Abstract
Custom-designed nucleases (CDNs) greatly facilitate genetic engineering by generating a targeted DNA double-strand break (DSB) in the genome. Once a DSB is created, specific modifications can be introduced around the breakage site during its repair by two major DNA damage repair (DDR) mechanisms: the dominant but error-prone nonhomologous end joining (NHEJ) pathway, and the less-frequent but precise homologous recombination (HR) pathway. Here we describe ObLiGaRe, a new method for site-specific gene insertions that uses the efficient NHEJ pathway and acts independently of HR. This method is applicable with both zinc finger nucleases (ZFNs) and Tale nucleases (TALENs), and has enabled us to insert a 15-kb inducible gene expression cassette at a defined locus in human cell lines. In addition, our experiments have revealed the previously underestimated error-free nature of NHEJ and provided new tools to further characterize this pathway under physiological and pathological conditions.
The development of custom-designed nucleases (CDNs), including zinc finger nucleases (ZFNs) and Tale nucleases (TALENs), has made it possible to perform precise genetic engineering in many cell types and species (Kim et al. 1996; Bibikova et al. 2003; Porteus and Baltimore 2003; Moehle et al. 2007; Hockemeyer et al. 2009; Christian et al. 2010; Meyer et al. 2010; Urnov et al. 2010). CDNs are hybrid endonucleases consisting of a FokI nuclease domain and a DNA binding domain assembled from optimized DNA binding modules that are specific for either single-nucleotide (for TALENs) or trinucleotide motifs (for ZFNs). Once introduced into cells, CDNs generate a double-strand break (DSB) in the genome at or near the desired modification site and induce DNA damage repair (DDR) to mend the break (Rouet et al. 1994). Repair is largely accomplished by error-prone nonhomologous end joining (NHEJ), in which the two ends are processed and ligated together in a way that is frequently accompanied by nucleotide insertions and deletions. Though highly efficient, NHEJ produces knockout alleles that are often heterogeneous, and individual cell clones must be isolated for characterization. Currently, specific gene modification relies on homologous recombination (HR), in which exogenous DNA fragments flanked by homologous sequences around the DSB site are copied faithfully from a template with defined boundaries (Rouet et al. 1994).
We have successfully applied ZFNs to generate knockout and knock-in alleles directly in mouse zygotes (Meyer et al. 2010; Cui et al. 2011). While optimizing gene targeting conditions, we observed that a donor plasmid can be “ligated” into the genome if it contained the same ZFN recognition site as the targeted genomic locus. It has been reported that short, double-stranded DNAs with 5′ overhangs could be ligated to complementary ends generated after ZFN digestion (Orlando et al. 2010). This observation has not been further explored, probably because it requires the knowledge of the overhangs generated by ZFNs, and only insertions of small oligonucleotide have been described (Orlando et al. 2010). Furthermore, it has also been shown that donor molecules, including single-strand oligodeoxynucleotides (ssODNs) (Radecke et al. 2010; Chen et al. 2011) and larger external linear sequences, can be captured at DSB sites generated by ZFNs (Mittelman et al. 2009; Fung and Weinstock 2011; Gabriel et al. 2011; Li et al. 2011). This feature has been harnessed to track “off-target” effects of the homing endonuclease I-SceI and ZFNs (Petek et al. 2010). Based on these reports and our own observation in mouse embryos, we surmised that it should be possible to directly ligate an external DNA fragment linearized in situ by the same ZFNs that target the genome.
We took advantage of the obligated heterodimeric property of the CDNs (Miller et al. 2007; Szczepek et al. 2007; Doyon et al. 2011; Ramalingam et al. 2011) and designed a strategy to achieve efficient and precise gene targeting without homology in the donor plasmid. We named this method ObLiGaRe (Obligate Ligation-Gated Recombination) to reflect the etymologic meaning of the Latin verb obligare (“to bind,” “to join to”). ObLiGaRe should be broadly applicable across different cell types and provides an additional approach for genetic engineering.
Results
ObLiGaRe mediated precise end-joining
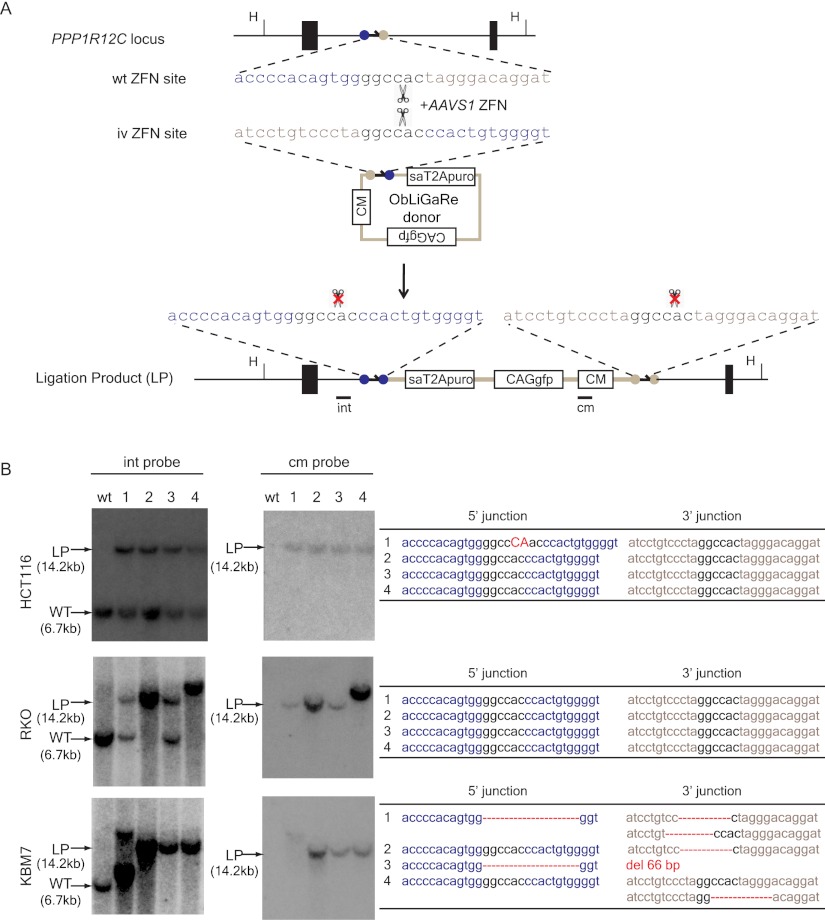
In order to directly ligate an exogenous DNA fragment into the genome, we initially introduced ZFN binding sites into a donor plasmid with the same orientation as in the genome. However we found this strategy often yielded unpredictable products, presumably because the same ZFN binding sites were produced after ligation, which could then be repetitively digested by the ZFNs, a process that could stimulate end recessing before joining (Pruett-Miller et al. 2008). One essential requirement for ZFN-mediated site-specific digestion using obligated heterodimers is that a pair of ZFNs needs to form heterodimers through the modified FokI nuclease domain after binding to their targeted DNA sequences on the opposite strands. We reasoned that if we altered the orientation of the ZFN recognition sequences in the donor plasmid, we could lock the ligation product in a palindrome of identical half ZFN recognition sites that would no longer be sensitive to the same obligated heterodimeric ZFN pairs. We used the well-characterized AAVS1 ZFN binding site (located in the first intron of PPP1R12C) (Hockemeyer et al. 2009) to illustrate this design principle (Fig. 1A). Here we inverted the two half AAVS1 ZFN binding sites in the vector without changing the orientation of the linker region (Fig. 1A, ObLiGaRe donor). The AAVS1 ZFNs cut both the genome and the donor plasmid to produce complementary overhangs. After ligation, the newly formed junctions are resistant to further digestion by AAVS1 ZFNs.
Figure 1.
ObLiGaRe at the AAVS1 locus. (A) Schematic illustration of the ObLiGaRe strategy. ZFN recognition sites are indicated as gray and blue circles with the corresponding sequences displayed in the same color scheme. The predicted joining sequences between the vector and the genome are shown. The insertion of the vector in the AAVS1 locus will cause a size shift from 6.7 kb (wild type [WT]) to 14.2 kb (ligation product [LP]) of a HindIII (H) digested fragment. AAVS1 internal (int) and vector-specific (cm) probes are indicated in the map at the hybridization sites. (B) Southern blot of four puro-resistant colonies after ObLiGaRe at AAVS1 locus in HCT116 (upper), RKO (middle), and KBM-7 (lower) with int probe (left) and cm probe (right) with sequences at the 5′ and 3′ junctions corresponding to each clone listed on the right. (Red) Insertions; (red dotted lines) deletions. (sa) Splicing acceptor; (CM) chloramphenicol-resistant marker.
To test our hypothesis, we constructed a vector containing the modified AAVS1 ZFN site followed by a promoter-less T2A-puro cassette. Precise ligation of this cassette into the AAVS1 site in the genome would render the cells resistant to puromycin. We also included a constitutive GFP reporter driven by CAG promoter (Xu et al. 2001) in the vector to track the cells transfected with the plasmid. We transfected the donor plasmid into HCT116 cells and observed an increase in the number of puromycin-resistant colonies when the donor plasmid was cotransfected with the plasmid encoding AAVS1 ZFNs (Supplemental Fig. 1). We isolated four randomly chosen puromycin-resistant, GFP-positive clones and examined the vector integration by Southern blot. All four clones were heterozygous insertions of the vector, as indicated with an AAVS1 locus-specific probe (Fig. 1B, int probe). In addition, we did not observe random insertions of the vector in the genome when using a vector-specific probe (Fig. 1B, cm probe). We then amplified the integration junctions by PCR and directly sequenced the PCR products. We found that three out of four clones had perfectly ligated junctions as predicted (Fig. 1B). Clone 1 had a 2-nucleotide insertion in the linker region at the 5′ junction, which was not predicted to affect the downstream splicing acceptor (sa) and the expression of puromycin-resistant gene (Fig. 1B). Similar results were obtained in RKO cells (Fig. 1B). Interestingly, one of the RKO clones was homozygous for a single-copy vector insertion (clone 2). One clone (clone 4) was homozygous for a larger insertion that may represent more than one copy of the vector.
We then tested ObLiGaRe in KBM7 cells. KBM7 is a near haploid human myeloid leukemia cell line in which gene targeting has not been reported (Kotecki et al. 1999; Carette et al. 2009). We cotransfected KBM7 cells with the ObLiGaRe donor plasmid and AAVS1 ZFN plasmid. Because the transfection efficiency was low, we sorted GFP-positive cells by FACS and subjected them to puromycin selection. We characterized four puromycin-resistant clones and found three had the expected insertions at the AAVS1 locus with no detectable random insertions as judged by Southern blot (Fig. 1B). All clones (clones 2–4) were hemizygous for the insertion, as expected, due to the haploid nature of the AAVS1 locus in KBM7 cells. We then sequenced the PCR amplified 5′ and 3′ insertion junctions of these clones. In the cases that we were unable to directly sequence the PCR products (3′ junction in clones 1 and 4, Fig. 1B), amplified junctions were cloned and sequenced. In contrast to experiments in RKO and HCT116 cells, the sequences of the junctions in KBM7 cells were heterogeneous and often involved deletions, probably between potential microhomologies (Fig. 1B). This high proportion of aberrant ligation might be due to the unique DDR mechanisms associated with haploid KBM7 cells, which are also karyotypically unstable (Andersson et al. 1995; Kobayashi et al. 1998; Kotecki et al. 1999; Skorski 2008).
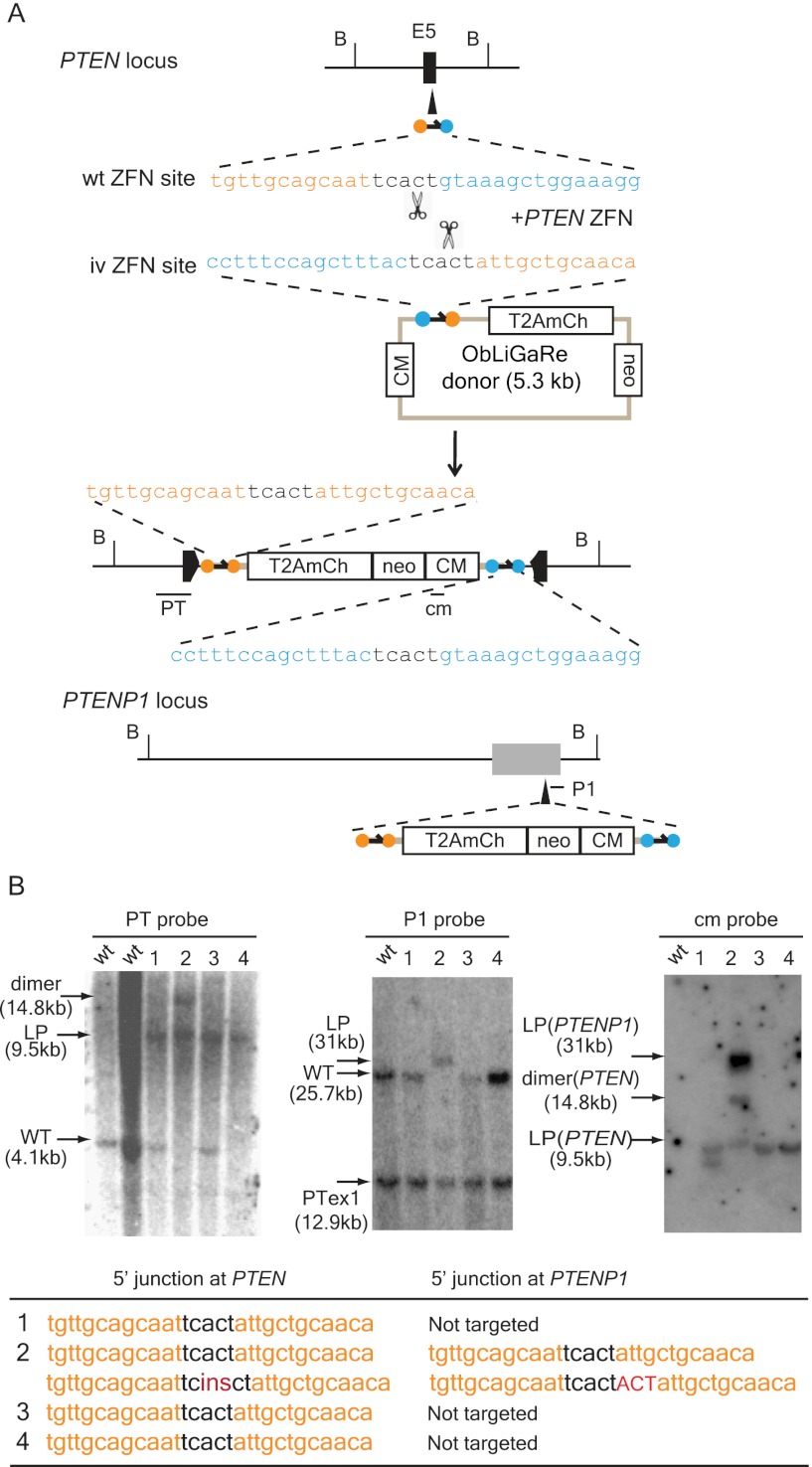
To determine whether ObLiGaRe is restricted to the AAVS1 locus and/or AAVS1 ZFNs, we used PTEN-specific ZFNs to insert a vector containing a promoter-less T2A-mCherry cassette and a CAG-neo selection marker into the PTEN locus in HCT116 cells. Because the human genome contains a PTEN pseudogene, PTENP1 (Poliseno et al. 2010), that is also targeted by the PTEN ZFNs, we expected to observe insertions at both PTEN and PTENP1. We isolated four G418-resistant clones and examined integration sites at both PTEN and PTENP1 loci by Southern blot. Because we used an autonomous selection marker, we expected that a proportion of G418-resistant clones would result from random insertion of the donor plasmid into the genome. Surprisingly, all clones had targeted insertions in the PTEN locus. Clones 1 and 3 were heterozygous, and clone 4 was homozygous for the insertion as judged by Southern blot (Fig. 2B). Interestingly, clone 2 had targeted homozygous insertions in both PTEN and PTENP1 loci (Fig. 2B). However, analysis of clone 2 DNA also showed a larger than expected band in the Southern blot probed with a PTEN allele-specific probe (Fig. 2B). The size of this band implied it was the result of incorporating two copies of donor plasmid ligated head-to-tail, which was confirmed by further characterization (Supplemental Fig. 2). Clone 1 had an extra insert in the genome as detected by the vector-specific probe (cm probe), which could be the result of either random insertion or insertion in other unidentified locus cleaved by PTEN ZFNs. We sequenced the 5′ junctions at PTEN locus in clones 1, 3, and 4 and found they all had precise predicted sequences at the junction between PTEN and T2A-mCherry (Fig. 2B). We used a TOPO TA Cloning (Life Technologies) strategy to isolate and sequence 5′ junctions at both PTEN and PTENP1 loci in clone 2 and identified additional ligation products in this clone, suggesting the two alleles of both genes might have been modified differently (Fig. 2B, table).
Figure 2.
ObLiGaRe at the PTEN locus. (A) Strategy for targeting human PTEN and PTENP1. PTEN ZFN recognition sites are indicated as orange and cyan circles with the corresponding sequences displayed in the same color scheme. The predicted joining sequences between the vector and the genome are indicated. The insertion of the vector will cause a size shift of a BsrGI (B) digested fragment from wild type of 4.1 kb (WT) to the ligation product of 9.5 kb (LP) at PTEN locus and 25.7 kb (WT) to 31.0 kb (LP) at PTENP1 locus. PTEN internal probe (PT), PTENP1 (P1), and vector-specific probe (cm) are indicated in the map at the hybridization sites. (B) Southern blots using probes specific for PTEN (PT, left), PTENP1 (P1, middle), and the vector (cm, right) for four G418-resistant colonies. P1 probe also could detect exon 1 of PTEN, which is the lowest signal across all lanes (middle blot). The sequence of the 5′ junction between PTEN or PTENP1 and mCherry are indicated in the table. Insertions are indicated in red, where “ins” represents a 224-bp insertion.
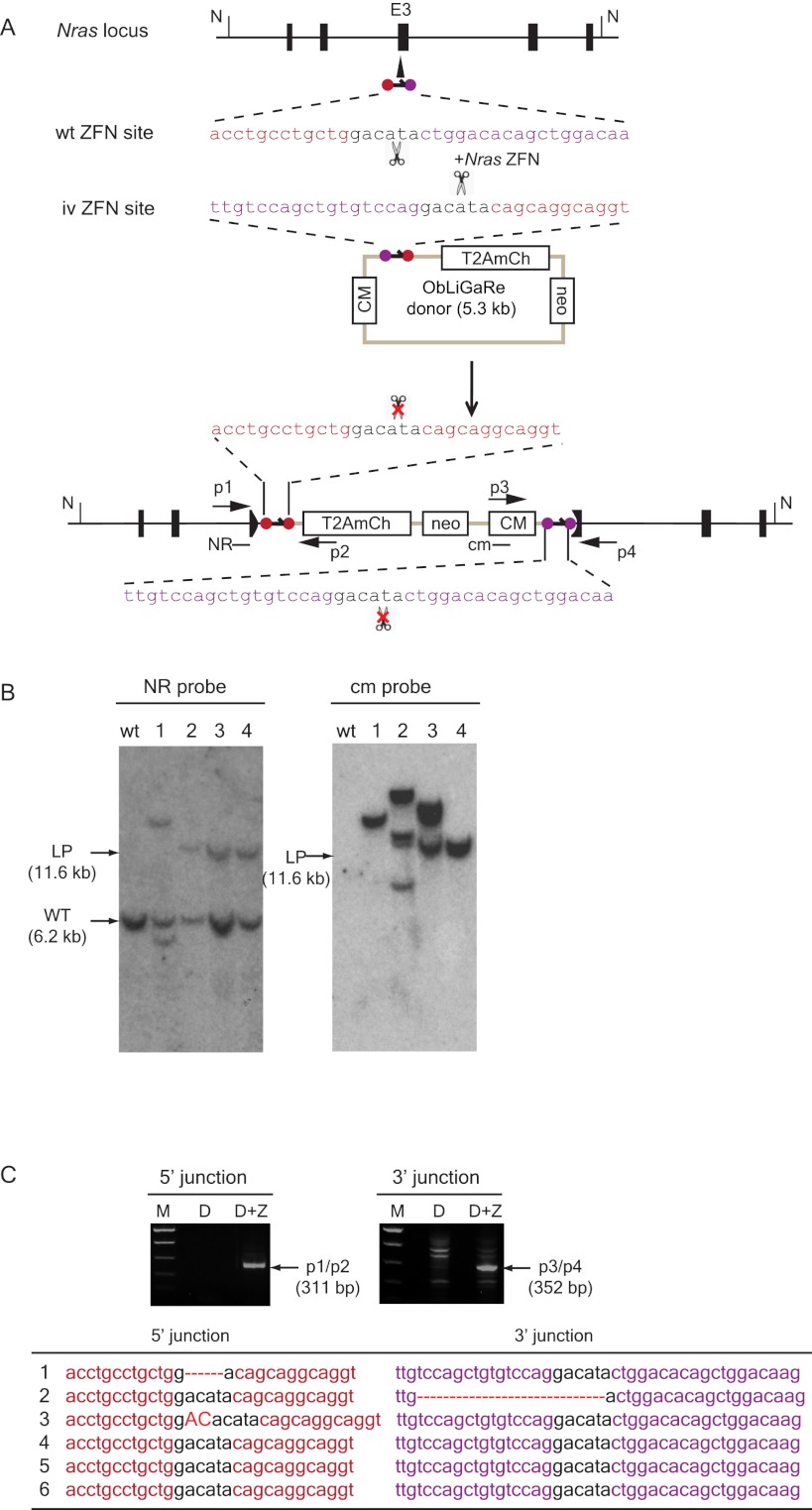
In order to confirm that ObLiGaRe does not rely on any specific mutations in HCT116 and RKO cells, which are known to be defective in DNA mismatch repair mechanisms (Brown et al. 2003), we decided to use a pair of ZFNs to target exon 3 of Nras gene in mouse myoblast C2C12 and primary mouse embryonic fibroblast (MEF) cells (Fig. 3). We cotransfected C2C12 cells either with the Nras ZFN plasmid alone or with an ObLiGaRe vector containing a promoter-less T2AmCherry cassette and a PGKneo selection marker. We then studied the integration in four randomly selected G418-resistant, mCherry fluorescent clones by Southern blot. Although all the clones were mCherry positive, suggesting correct in-frame fusion of T2AmCherry into Nras exon 3, only clone 4 had the predicted targeted integration of the vector at the Nras locus without additional genomic insertion events (Fig. 3B). Clone 1 had a smaller band hybridizing to the Nras-specific probe (NR), which might represent a deleted allele present in their largely tetraploid genome (Casas-Delucchi et al. 2011). We sequenced the junctions between Nras and mCherry in the genome of clone 4 and confirmed they were perfectly ligated (data not shown). In applying the same strategy to target primary MEF cells, we could not perform clonal selection, so we genotyped pooled cells 3 d after transfection using genomic PCR with primers specific for either the 5′ or 3′ junctions. We obtained the PCR products expected for correct integration only in cells transfected with the ObLiGaRe vector and Nras ZFN plasmid (Fig. 3C, D+Z). We sequenced individually cloned PCR products and found that the majority of the sequences were the precise end joining products at both the 5′ and 3′ ends (Fig. 3C).
Figure 3.
ObLiGaRe in C2C12 and MEF cells. (A) Strategy for targeting Nras locus in MEF and C2C12 cells. Nras ZFN sites are indicated as violet and red circles with the corresponding sequences displayed in the same color scheme. The predicted joining sequences between the vector and the genome are shown. Primers for PCR detection of the junctions and probes (cm and NR) are indicated. The insertion of the vector will cause a size shift of an NdeI (N) digested fragment from wild type of 6.2 kb (WT) to the ligation product of 11.6 kb (LP) at Nras locus. Nras-specific probe (NR) and vector specific probe (cm) are indicated in the map at the hybridization sites. (B) Southern blot for four C2C12 clones expressing mCherry. (LP) Expected ligation product band upon integration of the vector in the genome. The band lower than the WT band in clone 1 might be a deleted Nras allele since C2C12 cells are tetraploid. (C) Genomic PCR products amplified from pools of MEFs after transfection with ObLiGaRe donor alone (D) or with Nras ZFN plasmid (D+Z). p1-p2 primers amplify the 5′ junction and p3-p4 primers amplify the 3′ junction. The table lists the sequences of individually isolated PCR fragments. Deletions are indicated as red dotted lines; insertions, in red capital letters.
ObLiGaRe mediated insertion of a 15-kb inducible gene expression cassette at the AAVS1 locus
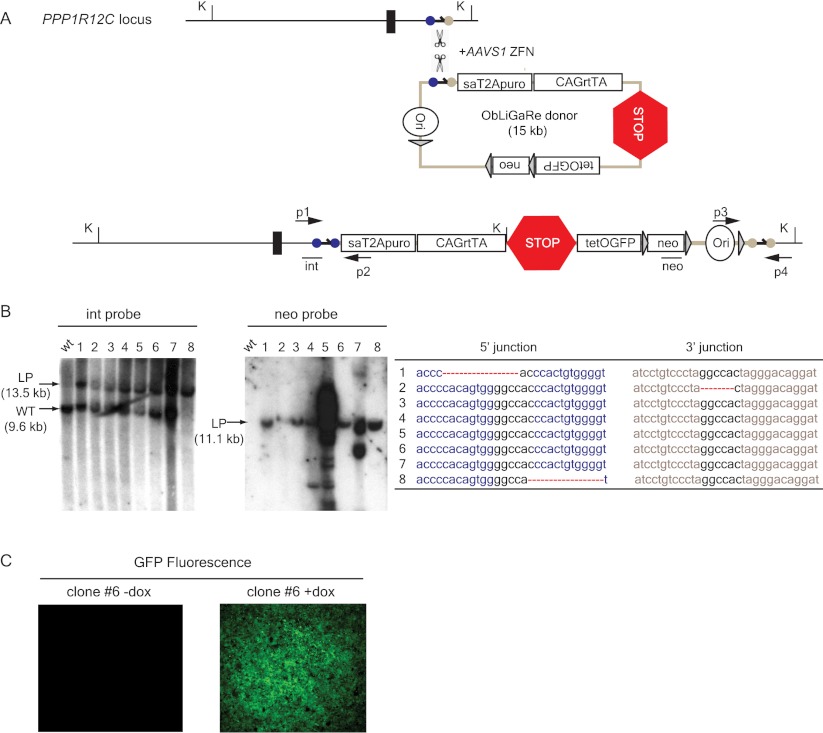
Previously, inducible transgene expression from the AAVS1 locus was achieved by targeting a tetracycline controlled responder in the AAVS1 locus followed by delivery of the reverse tetracycline transcription activator (rtTA) via lentiviral transduction or gene targeting in a second allele of AAVS1 (Hockemeyer et al. 2009; DeKelver et al. 2010). We determined whether ObLiGaRe could facilitate insertion of the entire 15-kb inducible transgenic cassette into the AAVS1 locus in a single step. We inserted the modified AAVS1 ZFN site at the 5′ of an inducible cassette that contains all the components for doxycycline-induced transgene expression in mice (Y Yang, unpubl.) and cotransfected it with AAVS1 ZFN plasmid in HCT116 cells. We screened 18 puromycin-resistant clones for correct integration by PCR and identified 14 with the expected PCR product (data not shown). We chose eight positive clones for further analysis by Southern blot and showed that they had either heterozygous (clones 1–7) or homozygous (clone 8) insertions of the vector at the AAVS1 locus (Fig. 4B, int probe). We observed precise end joining at both the 5′ and 3′ ends in the majority of these eight clones (Fig. 4B). We also noted that clones 4, 5, and 7 had additional integration events (Fig. 4B, neo probe). Furthermore, we could induce GFP expression by adding doxycycline in all eight clones as shown with clone 6 (Fig. 4C).
Figure 4.
Introduction of a 15-kb inducible gene expression cassette in the AAVS1 locus by single-step ObLiGaRe. (A) Strategy to introduce an inducible gene expression cassette. ZFN cutting sites are the same as in Figure 1. The expression of rtTA is controlled by the constitutive CAG promoter, while GFP is under the doxycycline responsive promoter (tetO) (Gossen and Bujard 1992). The STOP sign indicates a transcription termination cassette. Int and neo represent the probes for Southern blot. p1-p2 and p3-p4 are PCR primers used to amplify the 5′ and 3′ junctions. K indicates KpnI. (B) Southern blot of eight positive clones is shown with both internal (int) and vector-specific probe (neo). (LP) Expected ligation product. The sequences for 5′ and 3′ junctions of the eight clones are reported in the table on the right with dotted lines indicating deletions. (C) GFP fluorescence is detected by fluorescence microscopy in cells from clone 6 without (−) and with (+) 48 h treatment of 1 μg/mL doxycycline (dox).
Mechanism of ObLiGaRe
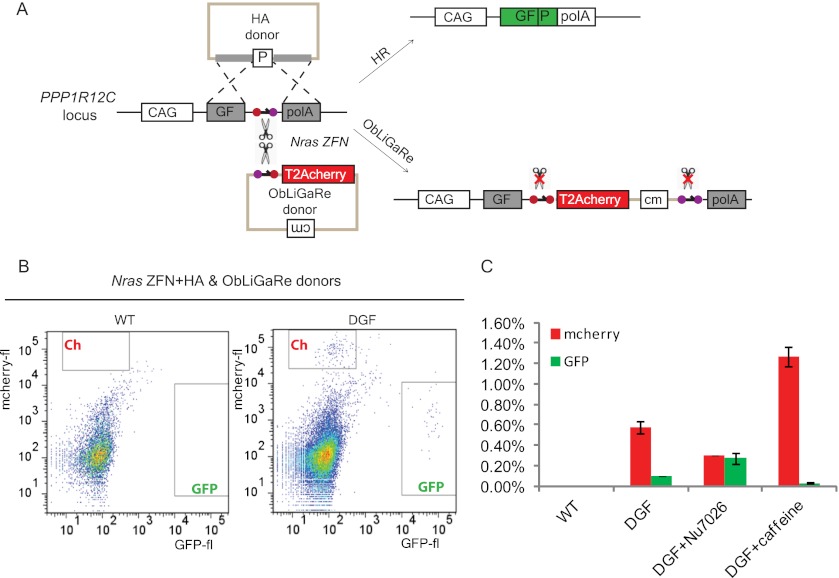
We speculated that ObLiGaRe might be mediated by NHEJ since it does not require any homology between the donor and the target. To test this hypothesis, we inserted a defective GFP (DGF) harboring a mouse Nras ZFN recognition site at the AAVS1 locus by HR in HCT116 cells (Fig. 5A). We chose one of the correctly targeted heterozygous clones as a reporter (clone 10, Supplemental Fig. 3). We designed two donor plasmids, one had 500-bp homology arms to the DGF and could reconstitute functional GFP by HR; the other had a modified Nras ZFN site 5′ to a promoter-less T2A-mCherry cassette, which would lead to mCherry expression upon insertion by ObLiGaRe (Fig. 5A). We transfected Nras ZFN plasmid with both ObLiGaRe and HR donors into the reporter cell line and measured GFP and mCherry-positive cells by FACS. We detected six times more mCherry-positive cells than GFP-positive cells in the reporter line but not the parental line (Fig. 5B). All clones derived from the mCherry fluorescent cells presented precise end joining of the mCherry with the defective GFP (data not shown). Interestingly, when we added a potent inhibitor of DNA-PK (a key component of NHEJ) (Hollick et al. 2003) to the medium, the number of mCherry fluorescent cells was significantly reduced, whereas that of GFP fluorescent cells was increased (P < 0.001) (Fig. 5C). Conversely, a significant increase of mCherry fluorescent cells with a concomitant decrease of GFP fluorescent cells (P < 0.001) (Fig. 5C) was observed after adding 4 mM caffeine (a nonspecific inhibitor of the ATM and ATR kinases involved in HR) to the medium (Sarkaria et al. 1999). Taken together, these data suggest that ObLiGaRe is mediated via the NHEJ pathway.
Figure 5.
ObLiGaRe is mediated via NHEJ. (A) Illustration of the reporter DGF cassette in the AAVS1 locus. ZFN sites are the same as in Figure 3. A functional GFP is generated after HR using the HA donor, while a functional mCherry is inserted upon ObLiGaRe using the ObLiGaRe donor. The two donors are about the same size. (B) Determination of fluorescent cell number after cotransfection of Nras ZFN, ObLiGaRe, and HA donors by FACS. Detection of GFP and mCherry cells were gated using parental cells (WT, left), which did not show any fluorescent cells upon cotransfection. When the reporter cell line was cotransfected with the three constructs, GFP and mCherry cells were detected in their corresponding gates (DGF, right). (C) Representation of the percentage of GFP- and mCherry-positive cells out of total live cells in WT HCT116 cells, of reporter HCT116 cells without treatment (DGF), or after treatment with 10 μM Nu7026 or 4 mM caffeine. Error bars, SD; n = 3.
Discussion
Targeted gene modification in mammalian cells has been achieved almost exclusively through laborious homologous recombination techniques and has been limited to cells where this pathway seems particularly active. Though CDNs have made gene targeting feasible in cell lines that have low intrinsic HR efficiency, NHEJ is still the dominant DSB repair pathway. As a result, knockout (KO) alleles generated by NHEJ are obtained at a much higher frequency than knock-in (KI) alleles generated by HR. A deficiency in NHEJ can significantly promote HR as seen in Escherichia coli in which NHEJ is negligible (Liang et al. 1996; Beumer et al. 2008; Maresca et al. 2010).
In this study we developed ObLiGaRe, a strategy of site-specific gene insertion utilizing the NHEJ pathway. It applies a similar logic to the one used in unidirectional loxP sites (Oberdoerffer et al. 2003) but maintains all the advantages and flexibility of CDNs. The alternate design of the ZFNs site that we insert in the vector is necessary and sufficient to achieve precise end joining of the vector in the genomic ZFN site. Though it is less frequent, we also observed aberrant joining products involving small deletions or insertions at the junctions in HCT116 cells (Figs. 1, 4; Supplemental Table 1). We speculated that these might result from processing alternative overhang types generated by the same ZFNs that were mismatched (Smith et al. 2000; Zeevi et al. 2008; Hockemeyer et al. 2009; Orlando et al. 2010). Furthermore, genetic and cellular context might influence the ligation precision, as observed in KBM7 cells (Fig. 1B). Nevertheless, we never observed precise insertion of the vector at targeted genomic locus when a ZFNs site identical to the one present in the genome is introduced in the vector. In fact no integration of a surrogate reporter plasmid bringing a ZFN site identical to the one in the genome was observed in a recent study (Kim et al. 2011). While our manuscript was under revision, Cristea et al. (2012) reported that introducing a ZFN site in a donor plasmid could promote its integration into the ZFN targeting site in the genome. Because the same ZFN sites were used in donor plasmids, only a minority of genomic insertions had precise 5′ and 3′ junctions, presumably due to repetitive digestions by ZFNs (Cristea et al. 2012). Interestingly, they found that inefficient donor integration could also happen when the ZFNs used for donor linearization and chromosomal cleavage were not identical. This is in agreement with what Orlando et al. (2010) reported previously that double-stranded oligos with mismatched overhangs to the DSB generated by ZFNs can be captured though at much lower frequency compared with those with complementary overhangs. ObLiGaRe should be more efficient under circumstances where precise ligation is critical, for example, to make an in-frame fusion between a reporter and the endogenous gene.
ObLiGaRe eliminates cloning homology arms into the donor vector and does not require any previous knowledge of the overhangs generated by specific CDNs. We are currently modifying the donor plasmids so that the vector backbone can be removed after integration using Cre or Flpe recombinases. Alternatively the ObLiGaRe donors lacking the vector backbone can be generated from minicircle DNA vectors (Kay et al. 2010).
Although we have tested ObLiGaRe mostly with ZFNs in this study, we found it also worked with AAVS1 TALENs (Supplemental Fig. 4). We also showed that it worked in a variety of cell types, including KBM7 and C2C12 cells, in which precise gene insertion has not previously been demonstrated. We are currently testing whether it can be used in primary, nonreplicating cells in which NHEJ seems to be the predominant pathway to repair DSBs. We are also exploring whether it can work in mouse and zebrafish embryos. Finally we used ObLiGaRe to insert the largest construct in the genome to date by ZFNs (Fig. 3), and we predict that we can use it to deliver even larger constructs, such as BACs, into a predefined genomic locus, which is very challenging, if not impossible, to achieve via HR.
Beyond the practical value of our method, ObLiGaRe can be used as a new tool in the study of NHEJ and the crosstalk between NHEJ and HR. Although similar strategies were reported using the I-SceI homing endonuclease (Certo et al. 2011), our strategy allows us to track any endogenous locus that can be targeted by ZFNs. We are interested in using this reporter system to monitor differential utilizations of NHEJ and HR in cells before and after oncogenic transformation. We are also continuing to identify chemical and genetic modulators that influence the cells to choose NHEJ or HR for DDR.
Methods
Cell culture and transfection
HCT116 and RKO (American Type Culture Collection) were cultured in McCoy's 5A medium (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies). C2C12 (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Life Technologies). KBM7 cells were cultured in IMDM Glutamax (Life Technologies) supplemented with 10% fetal bovine serum. HCT116, RKO, C2C12, and MEFs cells were plated in six-well plates (50%–80% confluence) and transfected with Lipofectamine LTX according to the manufacturer's instructions. Briefly, 2 μg of ZFNs plasmid and 0.5 μg of ObLiGaRe vector were mixed with 12 μL of Lipofectamine LTX reagent and cotransfected into 5 × 105 cells. Two days after transfection, cells were transferred to 15-cm plates and subjected to either puromycin (Life Technologies) selection (0.5 μg/mL) or G418 (Life Technologies) selection (100 μg/mL) the following day. GFP-positive, puromycin-resistant clones were harvested after 15 d of selection. We electroplated 107 KBM7 cells with 8 μg of ZFN plasmids and 2 μg of ObLiGaRe donor using a Gene PulserII electroporation system (Bio-Rad) with electrical settings of 250 V and 950 μF. GFP-positive cells were sorted by FACS (FACAria, Becton Dickinson) 2 d after transfection, and 100 GFP-positive cells were seeded in 12 wells and selected with puromycin (1.0 μg/mL); GFP-positive, puromycin-resistant clones (pools) were harvested after 10 d of selection.
Comparison of HR to ObLiGaRe
We transfected 2 × 105 reporter cells with 0.2 μg ZFN plasmid, 0.3 μg ObLiGaRe donor (Ob), and 0.5 μg HA donor plasmids (HA), using 4 μL Lipofectamine LTX in 12 wells.
The percentage of GFP and mCherry-positive cells was measured using a FACS cantoII flow cytometer (Becton Dickinson) 4 d after transfection.
Treatment with Nu7026 (Sigma Aldrich) was started 1 d before transfection with a final concentration of 10 μM Nu7026 (from a 10 mM stock solution in DMSO) and was continued for 2 d after transfection. ANOVA and Tukey's HSD tests were adopted to perform statistical analysis using R statistics software.
ZFN expression plasmids
ZFNs against the PTEN and Nras loci were designed and manufactured by Sigma-Aldrich. ZFNs and TALENs used in this work carried obligate heterodimer forms of the FokI endonuclease. AAVS1 ZFNs and TALENs were made according to the method described by Hockemeyer et al. (2009, 2011). The ZFN expression constructs were obtained by Sigma-Aldrich but modified to insert both ZFNs in one plasmids using ad hoc recombination (M Maresca, unpubl.). The sequence of linking the AAVS1 ZFN pair is shown in Supplemental Figure 5. SURVEYOR assay was performed according to the manufacturer's instructions (Transgenomic) using the primers listed in Table 1.
Table 1.
PCR primers
Acknowledgments
We thank Yan Feng and Yegor Smurnyy for providing KBM7 cells and growth conditions; Matthias Mueller and Bernd Kinzel for providing the STOP cassette; Alan Ho and Akos Szilvasi for FACS assistance; Hong Lei for providing MEF cells; Qing Fang for confirming karyotypes of C2C12 cells; John Slonimsky for negotiating contracts with Sigma-Aldrich; and Leslie Pond, Alan Buckler, Bernd Kinzel, William Dietrich, Peter Finan, Mark Labow, and Jeff Porter for scientific discussions and providing institutional support. We especially thank William Dietrich and Mark Labow for critical reading of the manuscript.
Author contributions: M.M. and Y.Y. conceived the study and wrote the manuscript. M.M. performed most experiments. V.G.L. and N.G. performed individual experiments. V.G.L. performed statistical analysis.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.145441.112.
References
- Andersson BS, Collins VP, Kurzrock R, Larkin DW, Childs C, Ost A, Cork A, Trujillo JM, Freireich EJ, Siciliano MJ, et al. 1995. KBM-7, a human myeloid leukemia cell line with double Philadelphia chromosomes lacking normal c-ABL and BCR transcripts. Leukemia 9: 2100–2108 [PubMed] [Google Scholar]
- Beumer KJ, Trautman JK, Bozas A, Liu JL, Rutter J, Gall JG, Carroll D 2008. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci 105: 19821–19826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D 2003. Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. [DOI] [PubMed] [Google Scholar]
- Brown KD, Rathi A, Kamath R, Beardsley DI, Zhan Q, Mannino JL 2003. The mismatch repair system is required for S-phase checkpoint activation. Nat Genet 33: 80–84 [DOI] [PubMed] [Google Scholar]
- Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, et al. 2009. Haploid genetic screens in human cells identify host factors used by pathogens. Science 326: 1231–1235 [DOI] [PubMed] [Google Scholar]
- Casas-Delucchi CS, Brero A, Rahn HP, Solovei I, Wutz A, Cremer T, Leonhardt H, Cardoso MC 2011. Histone acetylation controls the inactive X chromosome replication dynamics. Nat Commun. 2: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo MT, Ryu BY, Annis JE, Garibov M, Jarjour J, Rawlings DJ 2011. Tracking genome engineering outcome at individual DNA breakpoints. Nat Methods 8: 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Frodin M, Davis GD 2011. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods 8: 753–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea S, Freyvert Y, Santiago Y, Holmes MC, Urnov FD, Gregory PD, Cost GJ 2012. In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol Bioeng doi: 10.1002/bit.24733 [DOI] [PubMed] [Google Scholar]
- Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ 2011. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol 29: 64–67 [DOI] [PubMed] [Google Scholar]
- DeKelver RC, Choi VM, Moehle EA, Paschon DE, Hockemeyer D, Meijsing SH, Sancak Y, Cui X, Steine EJ, Miller JC, et al. 2010. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res 20: 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Gregory PD, Holmes MC 2011. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8: 74–79 [DOI] [PubMed] [Google Scholar]
- Fung H, Weinstock DM 2011. Repair at single targeted DNA double-strand breaks in pluripotent and differentiated human cells. PLoS ONE 6: e20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, et al. 2011. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol 29: 816–823 [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci 89: 5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. 2009. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Santiago Y, Miller JC, Zeitler B, et al. 2011. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29: 731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick JJ, Golding BT, Hardcastle IR, Martin N, Richardson C, Smith GCM, Griffin RJ 2003. 2,6-Disubstituted pyran-4-one and thiopyran-4-one inhibitors of DNA-dependent protein kinase (DNA-PK). Bioorg Med Chem Lett 13: 3083–3086 [DOI] [PubMed] [Google Scholar]
- Kay MA, He CY, Chen ZY 2010. A robust system for production of minicircle DNA vectors. Nat Biotechnol 28: 1287–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-G, Cha J, Chandrasegaran S 1996. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93: 1156–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Um E, Cho SR, Jung C, Kim H, Kim JS 2011. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat Methods 8: 941–943 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ruan S, Jabbur JR, Consoli U, Clodi K, Shiku H, Owen-Schaub LB, Andreeff M, Reed JC, Zhang W 1998. Differential p53 phosphorylation and activation of apoptosis-promoting genes Bax and Fas/APO-1 by irradiation and ara-C treatment. Cell Death Differ 5: 584–591 [DOI] [PubMed] [Google Scholar]
- Kotecki M, Reddy PS, Cochran BH 1999. Isolation and characterization of a near-haploid human cell line. Exp Cell Res 252: 273–280 [DOI] [PubMed] [Google Scholar]
- Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, et al. 2011. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475: 217–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Romanienko PJ, Weaver DT, Jeggo PA, Jasin M 1996. Chromosomal double-strand break repair in Ku80-deficient cells. Proc Natl Acad Sci 93: 8929–8933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca M, Erler A, Fu J, Friedrich A, Zhang Y, Stewart AF 2010. Single-stranded heteroduplex intermediates in Red homologous recombination. BMC Mol Biol 11: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, De Angelis MH, Wurst W, Kuhn R 2010. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci 107: 15022–15026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee Y-L, Rupniewski I, Waite AJ, Wang NS, Kim KA, Gregory PD, et al. 2007. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785 [DOI] [PubMed] [Google Scholar]
- Mittelman D, Moye C, Morton J, Sykoudis K, Lin Y, Carroll D, Wilson JH 2009. Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc Natl Acad Sci 106: 9607–9612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Holmes MC 2007. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci 104: 3055–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Otipoby KL, Maruyama M, Rajewsky K 2003. Unidirectional Cre-mediated genetic inversion in mice using the mutant loxP pair lox66/lox71. Nucleic Acids Res 31: e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando SJ, Santiago Y, DeKelver RC, Freyvert Y, Boydston EA, Moehle EA, Choi VM, Gopalan SM, Lou JF, Li J, et al. 2010. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res 38: e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petek LM, Russell DW, Miller DG 2010. Frequent endonuclease cleavage at off-target locations in vivo. Mol Ther 18: 983–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D 2003. Chimeric nucleases stimulate gene targeting in human cells. Science 300: 763. [DOI] [PubMed] [Google Scholar]
- Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH 2008. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther 16: 707–717 [DOI] [PubMed] [Google Scholar]
- Radecke S, Radecke F, Cathomen T, Schwarz K 2010. Zinc-finger nuclease-induced gene repair with oligodeoxynucleotides: Wanted and unwanted target locus modifications. Mol Ther 18: 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam S, Kandavelou K, Rajenderan R, Chandrasegaran S 2011. Creating designed zinc-finger nucleases with minimal cytotoxicity. J Mol Biol 405: 630–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M 1994. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol 14: 8096–8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res 59: 4375–4382 [PubMed] [Google Scholar]
- Skorski T 2008. BCR/ABL, DNA damage and DNA repair: Implications for new treatment concepts. Leuk Lymphoma 49: 610–614 [DOI] [PubMed] [Google Scholar]
- Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D 2000. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res 28: 3361–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T 2007. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol 25: 786–793 [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD 2010. Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11: 636–646 [DOI] [PubMed] [Google Scholar]
- Xu Z-L, Mizuguchi H, Ishii-Watabe A, Uchida E, Mayumi T, Hayakawa T 2001. Optimization of transcriptional regulatory elements for constructing plasmid vectors. Gene 272: 149–156 [DOI] [PubMed] [Google Scholar]
- Zeevi V, Tovkach A, Tzfira T 2008. Increasing cloning possibilities using artificial zinc finger nucleases. Proc Natl Acad Sci 105: 12785–12790 [DOI] [PMC free article] [PubMed] [Google Scholar]