Abstract
The nematode Caenorhabditis elegans contains each of the broad classes of eukaryotic small RNAs, including microRNAs (miRNAs), endogenous small-interfering RNAs (endo-siRNAs), and piwi-interacting RNAs (piRNAs). To better understand the evolution of these regulatory RNAs, we deep-sequenced small RNAs from C. elegans and three closely related nematodes: C. briggsae, C. remanei, and C. brenneri. The results reveal a fluid landscape of small RNA pathways with essentially no conservation of individual sequences aside from a subset of miRNAs. We identified 54 miRNA families that are conserved in each of the four species, as well as numerous miRNAs that are species-specific or shared between only two or three species. Despite a lack of conservation of individual piRNAs and siRNAs, many of the features of each pathway are conserved between the different species. We show that the genomic distribution of 26G siRNAs and the tendency for piRNAs to cluster is conserved between C. briggsae and C. elegans. We also show that, in each species, 26G siRNAs trigger stage-specific secondary siRNA formation. piRNAs in each species also trigger secondary siRNA formation from targets containing up to three mismatches. Finally, we show that the production of male- and female-specific piRNAs is conserved in all four species, suggesting distinct roles for piRNAs in male and female germlines.
Small noncoding RNAs, including microRNAs (miRNAs), piwi-interacting RNAs (piRNAs), and endogenous small-interfering RNAs (endo-siRNAs), regulate developmental and defense pathways in animals. Each class of small RNAs has unique roles and genetic requirements but invariably binds to Argonaute proteins to form effector complexes that target nucleic acids containing partial or complete complementarity to the small RNA guide. Caenorhabditis elegans contains 25 Argonautes belonging to three different clades and multiple distinct subfamilies (Yigit et al. 2006).
miRNAs are ∼22 nt and repress gene expression through mRNA decay and translational repression (Bartel 2004). miRNAs are typically processed from long primary transcripts through the sequential activities of the ribonucleases Drosha and Dicer. Mature miRNAs associate with the AGO clade Argonautes ALG-1 and ALG-2 in C. elegans (Grishok et al. 2001). Many miRNAs are conserved between C. elegans and humans, including miR-1 and let-7, which share 100% sequence identity across species (Pasquinelli et al. 2000; Lee and Ambros 2001).
C. elegans piRNAs (also called 21U-RNAs) are 21 nt long and contain a uracil at their 5′ termini (Ruby et al. 2006; Batista et al. 2008; Das et al. 2008). piRNAs bind to the two PIWI-clade Argonautes PRG-1 and PRG-2. Mutations in prg-1 result in defects in the production and/or functionality of germ cells and reduced fertility at elevated temperatures (Batista et al. 2008; Wang and Reinke 2008). piRNAs are thought to interact through imperfect complementarity with target mRNAs in the germline to trigger secondary siRNA production (Bagijn et al. 2012; Lee et al. 2012; Shirayama et al. 2012)
The majority of C. elegans siRNAs are either 22 or 26 nt long and start with a 5′ guanine and are thus referred to as 22G-RNAs (22G siRNAs) or 26G-RNAs (26G siRNAs), respectively. The 22G siRNAs are produced by the RNA-dependent RNA polymerases (RdRPs) RRF-1 and EGO-1 and bind to the WAGO clade Argonautes WAGO-1-12 (WAGO class siRNAs) to silence certain protein-coding genes, transposons, pseudogenes, and cryptic loci (Gu et al. 2009). A subset of 22G siRNAs produced by EGO-1 associate with the Argonaute CSR-1 (CSR-1 class siRNAs) to guide chromosome segregation (Claycomb et al. 2009; van Wolfswinkel et al. 2009). It was proposed that CSR-1 class siRNAs may also provide a memory of self to protect endogenous genes from being routed into the piRNA and WAGO class siRNA pathways (Lee et al. 2012; Shirayama et al. 2012).
The 26G siRNAs fall into two classes: a spermatogenesis-enriched class which associates with the AGO clade Argonautes, ALG-3 and ALG-4 (Han et al. 2009; Conine et al. 2010); and an oocyte- and embryo-enriched class which associates with the divergent PIWI-clade Argonaute, ERGO-1 (Han et al. 2009; Vasale et al. 2010; Fischer et al. 2011). Both classes of 26G siRNAs are produced by the RdRP RRF-3 and are thought to trigger secondary 22G siRNA production. However, the majority of 22G siRNAs are produced independently of a 26G siRNA trigger.
Many miRNAs are highly conserved in Caenorhabditis species (de Wit et al. 2009). In contrast, individual piRNA sequences are entirely nonconserved between C. elegans and C. briggsae (Ruby et al. 2006; de Wit et al. 2009). It is unclear if other features of the piRNA pathway are conserved among nematode species. It is also unknown whether or not the various classes of endogenous siRNAs are conserved in Caenorhabditis species. Here, we sequenced small RNAs from C. elegans, C. briggsae, C. remanei, and C. brenneri, and using a comparative genomics approach, we examined the conservation and evolution of each of the small RNA pathways. We identified hundreds of new miRNAs in C. briggsae, C. remanei, and C. brenneri, as well as thousands of piRNAs and each of the classes of endogenous siRNAs present in C. elegans. Although there is essentially no conservation of individual small RNAs among any of the four species aside from miRNAs and a very limited number of CSR-1 class siRNAs, many features such as their genomic distribution, expression patterns, and tendency for 26G siRNAs and piRNAs to trigger secondary 22G siRNA production are conserved. We also identified a new subfamily of RdRPs absent in C. elegans but conserved in each of the other three species. Finally, we discovered that production of distinct sex-specific populations of piRNAs is conserved across nematode species, suggesting different roles for piRNAs in male and female germlines.
Results
Expansion and diversification of Argonautes and RNA-dependent RNA polymerases
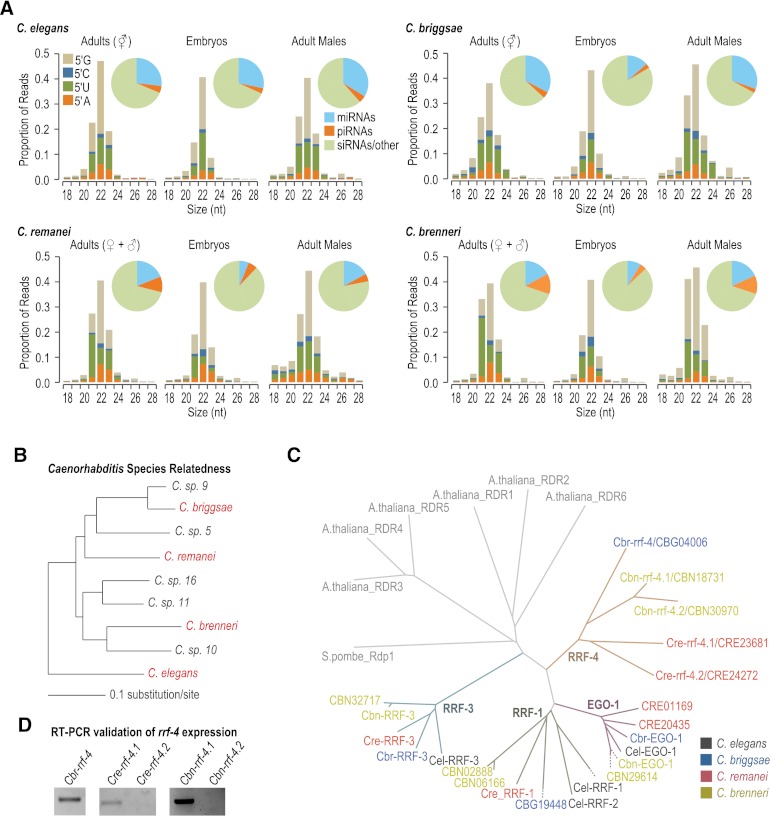
To examine small RNA evolution in nematodes, we subjected small RNAs from C. elegans and three other nematode species, C. briggsae, C. remanei, and C. brenneri, to high-throughput sequencing (Fig. 1A; Supplemental Table S1). Each of the four species is morphologically similar; however, their genomic sequences are highly divergent, with common ancestry ∼110 million generations ago (Fig. 1B; Cutter et al. 2009). We constructed 18- to 28-nt small RNA libraries from synchronized populations of young gravid adult hermaphrodites for the androdioecious (male/hermaphrodite) species C. elegans and C. briggsae and from mixed populations of adult males and females for the gonochoristic (male/female) species C. remanei and C. brenneri. For each species, we also sequenced small RNAs from embryos and young adult males. In each library ∼80%–90% of small RNAs were 21–23 nt and contained either a 5′ U or 5′ G, features indicative of piRNAs, miRNAs, and 22G siRNAs (Fig. 1A). Each of the libraries, but particularly male and embryo libraries, also had a small fraction of 26-nt 5′ G-containing small RNAs (26G siRNAs) (Fig. 1A).
Figure 1.
Identification of small RNAs and associated proteins in Caenorhabditis species. (A) Bar graphs show the size and 5′ first nucleotide distributions of small RNAs. (Insets) Pie charts display the proportion of reads belonging to different classes of small RNAs: miRNAs, piRNAs, and siRNAs, or unclassified. (B) Phylogenetic relationship of the elegans group of Caenorhabditis, adapted from Kiontke et al. (2011). C. briggsae, C. remanei, C. brenneri, and C. elegans (highlighted in red), having had their genomes sequenced and partially annotated, were chosen for this study. (C) Phylogeny of RNA-dependent RNA polymerases. Genes from different species are color-coded, and different subfamilies are indicated by colored branches. (Cel) C. elegans, (Cbr) C. briggsae, (Cre) C. remanei, (Cbn) C. brenneri. (D) RT-PCR of rrf-4 from mixed-stage C. briggsae, C. remanei, and C. brenneri.
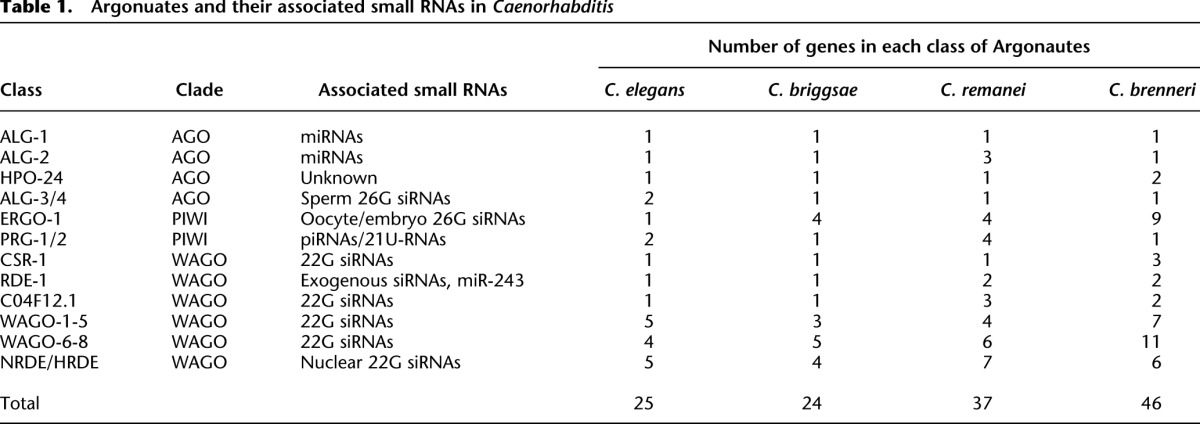
In C. elegans, small RNAs can be classified by their genetic requirements, particularly their Argonaute binding partners, but for siRNAs, the RNA-directed RNA polymerase that produces them is also indicative of their function. To determine the potential for each class of C. elegans small RNAs to be present in other nematodes and, thus, in our small RNA libraries, we identified all Argonautes and RdRPs in C. elegans, C. briggsae, C. remanei, and C. brenneri and examined their phylogenetic relatedness. For each of the 25 Argonaute proteins in C. elegans, we identified at least one, but often multiple, likely orthologs in each of the other three nematode species, suggesting that a similar repertoire of small RNA pathways exists in each species (Table 1; Supplemental Fig. S1).
Table 1.
Argonuates and their associated small RNAs in Caenorhabditis
There are four RdRPs in C. elegans: RRF-3 functions in the production of 26G siRNAs (Gent et al. 2010), EGO-1 and RRF-1 have partially overlapping roles in 22G siRNA formation (Gu et al. 2009), and RRF-2, which is closely related to RRF-1, has an unknown role (Sijen et al. 2001). From a phylogenetic analysis of nematode RdRPs, we identified likely orthologs of the three RdRP subfamilies, RRF-1, RRF-3, and EGO-1 in each of the other three nematode species (Fig. 1C). We also identified a fourth subfamily of RdRPs, RRF-4, in each of the other three nematode species but not in C. elegans (Fig. 1C). By RT-PCR assays from mixed-stage animals, we could readily detect expression of Cbr-rrf-4/CBG04006, Cre-rrf-4.1/CRE23681, and Cbn-rrf-4.1/CBN18731, but not Cre-rrf-4.2/CRE24272 or Cbn-rrf-4.2/CBN30970 (Fig. 1D), suggesting that at least one gene in the rrf-4 family is expressed in each species except C. elegans.
The majority of Caenorhabditis miRNA families within each species are conserved
miRNAs in C. elegans are well-characterized (Lau et al. 2001; Lee and Ambros 2001; Grad et al. 2003; Lim et al. 2003; Ruby et al. 2006; Kato et al. 2009; Gerstein et al. 2010); however, miRNAs in C. briggsae and C. remanei have been only partially characterized (de Wit et al. 2009), and miRNAs in C. brenneri have not been examined. To identify miRNAs in each of the four species, we used the miRDeep2 program (Mackowiak 2011; Friedlander et al. 2012). To conclusively call a miRNA, we required a candidate predicted by miRDeep2 to also be predicted by MIReNA (Mathelier and Carbone 2010) and/or contain a seed sequence (positions 2–7) conserved between Caenorhabditis species. As validation of the approach, we identified the majority of annotated Caenorhabditis miRNAs from our deep sequencing libraries (Supplemental Table S2). We identified 37 new miRNAs in C. briggsae, 48 new miRNAs in C. remanei, and 215 new miRNAs in C. brenneri (Supplemental Table S3) but did not identify any new miRNAs in C. elegans. We grouped all known and newly identified miRNAs into families, based on seed sequence. To date, there are 106 miRNA families annotated in C. elegans, 84 in C. briggsae, 85 in C. remanei, and 87 in C. brenneri, which constitute 176 distinct miRNA families (Fig. 2A,B; Supplemental Table S4). Fifty-four miRNA families are conserved in all four species, 61 families are present in three or more species, and most miRNA families within each species have homologs in at least two other species (Fig. 2C). Few (<5%) miRNA families are conserved between only two or three nematode species (Fig. 2C). However, in each species, at least 20% of miRNA families are unique, suggesting that miRNAs are born at relatively high rates (Fig. 2C).
Figure 2.
Conservation of miRNAs in nematodes. (A) Table of conserved miRNAs classified by family. Seed sequences are positions 2–7, relative to the 5′ end of the miRNA. The number in each row represents the number of miRNAs in each family in each species. Shading indicates presence of at least one member of a family. (B) As in A, but nonconserved miRNAs. (C) Venn diagram shows the number of miRNA families and their overlap in each of the four nematode species. (D) The percentage of families having the indicated number of members is shown for conserved and nonconserved miRNAs in each species. (E) Table of mirtrons classified by the gene hosting the mirtron.
Among conserved miRNA families, ∼40%–60% have multiple members within a species, whereas <13% of nonconserved families contain multiple members, suggesting that ancient miRNA families expand to confer robustness in gene regulatory networks (Fig. 2D). A striking example of this is the miR-35 family which has expanded to contain at least eight members in each species and as many as 27 members in C. brenneri (Fig. 2A). The miR-35 family is one of the few miRNA families essential for development (Alvarez-Saavedra and Horvitz 2010). Each of the other families essential for development, including miR-51, miR-58, and let-7, also contain multiple (≥4) members in each species (Fig. 2A).
Most miRNAs are processed from primary transcripts in sequential steps involving the ribonucleases Drosha and Dicer. However, some miRNAs are, instead, derived from short intronic hairpins called mirtrons during splicing, thereby bypassing Drosha cleavage (Okamura et al. 2007; Ruby et al. 2007). We found that out of the 15 C. elegans annotated mirtrons (Chung et al. 2011), only miR-62, embedded in the third intron of ugt-50, is conserved in the other three nematodes (Fig. 2E). miR-62 has 100% sequence conservation in all four nematodes, and the conservation of the intron sequence itself is much higher than that of other ugt-50 introns (Supplemental Fig. S2). Although the role of miR-62 is unknown, the strong selective pressure to maintain it hints at an important function. In addition to miR-62, we identified one nonconserved mirtron in C. briggsae, one in C. remanei, and two present in C. briggsae, C. remanei, and C. brenneri but not in C. elegans (Fig. 2E).
piRNAs share common features in their biogenesis and mechanism of action
C. elegans piRNAs (21U-RNAs) are 21 nt long and contain a 5′ U. Although the mechanism by which piRNAs are formed is unclear, they map downstream from conserved sequence motifs that may function as transcriptional promoters (Ruby et al. 2006; de Wit et al. 2009; Cecere et al. 2012). To identify piRNAs from C. briggsae, C. remanei, C. brenneri, and C. elegans, we filtered our data sets for small RNAs that were not identified in our miRNA analysis and that were 21 nt long and contained a 5′ U. Consistent with previous studies (Ruby et al. 2006; de Wit et al. 2009), we found that a GTTTC core motif was strongly overrepresented between −37 to −47 nt upstream of the starting T in candidate piRNA coding sequences from each species. The nucleotide composition and the length of spacer sequence between the large and small motifs were nearly identical in all species, consistent with previous findings in C. briggsae and C. remanei (Supplemental Fig. S3; de Wit et al. 2009).
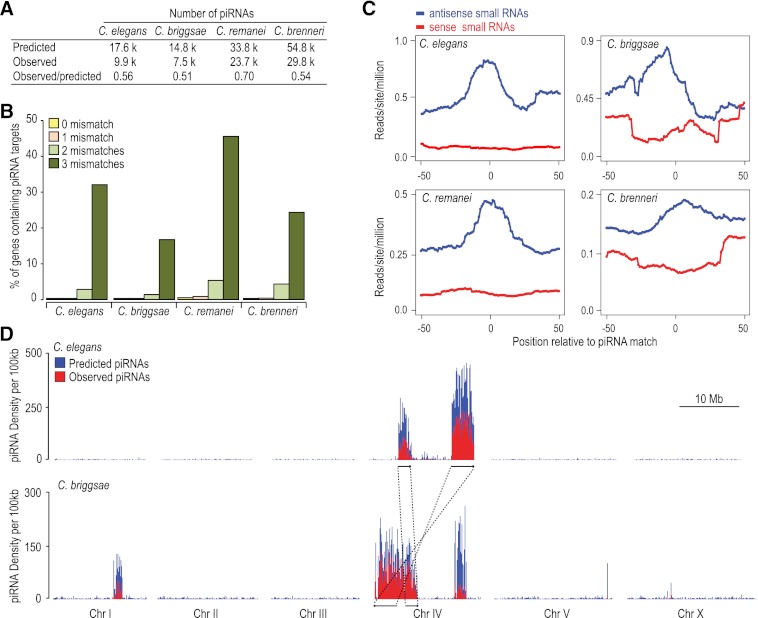
Using a scoring matrix based on the consensus motif, spacer sequence length, and 5′ U features of piRNAs, we scanned each of the four genomes for putative piRNA loci. The numbers of both predicted and observed piRNA loci were similar between C. elegans, with 17.6K predicted and 9.9K observed, and C. briggsae, with 14.8K predicted and 7.5K observed (Fig. 3A). C. remanei and C. brenneri possess far more piRNA loci than C. elegans and C. briggsae. In C. remanei, 33.8K piRNAs were predicted and 23.7K were observed, and in C. brenneri, 54.8K piRNAs were predicted and 29.8K were observed (Fig. 3A). A saturation analysis indicates that we captured the majority of piRNAs produced at these particular developmental stages (Supplemental Fig. S4). The genomes of C. remanei and C. brenneri are ∼35% larger than those of C. elegans and C. briggsae; however, this alone does not account for the 2–3 times more piRNAs identified in C. remanei and C. brenneri. Interestingly, C. elegans and C. briggsae are both androdioecious (male/hermaphrodite) species, whereas C. remanei and C. brenneri are both gonochoristic (male/female). This suggests that species that mate at every generation require a more complex repertoire of piRNAs for genome surveillance.
Figure 3.
Functional and genomic features of piRNAs are conserved. (A) The numbers of piRNA loci predicted based on regulatory motifs and the numbers identified from high-throughput sequencing. (B) The percentage of protein-coding genes in each genome that could be targeted by piRNAs if zero, one, two, or three mismatches are allowed. (C) Density of small RNAs within a 100-nt window centered on the predicted target sites of the top 20% most abundant piRNAs. (Blue) Small RNAs that are antisense to the predicted targets; (red) those that are sense to the targets. (D) Distribution of observed (red) and predicted (blue) piRNA loci per 100-kb window in C. elegans (top) and C. briggsae (bottom). There are two piRNA clusters on C. briggsae chromosome IV: the 0 to 6.9 Mb region largely in synteny with the two C. elegans piRNA clusters (highlighted in lines with arrows); and the 13.1 to 15.1 Mb cluster. In addition, C. briggsae has a third piRNA cluster on chromosome I at position 9.9 to 11.3 Mb.
Although we identified nearly 70,000 total piRNAs in our deep sequencing data sets, not a single piRNA sequence was present in more than one species. We also assessed piRNA sequence conservation when one, two, or three mismatches were allowed. Even with this less stringent criterion, only 0%, 0.01%, and 0.1% of C. elegans piRNAs have potential homologs in C. briggsae allowing for one, two, and three mismatches, respectively.
To identify potential piRNA targets, we aligned piRNA sequences with all annotated protein-coding genes from each species. piRNA-target recognition is thought to be permissive to around three mismatches (Bagijn et al. 2012; Lee et al. 2012); thus, we did the analysis allowing for zero, one, two, or three mismatches (Fig. 3B). When up to three mismatches are allowed, ∼30% of C. elegans and ∼20% of C. briggsae genes are potential targets (Fig. 3B). Although C. remanei and C. brenneri contain a substantially larger repertoire of piRNAs, the proportions of genes with potential piRNA targets are similar to that of C. elegans (Fig. 3B).
C. elegans piRNAs can trigger the production of RdRP-dependent secondary siRNAs centered on and antisense to piRNA target sites (Bagijn et al. 2012; Lee et al. 2012). To determine if piRNAs trigger siRNA formation in the other three nematodes, we assessed both sense and antisense siRNA abundance at candidate piRNA target sites. When all observed piRNAs were included in the analysis, there was only a slight enrichment of siRNAs at predicted piRNA target sites (data not shown). However, when only the top 20% most abundant piRNAs were considered, we observed a substantial enrichment of siRNAs antisense, but not sense, to the predicted piRNA target sites in each species (Fig. 3C). Of these siRNAs, 70%–80% are 22G siRNAs.
These results suggest that, although individual piRNAs are not conserved, the mechanism in which they are formed and their propensity to trigger secondary siRNA formation are conserved (Bagijn et al. 2012; Lee et al. 2012).
C. elegans piRNAs are primarily derived from two broad clusters on chromosome IV (Fig. 3D; Ruby et al. 2006). We asked whether piRNA loci are also clustered in other species. We restricted our analysis to C. briggsae because C. remanei and C. brenneri DNA sequences have not yet been assembled into chromosomes. There is extensive conservation of chromosome organization and synteny between C. elegans and C. briggsae (Stein et al. 2003; Hillier et al. 2007). In C. briggsae, the syntenic regions of the two major C. elegans piRNA clusters on chromosome IV also produced high levels of piRNAs (Fig. 3D; Ruby et al. 2006; de Wit et al. 2009). The two regions that give rise to the C. elegans piRNA clusters on chromosome IV are rearranged in C. briggsae such that they are separated from one another by only ∼1 Mb. Interestingly, the ∼1-Mb region separating the two clusters also contains a high abundance of piRNA loci. Together, the region forms a continuous 6.9-Mb piRNA cluster. We also identified a second piRNA cluster on chromosome IV (13.1–15.1 Mb) and another on chromosome I (9.9–11.3 Mb) specific to C. briggsae (Fig. 3D). The regions that give rise to these two C. briggsae-specific piRNA clusters lack continuous synteny with C. elegans, as determined by pairwise alignments. Two C. briggsae piRNA clusters previously identified on chromosomes I (7.8–9.5 Mb) and III (0–0.3 Mb) were represented by only average numbers of reads in our libraries and may have been artifacts of low sequencing depth (de Wit et al. 2009). That piRNAs in both C. elegans and C. briggsae tend to cluster and that conservation of these clusters appears to depend on long regions of continuous synteny suggests that the clusters are important for the birth of new piRNAs.
Distinct populations of piRNAs in males and females
In C. elegans, differences in the upstream regulatory motif drives male- and female-specific piRNA expression (M Freeberg and J Kim, pers. comm.). In each of the four Caenorhabditis species we analyzed, we observed a highly significant positive correlation between piRNA populations in early embryos and adult hermaphrodites or adult females+males (Fig. 4A). In contrast, piRNA levels between adult males and adult hermaphrodites or adult females+males are only modestly correlated and show a biphasic pattern of distribution indicative of two distinct classes (Fig. 4B). In C. elegans, we identified 9307 distinct piRNAs in hermaphrodites and 6065 distinct piRNAs in males. Of these, 5044 were enriched greater than threefold in hermaphrodites and 3336 were enriched greater than threefold in males. Only 1493 piRNAs had similar expression levels in hermaphrodites and males (Fig. 4C). Each of the other species also had distinct sets of piRNAs that were enriched in either males or hermaphrodites/females+males (Fig. 4C), suggesting that the production of distinct classes of male and female piRNAs is conserved in nematodes. The upstream motifs and length of spacer between the large and small motifs are nearly identical between the two classes of piRNAs, although several positions in the large motif and surrounding sequence show a stronger bias for a particular nucleotide in one class relative to the other (Supplemental Fig. S5).
Figure 4.
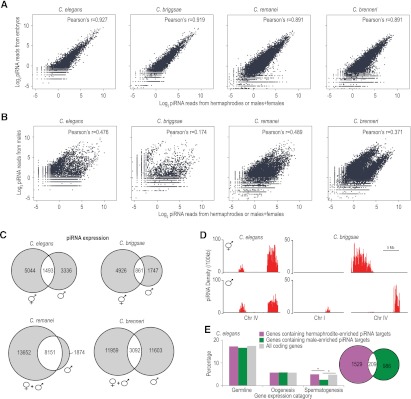
Two distinct classes of piRNAs in each species. (A) Scatter plots display the levels of piRNAs in adult hermaphrodites or adult females+males (x-axis) and early embryos (y-axis). (B) Scatter plots display the levels of piRNAs in adult hermaphrodites or adult females+males (x-axis) and adult males (y-axis). (C) Venn diagrams show the number of piRNAs that are greater than threefold enriched in hermaphrodites/females+males (left) or males (right). piRNAs shown in the overlapping section have similar expression levels in hermaphrodites/females and males. (D) Distribution of hermaphrodite- or male-enriched piRNA loci per 100 kb in C. elegans and C. briggsae. (E) The bar diagram displays the percentage of C. elegans genes containing hermaphrodite- or male-enriched piRNA target sites that are expressed in the germline, during oogenesis or during spermatogenesis. Asterisks indicate that the difference is statistically significant (P < 0.001, χ2 test). The Venn diagram shows the numbers of C. elegans genes containing target sites for hermaphrodite- or male-enriched piRNAs in purple and green, respectively.
The genomic distributions of C. elegans hermaphrodite- and male-enriched piRNAs are similar, although hermaphrodite-enriched piRNAs are somewhat more biased toward the cluster on the right arm of chromosome IV than are male-enriched piRNAs (Fig. 4D; Kato et al. 2009). In contrast to C. elegans, all C. briggsae piRNAs enriched in hermaphrodites are clustered on chromosome IV 0–6.9 Mb, the region in synteny with C. elegans piRNA clusters. However, almost all C. briggsae male-enriched piRNAs are located in the chromosome IV 13.1–15.1 Mb cluster or the chromosome I 9.9–11.3 Mb cluster (Fig. 4D). The nonoverlapping genomic distributions of C. briggsae hermaphrodite- and male-enriched piRNAs not only suggest that they have distinct evolutionary trajectories but also points to the possibility that differences in chromosomal-scale epigenetic marks may contribute to the sex-biased expression of piRNAs.
Given that the majority of piRNAs are differentially expressed, we asked whether the hermaphrodite/female- and male-enriched piRNAs have different sets of target genes. Indeed, when we allowed for up to three mismatches, genes containing target sites for hermaphrodite- or male-enriched piRNAs are largely nonoverlapping in C. elegans (Fig. 4E). In microarray-based studies, ∼5000 genes have been identified that are highly expressed in the germline, during either oogenesis or spermatogenesis (Reinke et al. 2000, 2004). We found that genes containing target sites for hermaphrodite- or male-enriched piRNAs were neither enriched nor depleted for germline or oogenesis genes. However, genes containing target sites for male-enriched piRNAs were significantly depleted of spermatogenesis genes (Fig. 4E). This suggests a selective pressure on male-enriched piRNAs to avoid targeting genes that are important for spermatogenesis. It is unclear why we do not observe a similar depletion of hermaphrodite-enriched piRNA target sites in genes important for oogenesis.
Conservation of 22G siRNA pathways
There are two distinct classes of 22G siRNAs in C. elegans which can be classified by their Argonaute binding partners. WAGO class 22G siRNAs silence certain protein-coding genes, transposons, pseudogenes, and cryptic loci to protect genome integrity (Gu et al. 2009). CSR-1 class 22G siRNAs have a role in regulating chromosome segregation (Claycomb et al. 2009; van Wolfswinkel et al. 2009) and might provide a memory of self to prevent endogenous genes from being silenced by the piRNA surveillance pathway, although data supporting this is lacking (Lee et al. 2012; Shirayama et al. 2012).
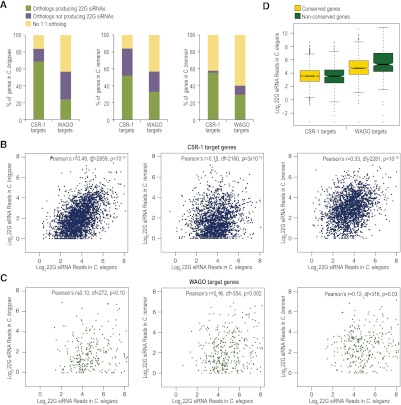
Approximately 85% of CSR-1 targets in C. elegans have a 1:1 ortholog in at least one of the other three Caenorhabditis species we analyzed, whereas ∼60% of the total coding genes in C. elegans have 1:1 ortholog in at least one other species. Similar to the average conservation of protein-coding genes, ∼60% of WAGO targets have a 1:1 ortholog in at least one of the other species analyzed (Fig. 5A). Approximately 50%–70% of C. elegans CSR-1 target genes also produce siRNAs in orthologs in each of the other species (Fig. 5A). We also observed a modest but significant (P < 10−11) positive correlation in 22G siRNA levels among orthologous CSR-1 target genes between C. elegans and each of the other species (Fig. 5B). In contrast, targeting by WAGO class siRNAs is more poorly conserved. Only ∼20%–30% of WAGO targets in C. elegans produce siRNAs in one of the other species analyzed, and there is lower correlation in siRNA levels among orthologous WAGO target genes between species (P = 0.002–0.1) (Fig. 5A,C). Because 22G siRNAs are biochemically indistinguishable from one another, we cannot rule out the possibility that in C. briggsae, C. remanei, and C. brenneri, a fraction of C. elegans CSR-1 targets are, instead, WAGO targets and vice versa. Nonetheless, these results point to relatively high conservation of targeting by CSR-1 class siRNAs and poor conservation of targeting by WAGO class siRNAs. WAGO class siRNAs, which are likely produced through successive rounds of siRNA amplification, are more abundant than CSR-1 class siRNAs in C. elegans (Claycomb et al. 2009; Gu et al. 2009; Phillips et al. 2012). We observed that WAGO target genes, on average, produce higher levels of 22G siRNAs than CSR-1 target genes, regardless of whether or not the genes are conserved. Moreover, nonconserved WAGO targets tend to produce more siRNAs than conserved WAGO targets (P < 0.0001), whereas CSR-1 targets tend to produce similar levels of siRNAs regardless of their conservation (P = 0.13) (Fig. 5D).
Figure 5.
Conservation of CSR-1 and WAGO class 22G siRNA pathways. (A) The percentages of C. elegans CSR-1 or WAGO target genes that contain an ortholog in C. briggsae, C. remanei, or C. brenneri, as well as the percentage of orthologous genes that produce 22G siRNAs in each species, is shown. (B) Scatter plots display conserved CSR-1 target genes having at least one siRNA read as a function of the log2 22G siRNA reads in C. elegans versus each of the other species. Pearson's r values and corresponding P-values for significant correlation are shown. Degree of freedom (df) = number of orthologs analyzed − 2 was used for P-value calculation. (C) Same as B, but conserved WAGO target genes are plotted. (D) Box plot displays the levels of 22G siRNAs generated from conserved and nonconserved CSR-1 and WAGO target genes in C. elegans.
Embryo and sperm class 26G siRNA pathways are conserved in Caenorhabditis species
There are two distinct classes of 26G siRNAs in C. elegans. One class is enriched in oocytes and embryos and associates with ERGO-1 (Han et al. 2009; Vasale et al. 2010; Fischer et al. 2011). The other class associates with ALG-3 and ALG-4 during spermatogenesis (Han et al. 2009; Conine et al. 2010). Both classes are thought to function by triggering 22G siRNA formation and subsequent silencing of target mRNAs. Because each class of 26G siRNAs is sex- and stage-specific, our embryo and adult male small RNA libraries are each enriched for one of the two classes.
From our C. elegans embryo small RNA library, we defined a set of 29 26G siRNA-yielding genes, which produced at least 10 26G siRNA reads per million total reads (RPM), 26 of which were previously annotated as ERGO-1 targets (Han et al. 2009; Vasale et al. 2010; Fischer et al. 2011). In addition, we identified 121 embryo 26G siRNA-yielding genes in C. briggsae, 71 in C. remanei, and 272 in C. brenneri. None of the 26G siRNA sequences are conserved among species. Although 2%–12% of embryo 26G siRNA-yielding genes are conserved (Fig. 6A), in each instance only one ortholog yields 26G siRNAs.
Figure 6.
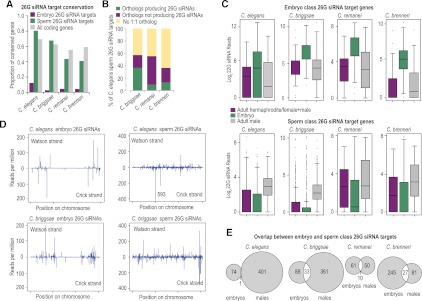
Features of 26G siRNAs are conserved. (A) The proportions of conserved embryo and sperm class 26G siRNA target genes are shown for each species. (B) The percentages of C. elegans sperm class 26G siRNA target genes that contain an ortholog in C. briggsae, C. remanei, or C. brenneri, as well as the percentage of orthologous genes that produce 26G siRNAs in each species, are shown. (C) Box plots show the level of 22G siRNAs generated from embryo and sperm class 26G siRNA targets in adult hermaphrodites or females+males (purple), embryos (green), and adult males (gray). (D) Distribution of embryo and sperm class 26G siRNAs across each C. elegans or C. briggsae chromosome is overlaid in one plot. Position is relative to the total length of each chromosome. (E) Venn diagrams show the numbers of embryo and sperm class 26G siRNA targets and their overlap in each of the four species.
In contrast to the poor conservation of embryo class 26G siRNA targets, ∼80% (∼50% 1:1 orthologs) of C. elegans sperm class 26G target genes have a potential ortholog in C. briggsae, compared to ∼70% (∼60% 1:1 orthologs) of all coding genes. This suggests that sperm class 26G target genes are not particularly conserved or fast evolving. Despite average conservation (∼70%) of sperm class 26G siRNA targets, we observed a disproportionally low tendency for more than one ortholog to produce 26G siRNAs. Only ∼38% of C. elegans genes that produce sperm class 26G siRNAs are conserved and also produce >1 RPM 26G siRNAs in C. briggsae (Fig. 6B). In C. remanei and C. brenneri, <15% of C. elegans genes are conserved and also produce 26G siRNAs (Fig. 6B). These results indicate that distinct cohorts of genes are targeted by 26G siRNAs during spermatogenesis and embryogenesis in each of the different nematode species.
In all four nematode species, embryo class 26G siRNA target genes also produce 22G siRNAs (Fig. 6C). In each species, these secondary 22G siRNAs are enriched in embryos relative to adults (Fig. 6C). Similarly, sperm class 26G siRNA target genes produce relatively high levels of 22G siRNAs that are enriched in adult males relative to embryos or hermaphrodites/males+females in each species (Fig. 6C). Thus, 26G siRNAs trigger stage-specific production of 22G siRNAs across nematode species.
Although 26G siRNA targets do not appear to be conserved, we nonetheless asked whether embryo class 26G siRNA chromosomal distribution is conserved. In C. elegans, oocyte/embryo class 26G siRNAs tend to be generated from gene-poor arms of chromosomes (Fig. 6D; Vasale et al. 2010; Fischer et al. 2011). In C. briggsae, for which the DNA sequence has been assembled on chromosomes, embryo class 26G siRNAs also tend to derive from chromosome arms (Fig. 6D). In contrast, sperm class 26G siRNAs are more or less evenly distributed along each chromosome in both C. elegans and C. briggsae (Fig. 6D). Thus, despite very little conservation of 26G siRNAs among species, the genomic distribution is very similar.
Embryo and sperm class 26G siRNAs are produced from nearly completely nonoverlapping targets in C. elegans (Fig. 6E; Han et al. 2009). However, in each of the other three nematodes, we observed substantial overlap between the embryo and sperm class 26G siRNA-yielding genes (Fig. 6E). Furthermore, the distributions of 26G siRNAs on the common targets were very similar in embryos and males within each species (data not shown). The common target genes are nonconserved, and in C. briggsae they are located on the chromosomal arms. Thus, the features of 26G siRNA targets that produce siRNAs in both males and embryos are more similar to ERGO-1 class 26G siRNA targets than to ALG-3/4 26G siRNA targets. It is interesting to note that ergo-1 appears to have undergone extensive gene duplications in C. briggsae, C. remanei, and C. brenneri after diverging from their common ancestor (Supplemental Fig. S1; Table 1). It is plausible that ergo-1 paralogs acquired new expression patterns or functions that account for the relatively high overlap between sperm and embryo 26G siRNA targets in species other than C. elegans.
Discussion
Using a deep-sequencing comparative genomics approach, we identified ∼25 million distinct small RNAs from gravid adults and developing embryos of the four nematode species C. elegans, C. briggsae, C. remanei, and C. brenneri. Our results indicate that each of the major classes of small RNAs is conserved across Caenorhabditis species, although the degree of conservation differs tremendously. miRNAs are by far the most highly conserved class of small RNAs based on sequence. CSR-1 class 22G siRNAs are typically not conserved at the sequence level, yet they derive from a largely overlapping panel of genes in each species, indicating that the features that route CSR-1 targets into the siRNA pathway are conserved. piRNAs, WAGO class 22G siRNAs, and each of the classes of 26G siRNAs are not individually conserved but share many similarities in their biogenesis and function among the different species.
Although piRNA sequences are not at all conserved, other features, including the upstream motifs (de Wit et al. 2009), genomic clustering, and the tendency to trigger secondary 22G siRNA production from target genes, are conserved. C. elegans piRNAs are thought to play a role in genome surveillance and protection against nonself nucleic acids (Bagijn et al. 2012; Lee et al. 2012; Shirayama et al. 2012). Our results suggest that the roles of piRNAs are conserved across nematodes. That there is essentially no overlap in piRNA sequences among different nematode species indicates that piRNAs themselves are under very little stabilizing selection. The sheer number of piRNAs (likely >15,000 in each species) would allow them to target most, if not all, genes if multiple mismatches are permissible and would, therefore, not require sequence conservation of individual piRNAs. Indeed, rapid evolution of piRNAs would be advantageous in an arms race against invading nucleic acids.
We found that gonochoristic species that mate at every generation possess much larger numbers of piRNAs than androdioecious species that primarily self-fertilize. In Drosophila, maternally deposited piRNAs can silence paternal transposons (Brennecke et al. 2008). Moreover, crosses between Drosophila strains that differ in the presence of a particular transposon result in sterile progeny only if the transposon is inherited from the paternal genome (Brennecke et al. 2008). Similarly, desilencing of paternal transposons was observed in interspecies Arabidopsis crosses (Josefsson et al. 2006). It is possible that gonochoristic nematodes require a larger repertoire of piRNAs than androdioecious nematodes in order to defend against the greater diversity of paternal transposons encountered during mating. Moreover, the gonochoristic nematodes also have nearly twice as many Argonautes (37 in C. remanei and 47 in C. brenneri) as the androdioecious nematodes (24 in C. briggsae and 25 in C. elegans) and have undergone extensive proliferation of the PIWI clade Argonautes ERGO and PRG (Table 1; Supplemental Fig. S1).
Most piRNAs are differentially expressed in hermaphrodites/females and males in each of the four species tested. Differences in the piRNA repertoire of C. elegans males and hermaphrodites may result from distinct upstream sequence motifs that drive sex-specific expression (M Freeberg and J Kim, pers. comm.). The male and female/hermaphrodite piRNAs target a largely nonoverlapping set of genes. Spermatogenesis genes are depleted of male-enriched piRNA targets, suggesting selective pressure on male-enriched piRNAs to avoid targeting genes that are highly expressed during spermatogenesis. It will be important to distinguish the specific roles of male and female piRNAs.
If the role of CSR-1 class siRNAs is to provide a memory of self as has been proposed (Bagijn et al. 2012; Lee et al. 2012; Shirayama et al. 2012), then one would expect targeting by CSR-1 to be highly conserved. This is, indeed, what we observed. Targeting by CSR-1 class 22G siRNAs does appear to be highly conserved despite the individual siRNAs lacking conservation. In contrast, WAGO class 22G siRNAs are thought to target aberrant and foreign RNAs, and targeting of annotated protein-coding genes by this class of small RNAs is more poorly conserved.
Embryo class 26G siRNAs primarily target recently evolved genes with very little sequence conservation, and thus, it is not surprising that their targets are completely distinct in each species. Sperm class 26G siRNAs target many conserved genes, yet orthologous genes often do not produce 26G siRNAs. In spite of this, 26G siRNAs share many features in common among different nematode species, including genomic distribution, low abundance, sex specificity, and the propensity to trigger stage-specific 22G siRNA production.
Through phylogenetic analysis, we identified a distinct subfamily of RdRPs, RRF-4, which is present in each of the Caenorhabditis species we analyzed except for C. elegans. We were unable to link a distinct class of siRNAs to RRF-4 from analysis of our small RNA libraries, likely because RRF-4-dependent siRNAs are biochemically indistinguishable from other classes of siRNAs present in these libraries. It will be important to dissect the role of RRF-4 once the necessary genetic resources are available to do so in rrf-4-containing species.
The evolutionary fluidity of the different classes of small RNAs and their effectors in Caenorhabditis underscores their importance in the regulation, surveillance, and evolution of the genome.
Methods
Nematode strains
Nematode strains used in this study: C. elegans N2, C. briggsae AF16, C. remanei PB4641, and C. brenneri PB2801. Worms were cultured with bacterial strain OP50 on modified nematode growth medium (Andersen et al. 2012) containing 1% agar and 0.7% agarose to prevent burrowing of C. brenneri. All strains were grown at 20°C.
Phylogenetic analyses of Argonautes and RdRPs
To obtain a comprehensive and nonredundant list of Argonaute proteins and RdRPs in C. briggsae, C. remanei, and C. brenneri, we did BLASTP searches in the National Center for Biotechnology Information (NCBI) database using the protein sequences of C. elegans alg-1 and rrf-1 as queries for Argonautes and RdRPs, respectively. We obtained Argonaute orthologs with E-values <1 × 10−10 and query coverage >35%, and RdRP orthologs with E-values <1 × 10−113 and query coverage >70%. Using different Argonautes or RdRPs as queries did not affect the outcome. Multiple sequence alignments were then performed using CLUSTALW slow/accurate alignments with default settings (Larkin et al. 2007). We constructed unrooted trees with the outputs of multiple sequence alignments, using the PHYLIP DRAWTREE program.
High-throughput sequencing and data analysis
For the construction of adult hermaphrodite/male+female, male or embryo small RNA libraries, animals were grown at 20°C for 72–74 h post-L1 synchronization and harvested as day one gravid adult hermaphrodites for C. elegans and C. briggsae and mixed populations of adult males and females for C. remanei and C. brenneri. For male isolation, 150∼200 day one adult males were hand-picked from the NGM plate. Embryos were harvested by bleach treatment of ∼15,000 gravid adults. Total RNA was isolated by dounce homogenization of worms in TRI Reagent, followed by chloroform extraction and isopropanol precipitation. Small RNA high-throughput sequencing libraries were prepared as described (Montgomery et al. 2012). Briefly, 18- to 28-nt small RNAs were size-selected and treated with 20 U Tobacco Acid Phosphatase (Epicenter) at 37°C for 2 h to digest 5′ tri- and diphosphates to monophosphates. Small RNAs were then ligated to the 3′ adapter using T4 RNA ligase 2 truncated (NEB) for 16 h at 16°C. 5′ ligations were done with T4 RNA ligase 1 (NEB) for 16 h at 16°C. Adapter-ligated RNAs were then reverse-transcribed and PCR-amplified using Illumina's TruSeq RNA Indexing PCR primers. Small RNA amplicons were size-selected by gel purification and subjected to Illumina HiSeq sequencing. Small RNA sequences were parsed using a custom Python program to remove adapter sequences and then mapped to the corresponding nematode reference genome (WormBase release WS230) allowing for 0 mismatches using Bowtie software (Langmead et al. 2009). For sequences mapping to multiple genomic loci, the total number of reads was divided by the number of genomic loci. Small RNA reads were then normalized to the total number of millions of mapped reads (i.e., reads per million).
Small RNA classification
C. elegans small RNAs were classified as described (Zhang et al. 2011). Briefly, published data sets were parsed for classifying targets of C. elegans WAGO, CSR-1, ERGO-1, and ALG-3/4 class siRNAs. WAGO class was defined as the siRNAs produced from 1112 genes depleted of siRNAs in WAGO-1-12, rde-3, and mut-7 (Gu et al. 2009). CSR-1 class was defined as siRNAs produced from the 4191 genes that yield siRNAs enriched in CSR-1 immunoprecipitates (Claycomb et al. 2009). ALG-3/4 (401 genes) class and ERGO-1 (75 genes) class were defined as 26G siRNAs enriched during spermatogenesis and oogenesis/embryogenesis, respectively (Han et al. 2009; Conine et al. 2010; Vasale et al. 2010). miRNAs were predicted using miRDeep2 with default settings (Mackowiak 2011; Friedlander et al. 2012). To identify new miRNAs with high confidence, we further required a candidate miRNA predicted by miRDeep2 to also be predicted by MIReNA (Mathelier and Carbone 2010) and/or possess a seed sequence conserved between Caenorhabditis species. piRNAs/21U-RNAs were predicted as described (Ruby et al. 2006), using a scoring matrix based on the consensus motif, spacer sequence length, and 5′ U features of piRNAs. piRNA saturation analysis was performed by taking a random subset of sequencing reads with increasing size and calculating the percentage of piRNAs identified from this sublibrary.
Gene orthology analyses
Orthologous genes were identified using InParanoid4, which allows both one-to-one and many-to-many orthology cases (O'Brien et al. 2005). For clarity, we only kept one-to-one orthologs to analyze the conservation of different classes of small RNA target genes.
Statistical analyses
For analysis of C. elegans genes containing hermaphrodite- or male-enriched piRNA target sites in Figure 4E, the χ2 test (df = 1) was performed. For the comparison of 22G siRNA levels among orthologous CSR-1 or WAGO target genes between C. elegans and each of the other three species, Pearson's r correlation and corresponding P-values were calculated using R. P-values for comparing 22G siRNA levels produced from C. elegans CSR-1 and WAGO target genes were calculated using the Welch's t-test.
Data access
All high-throughput sequencing data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE41461.
Acknowledgments
We thank the Ruvkun lab members for constructive suggestions and thoughtful critiques of the work. We thank Allison Billi, Mallory Freeberg, and John Kim for sharing unpublished information. We thank the Caenorhabditis Genetics Center (CGC) for worm strains. Z.S. is supported by an American Heart Association predoctoral fellowship. T.A.M. was supported by the Damon Runyon Cancer Research Foundation (DRG 2029-09) and is a Massachusetts General Hospital Executive Committee of Research Tosteson postdoctoral fellow. Y.Q. is an Ellison Medical Foundation/AFAR postdoctoral fellow supported by the Life Sciences Research Foundation. This work was supported by a grant from the NIH to G.R. (R01-GM44619).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.149112.112.
References
- Alvarez-Saavedra E, Horvitz HR 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol 20: 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, Felix MA, Kruglyak L 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet 44: 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA 2012. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337: 574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere G, Zheng GX, Mansisidor AR, Klymko KE, Grishok A 2012. Promoters recognized by forkhead proteins exist for individual 21U-RNAs. Mol Cell 47: 734–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Agius P, Westholm JO, Chen M, Okamura K, Robine N, Leslie CS, Lai EC 2011. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res 21: 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC 2010. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci 107: 3588–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Dey A, Murray RL 2009. Evolution of the Caenorhabditis elegans genome. Mol Biol Evol 26: 1199–1234 [DOI] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell 31: 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Linsen SE, Cuppen E, Berezikov E 2009. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res 19: 2064–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SE, Montgomery TA, Zhang C, Fahlgren N, Breen PC, Hwang A, Sullivan CM, Carrington JC, Ruvkun G 2011. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet 7: e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ 2010. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell 37: 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE Project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad Y, Aach J, Hayes GD, Reinhart BJ, Church GM, Ruvkun G, Kim J 2003. Computational and experimental identification of C. elegans microRNAs. Mol Cell 11: 1253–1263 [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK 2009. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci 106: 18674–18679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, Miller RD, Baird SE, Chinwalla A, Fulton LA, Koboldt DC, Waterston RH 2007. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol 5: e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson C, Dilkes B, Comai L 2006. Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol 16: 1322–1328 [DOI] [PubMed] [Google Scholar]
- Kato M, de Lencastre A, Pincus Z, Slack FJ 2009. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol 10: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke KC, Felix MA, Ailion M, Rockman MV, Braendle C, Penigault JB, Fitch DH 2011. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol 11: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864 [DOI] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D Jr, Mello CC 2012. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150: 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP 2003. The microRNAs of Caenorhabditis elegans. Genes Dev 17: 991–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackowiak SD 2011. Identification of novel and known miRNAs in deep-sequencing data with miRDeep2. Curr Protoc Bioinformatics 36: 12.10.1–12.10.15 [DOI] [PubMed] [Google Scholar]
- Mathelier A, Carbone A 2010. MIReNA: Finding microRNAs with high accuracy and no learning at genome scale and from deep sequencing data. Bioinformatics 26: 2226–2234 [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SE, Ruvkun G 2012. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet 8: e1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien KP, Remm M, Sonnhammer EL 2005. Inparanoid: A comprehensive database of eukaryotic orthologs. Nucleic Acids Res 33: D476–D480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86–89 [DOI] [PubMed] [Google Scholar]
- Phillips CM, Montgomery TA, Breen PC, Ruvkun G 2012. MUT-16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Genes Dev 26: 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, et al. 2000. A global profile of germline gene expression in C. elegans. Mol Cell 6: 605–616 [DOI] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D Jr, Mello CC 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476 [DOI] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF 2009. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139: 135–148 [DOI] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D Jr 2010. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci 107: 3582–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Reinke V 2008. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol 18: 861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757 [DOI] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Gabel HW, Fischer SE, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G 2011. Inaugural article: mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc Natl Acad Sci 108: 1201–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]