (as noted) that were one-tailed except where stated as two-tailed.
Data access
All next-generation sequencing data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and are accessible with the accession no. GSE41903.
Acknowledgments
We thank P.V. Maillard and J. Marquis for advice, S. Verp and S. Offner for technical assistance through the EPFL Lentiviral Transgenesis platform, B. Khubieh and J. Rougemont for bioinformatics help (EPFL Biostatistics and Bioinformatics core facility), Y. Shinkai and D. Schübeler for the Ehmt2 knock-out and parental ES cells, and K. Harshman and A. Reymond (Center for Integrative Genomics, University of Lausanne) for high-throughput sequencing or use of a pyrosequencer, respectively. All computing for high-throughput sequencing was done on the Vital-it cluster. This work was supported through grants from the Swiss National Science Foundation and the European Research Council to D.T. and an NIH grant to S.L.P.
Author contributions: H.M.R. conceived the study, designed and performed the experiments, analyzed the data, and wrote the manuscript. A.K. performed bioinformatics analyses. A.C., L.F., T.S.M., and Y.T. performed experiments. J.J., S.V., and S.L.P. designed experiments. D.T. conceived the study, designed experiments, and wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.147678.112.
References
- Anders S, Huber W 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barde I, Salmon P, Trono D 2010. Production and titration of lentiviral vectors. Curr Protoc Neurosci 53: 4.21.1–4.21.23 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA 2009. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev 23: 2484–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkowska K, Aloisio F, Cassano M, Kapopoulou A, de Sio FS, Zangger N, Offner S, Cartoni C, Thomas C, Quenneville S, et al. 2012. Liver-specific ablation of KRAB associated protein 1 in mice leads to male-predominant hepatosteatosis and development of liver adenoma. Hepatology 56: 1279–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. 2008. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18: 1752–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas F, Mark M, Dolle P, Dierich A, Chambon P, Losson R 2000. Mice lacking the transcriptional corepressor TIF1β are defective in early postimplantation development. Development 127: 2955–2963 [DOI] [PubMed] [Google Scholar]
- Carat S, Houlgatte R, Bourdon J 2010. A parallel scheme for comparing transcription factor binding sites matrices. J Bioinform Comput Biol 8: 485–502 [DOI] [PubMed] [Google Scholar]
- Chikuma S, Suita N, Okazaki IM, Shibayama S, Honjo T 2012. TRIM28 prevents autoinflammatory T cell development in vivo. Nat Immunol 13: 596–603 [DOI] [PubMed] [Google Scholar]
- Conlon EM, Liu XS, Lieb JD, Liu JS 2003. Integrating regulatory motif discovery and genome-wide expression analysis. Proc Natl Acad Sci 100: 3339–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schubeler D, et al. 2008. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J 27: 2691–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS 1994. Neomorphic agouti mutations in obese yellow mice. Nat Genet 8: 59–65 [DOI] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH 2009. Adaptive evolution in zinc finger transcription factors. PLoS Genet 5: e1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134: 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ III 1996. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev 10: 2067–2078 [DOI] [PubMed] [Google Scholar]
- Frietze S, O'Geen H, Blahnik KR, Jin VX, Farnham PJ 2010. ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS ONE 5: e15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Q, Yoshida T, McDonald OG, Owens GK 2007. Concise review: Epigenetic mechanisms contribute to pluripotency and cell lineage determination of embryonic stem cells. Stem Cells 25: 2–9 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Young RA 2010. Transcription. Repressive transcription. Science 329: 150–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ 2009. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 23: 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, Gordon L, Branscomb E, Stubbs L 2006. A comprehensive catalog of human KRAB-associated zinc finger genes: Insights into the evolutionary history of a large family of transcriptional repressors. Genome Res 16: 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. 2007. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell 28: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson J, Cordero MI, Bisaz R, Groner AC, Busskamp V, Bensadoun JC, Cammas F, Losson R, Mansuy IM, Sandi C, et al. 2008. KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron 60: 818–831 [DOI] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The Human Genome Browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs CJ, Schultz DC, Robins DM 2012. The KRAB zinc finger protein RSL1 regulates sex- and tissue-specific promoter methylation and dynamic hormone-responsive chromatin configuration. Mol Cell Biol 32: 3732–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G 2010. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner MS, Begg GE, Speicher DW, Rauscher FJ III 2000. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: Direct chromoshadow domain–KAP-1 corepressor interaction is essential. Mol Cell Biol 20: 6449–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC 2008. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 15: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, et al. 2011. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev 25: 594–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- McDonel P, Costello I, Hendrich B 2009. Keeping things quiet: Roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int J Biochem Cell Biol 41: 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB 2012. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335: 1499–1502 [DOI] [PubMed] [Google Scholar]
- Mey A, Acloque H, Lerat E, Gounel S, Tribollet V, Blanc S, Curton D, Birot AM, Nieto MA, Samarut J 2012. The endogenous retrovirus ENS-1 provides active binding sites for transcription factors in embryonic stem cells that specify extra embryonic tissue. Retrovirology 9: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud EJ, van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP 1994. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev 8: 1463–1472 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okitsu CY, Hsieh CL 2007. DNA methylation dictates histone H3K4 methylation. Mol Cell Biol 27: 2746–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Ivanov AV, Oh HJ, Lau YF, Rauscher FJ III 2009. Epigenetic gene silencing by the SRY protein is mediated by a KRAB-O protein that recruits the KAP1 co-repressor machinery. J Biol Chem 284: 35670–35680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi W, Yang Z, Wang J, Ruan L, Yu X, Ling J, Krantz S, Isales C, Conway SJ, Lin S, et al. 2004. The LTR enhancer of ERV-9 human endogenous retrovirus is active in oocytes and progenitor cells in transgenic zebrafish and humans. Proc Natl Acad Sci 101: 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. 2011. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell 44: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D 2012. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Rep 2: 766–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2010. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Karimi MM, Bilenky M, Gagnier L, Miceli-Royer K, Zhang Y, Goyal P, Keane TM, Jones S, Hirst M, et al. 2011. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet 7: e1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, Behrens A, Bertone P, Hendrich B 2011. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J 31: 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Trono D 2011. Dynamic control of endogenous retroviruses during development. Virology 411: 273–287 [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240 [DOI] [PubMed] [Google Scholar]
- Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D 2013. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 140: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni de Sio F, Barde I, Offner S, Kapopoulou A, Corsinotti A, Bojkowska K, Genolet R, Thomas JH, Luescher IF, Pinschewer D, et al. 2012a. KAP1 regulates gene networks controlling T-cell development and responsiveness. FASEB J 26: 4561–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni de Sio FR, Massacand J, Barde I, Offner S, Corsinotti A, Kapopoulou A, Bojkowska K, Dagklis A, Fernandez M, Ghia P, et al. 2012b. KAP1 regulates gene networks controlling mouse B lymphoid cell differentiation and function. Blood 119: 4675–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT 2012. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ III 2001. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: The PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev 15: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ III 2002. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Kurisaki A, Watanabe-Susaki K, Nakajima Y, Nakanishi M, Arai Y, Shiota K, Sugino H, Asashima M 2010. TIF1β regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proc Natl Acad Sci 107: 10926–10931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy SP, Stevens J, Schultz DC 2006. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol 26: 8623–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y 2008. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J 27: 2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L, Firpi HA, Tan K 2011. Enhancers in embryonic stem cells are enriched for transposable elements and genetic variations associated with cancers. Nucleic Acids Res 39: 7371–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Schneider S 2011. Coevolution of retroelements and tandem zinc finger genes. Genome Res 21: 1800–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia R 2003. KRAB-containing zinc-finger repressor proteins. Genome Biol 4: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA 2012. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482: 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131: 46–57 [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A 2011. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol 13: 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B, Ng HH 2009. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev 23: 2507–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC 2011. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21: 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Sheng J, Lau HT, McDonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, et al. 2012. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J Biol Chem 287: 2107–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]