Abstract
Improved methods for engineering sequence-specific nucleases, including zinc finger nucleases (ZFNs) and TAL effector nucleases (TALENs), have made it possible to precisely modify plant genomes. However, the success of genome modification is largely dependent on the intrinsic activity of the engineered nucleases. In this study, we sought to enhance ZFN-mediated targeted mutagenesis and gene targeting (GT) in Arabidopsis by manipulating DNA repair pathways. Using a ZFN that creates a double-strand break (DSB) at the endogenous ADH1 locus, we analyzed repair outcomes in the absence of DNA repair proteins such as KU70 and LIG4 (both involved in classic nonhomologous end-joining, NHEJ) and SMC6B (involved in sister-chromatid-based homologous recombination, HR). We achieved a fivefold to 16-fold enhancement in HR-based GT in a ku70 mutant and a threefold to fourfold enhancement in GT in the lig4 mutant. Although the NHEJ mutagenesis frequency was not significantly changed in ku70 or lig4, DNA repair was shifted to microhomology-dependent alternative NHEJ. As a result, mutations in both ku70 and lig4 were predominantly large deletions, which facilitates easy screening for mutations by PCR. Interestingly, NHEJ mutagenesis and GT at the ADH1 locus were enhanced by sixfold to eightfold and threefold to fourfold, respectively, in a smc6b mutant. The increase in NHEJ-mediated mutagenesis by loss of SMC6B was further confirmed using ZFNs that target two other Arabidopsis genes, namely, TT4 and MPK8. Considering that components of DNA repair pathways are highly conserved across species, mutations in DNA repair genes likely provide a universal strategy for harnessing repair pathways to achieve desired targeted genome modifications.
Customizable, sequence-specific nucleases make it possible to precisely modify the genomes of many higher organisms, including diverse plant species (Voytas 2013). There are three primary types of sequence-specific nucleases, namely, zinc finger nucleases (ZFNs) (Kim et al. 1996; Carroll 2011), TAL effector nucleases (TALENs) (Christian et al. 2010; Bogdanove and Voytas 2011), and meganucleases (Smith et al. 2006; Paques and Duchateau 2007). All three nuclease types introduce targeted DNA double-strand breaks (DSBs), which activate the cell's DNA repair pathways, principally nonhomologous end-joining (NHEJ) and homologous recombination (HR) (Kanaar et al. 1998; Puchta 2005; Hartlerode and Scully 2009). NHEJ predominates in nonreplicating cells and repair is often imprecise, such that mutations are introduced at the cut site. HR, which enables gene replacement or gene targeting (GT), predominates in replicating cells. Because genome modifications made with customizable nucleases rely on DSB repair, it should be possible to manipulate DNA repair pathways to influence the type and frequency of targeted modifications attained. For example, promoting NHEJ should enhance nuclease-mediated site-specific mutagenesis; however, there are no reports of this approach being successful in plants. Strategies to enhance HR have been reported in a few plant studies; for example, high-frequency HR was obtained by overexpressing the yeast RAD54 gene (Shaked et al. 2005). Enhanced HR was also attained in Arabidopsis by knocking out RAD50 (Gherbi et al. 2001) or CAF1(Endo et al. 2006) or by overexpressing SMC6B (also known as MIM) (Hanin et al. 2000). Almost all of these studies used a GUS transgene reporter that measures intrachromosomal, single-strand annealing (SSA) and/or synthesis-dependent strand annealing (SDSA) in which sister chromatids or homologous chromosomes are used as repair templates (Orel et al. 2003; Puchta 2005). For GT, exogenous donors or chromosomally integrated, nonallelic donors are usually used. The repair mechanisms used in GT, therefore, could be quite different, and it remains to be determined whether previously demonstrated manipulations of DNA repair pathways will apply to GT of endogenous genes. One of the few attempts to influence GT in plants involved overexpression of the bacterial RecA protein, which enhanced the fidelity of DSB-induced GT in tobacco but not the overall GT frequency (Reiss et al. 2000).
In this study, we tested the consequence of manipulating key regulators of NHEJ and HR on targeted mutagenesis or GT mediated by sequence-specific nucleases. In classic NHEJ (C-NHEJ), KU70 dimerizes with KU80 to form KU protein complexes, which directly bind to DSBs to initiate repair (Pastwa and Blasiak 2003). In HR, SMC6 acts when sister chromatids are used as repair templates in both human (De Piccoli et al. 2006; Potts et al. 2006) and Arabidopsis cells (Watanabe et al. 2009). SMC6 dimerizes with SMC5 to form SMC6/5 complexes that initiate repair of DSBs using sister chromatids as templates (Potts 2009). The upstream positions of KU70 and SMC6 in the NHEJ or HR pathways, respectively, made them good candidates as regulators of DNA repair pathway choice. We also chose LIG4, because it carries out a well-known and conserved function downstream from KU (Pastwa and Blasiak 2003). A ZFN that targets the endogenous Arabidopsis ADH1 gene was used to create DSBs (Zhang et al. 2010), and NHEJ mutagenesis and GT were measured in whole seedlings and rosette leaf protoplasts in each of the three DNA repair mutant backgrounds. Our results clearly demonstrate that manipulating these key DNA repair genes in Arabidopsis can influence the efficiencies and outcomes of targeted mutagenesis and GT.
Results
Targeting a DSB at the ADH1 locus in different DNA repair mutants
We previously reported targeted mutagenesis at the Arabidopsis ADH1 locus using custom ZFNs expressed from an estradiol-inducible promoter (Supplemental Fig. S1A; Zhang et al. 2010). We generated a line (designated ADH1-ZFN-4) that has two ADH1 ZFN monomers expressed from a single mRNA and separated by the T2A translational skipping sequence. The ADH1 locus is wild type (WT) in this line and thus cleavable by the nuclease. Immunoblot analysis indicated that the ZFNs were induced by estrogen (Supplemental Fig. S1B), and the ADH1-ZFN transgene was mapped to chromosome 1 by TAIL-PCR (Supplemental Fig. S1C).
The ADH1-ZFN transgene was crossed into ku70, lig4, and smc6b backgrounds. We chose SMC6B for study because mutations in this gene (1) cause hypersensitivity to the DNA-damaging agent MMS (Mengiste et al. 1999); (2) show very slow DNA repair kinetics (Kozak et al. 2009); and (3) have more severe defects in DNA repair than the smc6a mutant (Watanabe et al. 2009). We obtained a new allele, smc6b-3, which was generated by a T-DNA insertion (Supplemental Fig. S1D). Full-length SMC6B mRNA was not detected by RT-PCR in the smc6b-3 mutant, suggesting it is a null allele. Because neither ku70, lig4, nor smc6b is closely linked to the ADH1-ZFN-4 transgene (Supplemental Fig. S1E), it was possible to obtain F3 plants that were homozygous for both the transgene and the mutations (Supplemental Fig. S1F).
High-frequency ZFN-mediated mutagenesis in the smc6b mutant
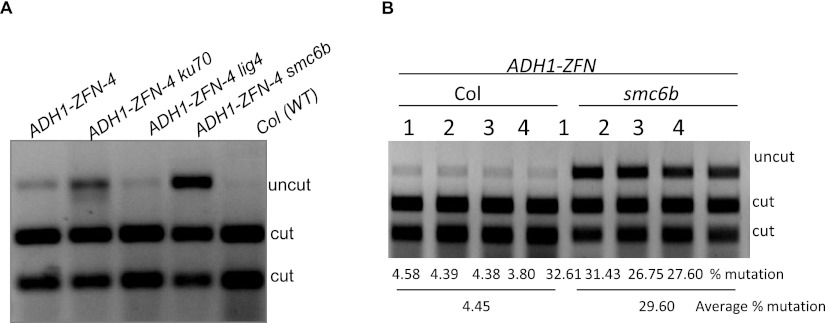
To compare ZFN-induced mutagenesis in the different genetic backgrounds, the mutant plant lines with the ADH1-ZFN transgene were grown on MS medium with estradiol to induce ZFN expression. Since an NlaIII restriction site overlaps with the ZFN cut site, PCR fragments that include the ZFN target site will be rendered undigestable by NlaIII if they sustain NHEJ-induced mutations (Zhang et al. 2010). All of the ZFN-expressing plants showed detectable levels of mutagenesis (Fig. 1A) with a strikingly higher level in smc6b mutants, approximating 30% of the PCR products or a sixfold increase over the WT (Fig. 1B).
Figure 1.
ZFN-mediated mutagenesis at the ADH1 locus in different DNA repair mutants. (A) ADH1-ZFN-mediated mutagenesis in Col (WT), ku70, lig4, and smc6 mutant backgrounds. DNA from 1-wk-old pooled seedlings grown on 20 μM estradiol MS medium was used for PCR amplification of the ZFN target site. The PCR products were digested with NlaIII. This experiment was repeated six independent times with similar results. (B) High-frequency mutagenesis in the smc6b mutant. Mutation frequencies were measured by dividing the signal intensity of the uncut band by the total signal from uncut and cut bands. Four biological replicates of ADH1-ZFN-4 and ADH1-ZFN-4 smc6b were used to calculate mutation frequencies.
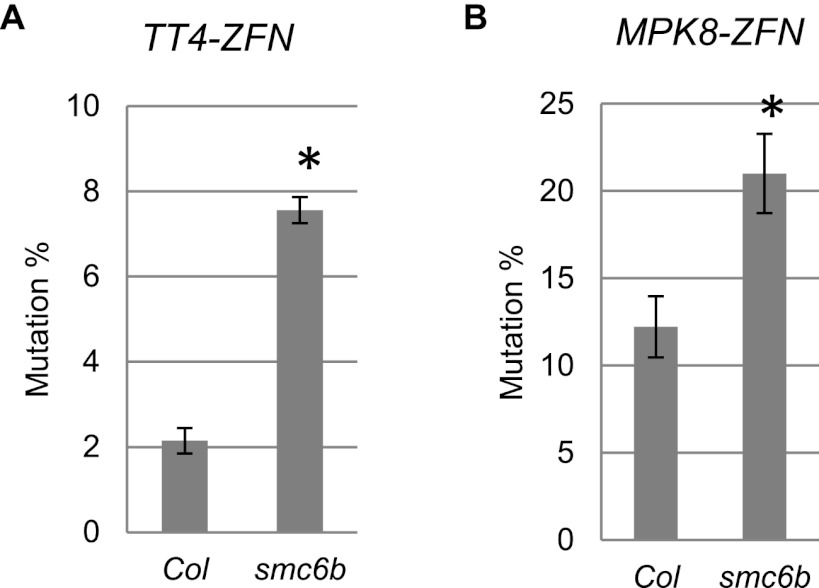
To test whether enhanced mutagenesis in the smc6b background occurs with other ZFNs, we introduced ZFNs targeting the Arabidopsis genes TT4 (At5g13930) (Zhang et al. 2010) and MPK8 (At1g18150) (Sander et al. 2010) into smc6b mutants by floral dip transformation. ZFN-mediated mutagenesis in pooled transgenic seedlings was measured, and a nearly fourfold increase in mutagenesis by the TT4-ZFN was observed in smc6b compared with WT (Fig. 2A; Supplemental Fig. S2A). The mutation frequency for the MPK8-ZFN increased approximately twofold in smc6b (Fig. 2B; Supplemental Fig. S2B). Differences in ZFN activity and chromosomal context may contribute to the observed differences in mutagenesis frequencies; nevertheless, these results clearly demonstrate that ZFN-mediated mutagenesis is significantly enhanced in the absence of SMC6B.
Figure 2.
High-frequency mutagenesis by other ZFNs in the smc6b background. (A) TT4-ZFN-mediated mutagenesis in Col and smc6 mutant backgrounds. (B) MPK8-ZFN-mediated mutagenesis in Col and smc6 mutant backgrounds. In both cases, DNA was prepared from 1-wk-old pooled T1 seedlings in Col or smc6 backgrounds. The seedlings were grown on 20 μM estradiol MS medium. Target sites were PCR-amplified and digested with NspI (for TT4-ZFN) or MslI (for MPK8-ZFN). Four biological replicates of each sample were used to calculate mutation frequencies (see Supplemental Fig. S2). (Error bars) Standard errors. (*) Statistically significant differences (P < 0.005, t-test).
ZFN-mediated mutagenesis and GT in leaf protoplasts
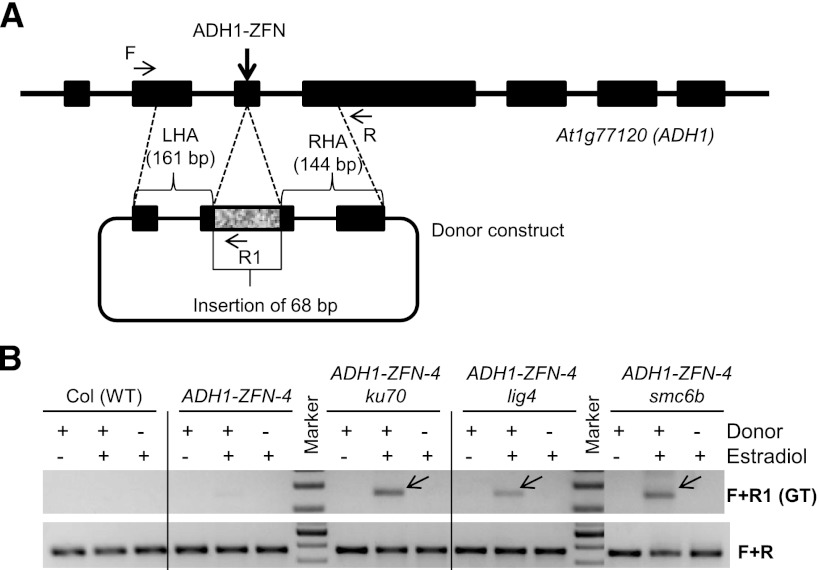
To compare ZFN-induced GT in different genetic backgrounds, we delivered a donor template concomitant with induction of ZFN expression by estradiol. The donor construct has homology arms each around 150 bp (Fig. 3A; Supplemental Fig. S3), and successful GT incorporates a 68-bp sequence and simultaneously deletes 12 bp at the ZFN cut site. GT was initially detected by PCR using a primer that anneals to a site within the 68-bp insertion and another that anneals to the flanking genomic DNA sequence outside of the homology arms (Fig. 3A). In samples with both the donor and the ZFN (induced by estradiol), a GT-specific product was reproducibly amplified from ADH1-ZFN-4 ku70, ADH1-ZFN-4 lig4, and ADH1-ZFN-4 smc6b protoplasts, but only faintly from ADH-ZFN-4 protoplasts (Fig. 3B). The GT-specific PCR bands were absent in all “donor only” samples when the ZFN was not induced (Fig. 3B), indicating that the observed PCR products are not likely due to DNA recombination during PCR amplification (Meyerhans et al. 1990). We conclude that GT is greatly enhanced by ZFN-induced DSBs, and that knocking out KU70, LIG4, or SMC6B attains an additional enhancement.
Figure 3.
ZFN-facilitated gene targeting at the ADH1 locus. (A) A schematic of the ADH1 locus is shown with exons depicted as black boxes. The donor plasmid shown below the locus contains a left homology arm (LHA) of 161 bp and a right homology arm (RHA) of 144 bp. Successful HR will result in an insertion of 68 bp and deletion of 12 bp at the ZFN cut site. (B) PCR-based detection of HR. Protoplasts derived from the five genotypes were treated with donor only, donor and estradiol, and estradiol only. As depicted in panel A, the ZY070-F and ZY073-R1 (F + R1) primer set was used to detect HR events. Amplification of the locus with the ZY070-F and ZY070-R (F + R) primer set was used as a PCR control. (Arrows) Amplification products due to HR. This experiment was repeated twice with similar results.
To confirm these results, PCR was conducted using primers that recognize sites completely outside of the homology arms of the donor; the population of amplified DNA fragments includes those with both NHEJ-induced mutations as well as GT events. Genomic DNA was digested with NlaIII prior to PCR amplification to increase the proportion of PCR products with alterations of the ZFN recognition site (and thereby the NlaIII restriction enzyme site; enrichment PCR, see Methods). Uncut PCR products, indicative of mutagenesis, were observed in samples derived from ADH1-ZFN-containing plants treated only with estradiol (Fig. 4A). Larger deletions were present in the ku70 and lig4 backgrounds relative to smc6b and WT, as indicated by the presence of shorter PCR products (Fig. 4A). Similar observations were made for samples treated with both the donor and estradiol (Fig. 4B). In addition, a larger DNA band was clearly detected in ADH1-ZFN-4 ku70, ADH1-ZFN-4 lig4, and ADH1-ZFN-4 smc6b protoplasts, but only faintly in the ADH-ZFN-4 protoplasts and not in the WT control (Fig. 4B). This DNA band was ∼50 bp larger than the WT PCR product, the size expected for GT events. Cloning and sequencing of this larger PCR product from the ADH1-ZFN-4 ku70 line confirmed that it was due to GT: 68 bp was inserted and 12 bp was deleted at the target site (Fig. 4C). Interestingly, GT seemed particularly enhanced in ku70 relative to the lig4 or smc6b backgrounds.
Figure 4.
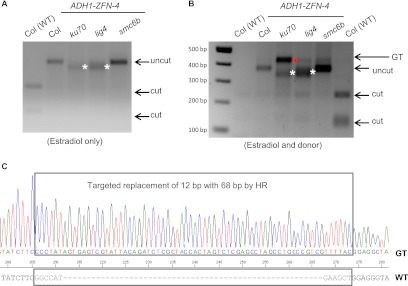
Enrichment PCR detection of ZFN-mediated NHEJ and GT at the ADH1 locus. (A) Detection of mutagenesis at the ADH1 locus. Protoplasts derived from the five genotypes were treated with estradiol. An enrichment PCR procedure (see Methods) was performed to detect ZFN-induced mutagenesis (uncut band). This experiment was repeated twice with similar results. (White asterisks) PCR products shorter than wild type (WT). (B) Simultaneous detection of mutagenesis and GT at the ADH1 locus. Protoplasts derived from the five genotypes were treated with both estradiol and the donor. An enrichment PCR procedure was performed to detect ZFN-induced mutagenesis (lower uncut band) and GT (upper uncut band) (red asterisk). (White asterisks) PCR products shorter than WT. Note that for one of the two WT controls (lane 1), the first round of NlaIII digestion of the WT DNA was so complete that very little PCR product was obtained for the second round of digestion. This experiment was repeated twice with similar results. (C) DNA sequence confirmation of gene targeting events. The larger PCR band indicated by a red asterisk in panel B in the ku70 background was cloned and sequenced. The upper box indicates a sequencing trace expected for an HR product. The lower box indicates the 12 bp in the WT target locus that was deleted through GT.
Mutation profiling by 454 sequencing
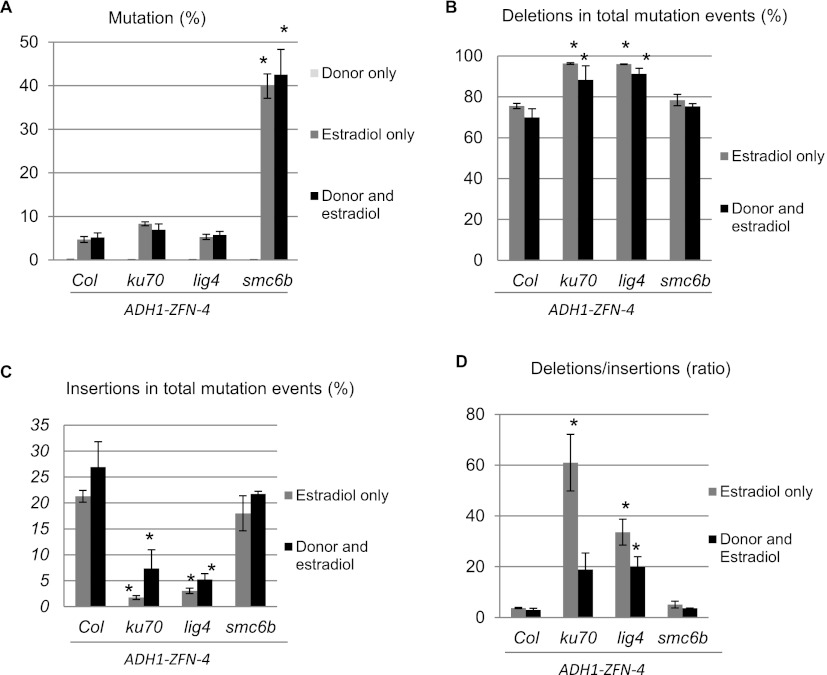
The PCR products obtained using primers outside of the homology arms were subjected to 454 Life Sciences (Roche) pyro-sequencing to assess outcomes of DSB repair at the nucleotide level. PCR products derived from protoplasts treated with donor only served as negative controls and established the baseline level of PCR and DNA sequencing errors, which ranged from 0.08% to 0.11% of the sequencing reads (Fig. 5A; Supplemental Table S1). As expected, the mutation frequency in smc6b background was very high—approximating 40% of the sequencing reads and representing an eightfold increase over WT. In contrast, the mutation frequency in ku70 or lig4 was not significantly different from WT, and presence of the donor did not appreciably impact mutation frequencies across all genotypes.
Figure 5.
Mutagenesis profiles revealed by 454 sequencing. (A) The total mutation (insertion, deletion, and nucleotide substitution) frequency for each sample was scored. (B) Proportion of deletions in total mutation events in different genotypes under different treatments. (C) Proportion of insertions in total mutation events in different genotypes under different treatments. (D) Relative ratio of deletion events to insertion events in different genotypes under different treatments. Protoplasts derived from Col, ku70, lig4, and smc6b plants and carrying the same ADH1-ZFN-4 transgene were treated with donor, estradiol, or donor and estradiol. Two (donor treatment) or three (estradiol, donor and estradiol) independent biological replicates were subjected to 454 high-throughput sequencing and analysis. (Error bars) Standard errors. In all panels, statistically significant differences between the mutants and Col (WT) are indicated by asterisks (P < 0.05, t-test).
The majority of mutations recovered in the various genetic backgrounds were deletions—roughly three quarters in WT and smc6b (Fig. 5B). The proportion of deletions in ku70 or lig4 was significantly higher and approximated 90% of all mutations (Fig. 5B). Conversely, the proportion of insertions in ku70 or lig4 was significantly lower than in WT or smc6b (Fig. 5C). As a consequence, the ratio of deletions to insertions increased approximately fourfold in WT to ∼61-fold in ku70 and ∼34-fold in lig4, clearly demonstrating that both mutants are more likely to have DSB-induced deletions than insertions (Fig. 5D). Notably, the ratio of deletions to insertions in ku70 dropped nearly 70% in the presence of the donor, whereas a more modest decrease was observed in lig4; smc6b and WT strains showed no statistical difference in the presence or absence of donor (Fig. 5D). As indicated below, this shift was likely due to an enhancement in GT when NHEJ is impaired and implies that KU70 plays a more important role than LIG4 in controlling DNA repair pathway choice.
Microhomology-mediated alternative NHEJ in ku70 and lig4
We further examined deletion profiles in all samples treated with either estradiol (Fig. 6) or both donor and estradiol (Supplemental Fig. S4). Overall, the results from both conditions were quite comparable. About 80% of deletions in WT and smc6b were <10 bp; the remaining 20% predominantly ranged from 10 to 49 bp (Fig. 6A; Supplemental Fig. S4A). In contrast, for ku70 or lig4, deletions from 1 to 9 bp were extremely rare and larger deletions predominated. For example, ∼20% of deletions in ku70 were longer than 100 bp. Differences were also observed in deletion profiles between ku70 and lig4: there were a greater number of 10- to 49-bp deletions in lig4 than in ku70, whereas ku70 had more deletions longer than 100 bp.
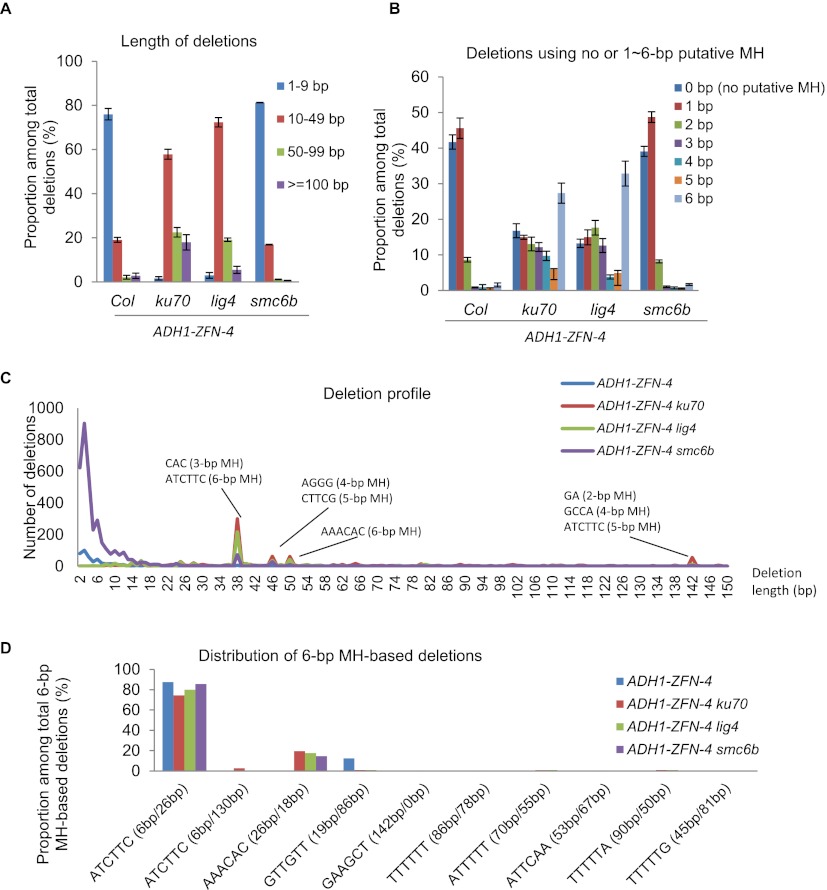
Figure 6.
Microhomology (MH)–based deletions predominate in ku70 and lig4. (A) Distribution of deletions by length. (Error bars) Standard errors. (B) Distribution of deletions using no or 1–6 bp of MH. (Error bars) Standard errors. (C) Profile of 2- to 150-bp deletions. MH sequences contributing to large deletion peaks are indicated. (D) Distribution of deletions that use different 6 bp of MH around the ZFN site. The distances of the left MH sequence and the right MH sequence to the DSB are indicated in the parentheses and separated by a slash. Data for all panels were generated from protoplasts derived from Col, ku70, lig4, and smc6b plants that carried the same ADH1-ZFN-4 transgene. Protoplasts were treated with estradiol, and three independent biological replicates were used for 454 sequencing and analysis.
The differences in deletion profiles between WT and smc6b versus ku70 and lig4 suggested a shift in the repair mechanism used to correct the DSB, and thus we looked for evidence of microhomology (MH) at repair sites indicative of alternative NHEJ (A-NHEJ). We found MH ranging from 1 to 6 bp (Fig. 6B; Supplemental Fig. S4B; Supplemental Table S2). In a rare case, 7 bp (TTTTAGA) of MH was used (Supplemental Table S2), resulting in a 147-bp deletion. In either WT or the smc6b background, nearly 90% of the deletions used no MH or MH of 1 bp in length, and the remaining 10% of deletions almost exclusively using MH of 2 bp (Fig. 6B; Supplemental Fig. S4B). Since deletions using 1 or 2 bp of MH could happen quite frequently by chance via classic KU70/LIG4-dependent NHEJ, they are less likely to represent events repaired by A-NHEJ. The fact that MH sequences of >2 bp, especially 6 bp, were frequently used in ku70 and lig4 strongly suggests that MH-dependent repair was used in these mutants.
To further explore whether the larger deletions resulted from MH-dependent repair, we examined the lengths of all deletions from 2 to 150 bp in all four genotypes (Fig. 6C; Supplemental Fig. S4C). Four frequently occurring deletion sizes were identified that were 38 bp, 46 bp, 50 bp, and 142 bp in length. Analysis of these deletions revealed in each case that there was one or a few base pairs of MH at the repair site (Fig. 6C; Supplemental Fig. S4C; Supplemental Table S3). This analysis not only revealed that MH was responsible for making the larger deletions, but also suggested that MH-based repair was used in WT and smc6b, although less often.
Stretches of 6 bp of MH were used most frequently among all deletions (Fig. 6B; Supplemental Fig. S4B), including the motifs “ATCTTC (6bp/26bp)” and “AAACAC (26bp/18bp)” (Fig. 6D; Supplemental Fig. S4D). In both cases, the 6 bp of MH is located very close to the ZFN cut site and resulted in deletion sizes of 38 bp and 50 bp, respectively. The frequency of using the “AAACAC (26bp/18bp)” MH was only about one quarter of that of “ATCTTC (6bp/26bp),” although there was only a 12-bp difference for their deletion sizes. This suggests that the mechanism of searching for MH from the DSB is very efficient, likely obeying a “first come, first use” rule. Further evidence comes in comparing deletions resulting from the same 6 bp of MH located at different distances from the cut site: ATCTTC (6bp/26bp) and ATCTTC (6 bp/130 bp). The 6 bp of MH closest to the DSB was used almost exclusively relative to the distal site, suggesting that the MH-dependent repair machinery searches for MH outward from the DSB in a very efficient manner, rarely skipping the first available region of MH.
High-frequency GT in ku70 mutants
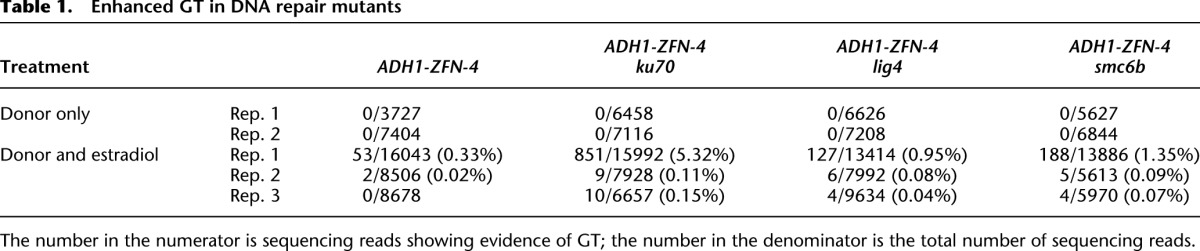
As revealed by both PCR (Fig. 3B) and enrichment PCR (Fig. 4B), GT was enhanced in all three DNA repair mutants, especially in ku70 (Fig. 4B). We used 454 pyro-sequencing to quantify these observed enhancements. No GT events were found for all “donor only” samples in both biological replicates, indicating that the frequency of GT was <0.01% (the number of sequencing reads ranged from 3727 to 7404) (Table 1). In contrast, HR events were detected in all samples treated with both the donor and estradiol with one exception: No HR was detected for the ADH1-ZFN-4 sample in replicate 3. The data clearly demonstrate that a DSB created by the ZFN greatly stimulates GT. We observed variation in GT frequencies between biological replicates, which was likely due to differences in the efficiency of donor delivery and donor plasmid quality. Nevertheless, the overall pattern was clear: GT was enhanced in all three mutants, and there was at least a threefold to fourfold increase in lig4 or smc6b across all three experiments. Application of the Cochran-Mantel-Haenszel test demonstrated that the enhancement in GT in the three independent replicates was reproducibly higher than WT (P = 8.7 × 10−164 for ku70, P = 8.6 × 10−13 for lig4, and P = 1.1 × 10−24 for smc6b). In replicate 1, which contained the highest number of sequencing reads, the GT frequency in ku70 was >5%, which was even higher than the NHEJ mutagenesis frequency (4.21%) in the same sample and represented a 16-fold increase in GT from WT (Table 1; Supplemental Table S1). Consistent with the enrichment PCR results (Fig. 4B), the 454 sequencing data indicate that loss of KU70 has a greater effect than deficiency in LIG4 or SMC6B in promoting HR-based GT.
Table 1.
Enhanced GT in DNA repair mutants
In plants, synthesis-dependent strand annealing (SDSA) is the primary mechanism by which DSBs are repaired (Formosa and Alberts 1986; Puchta 2005). In SDSA, one end of the break is typically repaired by HR and the other by NHEJ (Puchta et al. 1996). This would yield some events with insertions introduced by HR that are flanked by NHEJ-induced mutations. We analyzed the DNA sequences of all GT events and found examples in the mutant backgrounds indicative of repair by both HR and NHEJ (Supplemental Tables S4, S5). In ku70, for example, ∼11% of the GT events were associated with mutations on one side of the targeted insertion. It is likely that SDSA was also used to repair breaks in wild type, but the number of recovered events (i.e., 55) was likely too small to detect targeted insertions flanked by NHEJ-induced mutations.
Discussion
Precise genome modifications created by sequence-specific nucleases have considerable value for both basic and applied plant biology. To increase their efficacy, most effort has focused on improving the intrinsic activities of these nucleases. In this study, we demonstrate that by manipulating DNA repair pathways, ZFN-induced genome modification frequencies can be increased and the types of modifications that result from cleavage can be predicted and controlled.
Among the three DNA repair mutants tested, we found that a deficiency in SMC6B greatly enhanced ZFN-mediated mutagenesis at three different endogenous gene targets. Since SMC6B appears to have redundant functions with SMC6A (Watanabe et al. 2009), knocking down SMC6A in an smc6b mutant may lead to even higher frequencies of mutagenesis. Notably, ku70 or lig4 mutations did not significantly alter mutagenesis frequencies, consistent with a previous report that ZFN-mediated mutagenesis was not improved in a ku80 mutant (Osakabe et al. 2010).
Analysis of DNA repair products provided compelling evidence in Arabidopsis for an MH-dependent A-NHEJ pathway, which recent work suggests involves multiple compensatory subpathways (Charbonnel et al. 2011). A-NHEJ was used in WT cells infrequently, but very often when C-NHEJ was disrupted by knocking out KU70 or LIG4. The fidelity with which DNA breaks were repaired by C-NHEJ and A-NHEJ was comparable, because mutation frequencies did not differ when either pathway predominated. Thus, for the purpose of improving mutagenesis, it is unlikely to be useful to switch between these two NHEJ pathways. The use of ku70 or lig4 backgrounds, however, could facilitate screens for targeted mutations. Approximately 40% of the deletions were >50 bp in these mutants, and such deletions, when amplified by PCR, should be distinguishable from WT amplicons when separated using a high-percentage agarose or polyacrylamide gels. This would expedite and reduce expenses when screening for mutations over methods that use either PCR and restriction enzyme digestion (Guschin et al. 2010; Zhang et al. 2010) or DNA sequencing.
We previously reported efficient GT at an endogenous locus in tobacco using ZFNs (Townsend et al. 2009). Here, we demonstrate GT at an endogenous locus in Arabidopsis and that GT frequencies can be enhanced in ku70, lig4, and smc6b backgrounds. Loss of KU70 had the biggest effect—resulting in increases in GT as high as 16-fold over WT. Our data are consistent with studies in other higher organisms. For example, knocking out one copy of KU70 resulted in a fivefold to 10-fold increase in GT in human cells (Fattah et al. 2008), and in Drosophila, knocking out LIG4 promoted roughly a threefold increase in HR (Bozas et al. 2009). The significant differences that we observed between ku70 and lig4 in terms of the ratio of deletions to insertions, the size of the deletions, and the frequencies of GT all suggest that KU70 is the main regulator of pathway choice for DNA DSB repair in Arabidopsis. This is consistent with the finding that KU regulates NHEJ pathway choice in human somatic cells (Fattah et al. 2010).
Some DSBs were repaired by a combination of HR and NHEJ (Supplemental Figs. S4, S5), consistent with SDSA (Risseeuw et al. 1995; Puchta et al. 1996; Shalev and Levy 1997; Puchta 1998, 1999; Wright et al. 2005). Events repaired by both HR and NHEJ were observed in all three mutant backgrounds and accounted for 123 of the 1259 total GT products recovered. It is likely that SDSA is also used to repair breaks in WT; however, no evidence was observed among the few GT events recovered from WT strains. It is interesting to note that we observed more imprecise NHEJ repair on the left side of the DSB (Supplemental Tables S4, S5). The reason for this asymmetry is not clear, and it will be interesting to see if it holds up in experiments in which more GT events are analyzed.
Knocking out SMC6B enhanced all three DNA repair mechanisms: C-NHEJ, A-NHEJ, and HR. Blocking SMC6B function apparently hinders sister-chromatid-mediated HR, and, consequently, NHEJ is more frequent. When HR does occur, “substitute” DNA templates are used such as the exogenous donor. Our results are consistent with a study of SMC6 in human cells (Potts et al. 2006): Inactivation of the SMC5/6 complex led to a twofold enhancement in I-SceI-facilitated GT as well as a twofold increase in imprecise end-joining. The significant increase in NHEJ observed by knocking down the SMC5/6 complex in human cells (Potts et al. 2006) or by knocking out SMC6B in Arabidopsis (this study) suggests that sister-chromatid-based HR is efficient in eukaryotes. A recent study on Arabidopsis SMC5 and SMC6 genes also supports this conclusion: Sister-chromatid-based HR was 25% as efficient in the smc6b-1 mutant (Watanabe et al. 2009). This observation is consistent with separate studies on SMC6B in Arabidopsis that used similar reporter systems (Mengiste et al. 1999; Hanin et al. 2000). Collectively, these reported decreases in sister-chromatid HR in smc6b mutants contrast with our observed increase in GT, likely because the GT template used for repair is nonallelic.
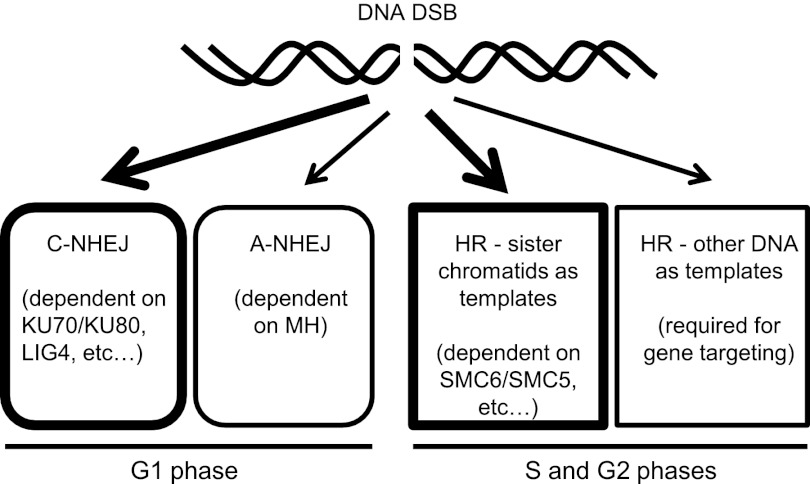
We propose a pathway choice model to explain our findings (Fig. 7). Our model considers four pathways used for DNA DSB repair in Arabidopsis, including KU/LIG4-dependent C-NHEJ, MH-dependent A-NHEJ, SMC6/SMC5-mediated sister-chromatid-based HR, and other types of HR (including nonallelic HR used for GT). The KU70/K80-mediated C-NHEJ pathway predominates in nonreplicating cells (G1 phase). SMC6/SMC5-mediated sister-chromatid-dependent HR is likely used by cells in the S and G2 phases of the cell cycle when DNA is replicated and sister chromatids are paired. Disabling C-NHEJ by knocking out KU70/KU80 or LIG4 leads to a shift to the other three pathways, with loss of KU70 or KU80 having stronger effects than LIG4, especially for switching to HR. Similarly, blocking the SMC6/SMC5-mediated HR pathway promotes a shift to both NHEJ pathways and other types of HR.
Figure 7.
A pathway choice model for repair of a DNA DSB in Arabidopsis. Four different pathways for repairing a DNA DSB in diploid Arabidopsis cells are depicted. The thick arrows and windows indicate dominant pathways (C-NHEJ and HR-sister chromatids as templates) in WT cells. Disruption of either of the major pathways (such as by knocking out KU70 or SMC6B in this study) will promote a pathway switch. Depending on the stage of the cell cycle, NHEJ pathways are dominant in the G1 phase, whereas HR pathways may be preferred in the S and G2 phases.
In conclusion, this study on endogenous Arabidopsis genes contributes a least three new insights into ZFN-mediated genome modification: (1) enhanced site-specific mutagenesis can be achieved by knocking out SMC6B; (2) an increase in GT can be realized by knocking out LIG4, SMC6B, and especially KU70; and (3) deletion mutants can be easily identified by knocking out KU70 or LIG4. Although null mutants were used in this study, we anticipate that suppressing these DNA repair genes using RNAi could attain similar results. Because ZFNs, TALENs, and meganucleases all make DNA DSBs, and because repair pathways for DSBs are highly conserved, it is expected that our findings can be adopted to facilitate the introduction of genome modifications in other higher plants, including crop species.
Methods
Plasmids and plant materials
Both TT4-ZFN (Zhang et al. 2010) and MPK8-ZFN (Sander et al. 2010) were expressed under an estrogen-inducible promoter in the binary vector pFZ19. Agrobacteria that carry these vectors were used to transform both Col (WT) and smc6b plants through the floral dip method (Clough and Bent 1998). T-DNA insertional mutants were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH) and include lig4-4 (SALK_044027) (Heacock et al. 2007), ku70 (SALK_123114) (Kannan et al. 2008), and smc6b-3 (SALK_124719). A transgenic homozygous ADH1-ZFN-4 plant was derived from a line previously described (Zhang et al. 2010). Homozygous plants that contain both the ADH1-ZFN-4 transgene and mutations in each DNA repair gene were generated by crossing and subsequent screening using primers summarized in Supplemental Table S4. The donor construct (Supplemental Fig. S2) was made by direct DNA synthesis.
Analysis of nucleic acids
Total RNA was extracted from 4-wk-old Col WT and smc6b-3 rosette leaves with TRIzol reagent (Invitrogen). The extracted RNA was used as a template for amplifying SMC6B and Actin2 with the QIAGEN OneStep RT-PCR kit (QIAGEN). The primer sequences are included in Supplemental Table S4. Genomic DNA was extracted from either seedlings or rosette leaves using the CTAB method (Stewart and Via 1993). For enrichment PCR, ∼500 ng of genomic DNA was digested by NlaIII (for ADH1-ZFN), NspI (for TT4-ZFN), or MslI (for MPK8-ZFN) in a 50-μL reaction overnight. Four microliters of digestion product was used for PCR amplification of the ZFN target site in a 25-μL reaction. Ten microliters of PCR product of each was then digested in a 40-μL reaction overnight. The digestion products were subsequently examined by electrophoresis.
NHEJ and GT frequency measurements
To measure NHEJ in seedlings, Arabidopsis seeds were first sterilized by a 5-min treatment with bleach (2.6% sodium hypochlorite) and subsequent washes with sterile water. After incubating in the dark for 4 d at 4°C, seeds in a 0.05% agar suspension were plated on 0.5× MS medium with 0.75% agar and 20 μM of β-estradiol (Sigma-Aldrich). One-week-old seedlings were then pooled (with six seedlings in each pool) for DNA extraction, and the DNA concentration was adjusted to ∼50 ng/μL. PCR and restriction digestions were performed as described for enrichment PCR (Zhang et al. 2010). Unsaturated images of the digestion products resolved by electrophoresis were quantified using LabWork 4.5, an image acquisition and analysis software (UVP). For measuring NHEJ and GT in protoplasts, detailed protocols for the protoplast transformation, sample preparation for 454-sequencing, and data analysis are all included in the Supplemental Methods.
Immunoblot analysis
Rosette leaves of 4-wk-old Arabidopsis plants were infiltrated with 20 μM β-estradiol with a needleless syringe. Estradiol-treated leaves were collected 24 h after infiltration and frozen with liquid nitrogen. Total protein samples were prepared by grinding 0.2 g of frozen leaf tissue with 400 μL of 2× SDS sample buffer with a small mortar and pestle. The tubes that contain the extracts were boiled for 5 min and spun at 16,000g for 10 min at room temperature. The supernatants were separated by SDS-PAGE and transferred to a PVDF membrane. ZFNs were detected with a mouse Anti-Flag antibody (Sigma-Aldrich) at 1:1000 dilution and the second antibody, goat anti-mouse horseradish peroxidase (HRP) conjugate (Pierce), at 1:5000 dilution. SuperSignal West Femto maximum sensitivity substrate (Pierce) was used for detection using a chilled CCD camera.
Data access
DNA sequence data from this study have been submitted to the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP017506.
Competing interest statement
Y.Q. and D.F.V. are inventors on a provisional patent application that is based on findings in this manuscript. D.F.V. serves as Chief Science Officer for Cellectis Plant Sciences, a biotechnology company that uses sequence-specific nucleases to create new crop varieties.
Acknowledgments
We thank all members of the Voytas Laboratory for helpful discussions and Dr. Fumiaki Katagiri for help with statistical analyses. This work is supported by grants (MCB 0209818 and DBI 0923827) from the National Science Foundation to D.F.V., as well as research funds from the Central Universities of China (ZYGX2009J084) and the National Natural Science Foundation of China (30900779) to Y.Z.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.145557.112.
References
- Bogdanove AJ, Voytas DF 2011. TAL effectors: Customizable proteins for DNA targeting. Science 333: 1843–1846 [DOI] [PubMed] [Google Scholar]
- Bozas A, Beumer KJ, Trautman JK, Carroll D 2009. Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics 182: 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D 2011. Genome engineering with zinc-finger nucleases. Genetics 188: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnel C, Allain E, Gallego ME, White CI 2011. Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair (Amst) 10: 611–619 [DOI] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Piccoli G, Cortes-Ledesma F, Ira G, Torres-Rosell J, Uhle S, Farmer S, Hwang JY, Machin F, Ceschia A, McAleenan A, et al. 2006. Smc5–Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat Cell Biol 8: 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K, Abe K, Ichikawa H, Valentine L, et al. 2006. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J 25: 5579–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattah FJ, Lichter NF, Fattah KR, Oh S, Hendrickson EA 2008. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc Natl Acad Sci 105: 8703–8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA 2010. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet 6: e1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T, Alberts BM 1986. DNA synthesis dependent on genetic recombination: Characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell 47: 793–806 [DOI] [PubMed] [Google Scholar]
- Gherbi H, Gallego ME, Jalut N, Lucht JM, Hohn B, White CI 2001. Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep 2: 287–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ 2010. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 649: 247–256 [DOI] [PubMed] [Google Scholar]
- Hanin M, Mengiste T, Bogucki A, Paszkowski J 2000. Elevated levels of intrachromosomal homologous recombination in Arabidopsis overexpressing the MIM gene. Plant J 24: 183–189 [DOI] [PubMed] [Google Scholar]
- Hartlerode AJ, Scully R 2009. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J 423: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock ML, Idol RA, Friesner JD, Britt AB, Shippen DE 2007. Telomere dynamics and fusion of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic Acids Res 35: 6490–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Hoeijmakers JH, van Gent DC 1998. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol 8: 483–489 [DOI] [PubMed] [Google Scholar]
- Kannan K, Nelson AD, Shippen DE 2008. Dyskerin is a component of the Arabidopsis telomerase RNP required for telomere maintenance. Mol Cell Biol 28: 2332–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S 1996. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93: 1156–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak J, West CE, White C, da Costa-Nunes JA, Angelis KJ 2009. Rapid repair of DNA double strand breaks in Arabidopsis thaliana is dependent on proteins involved in chromosome structure maintenance. DNA Repair (Amst) 8: 413–419 [DOI] [PubMed] [Google Scholar]
- Mengiste T, Revenkova E, Bechtold N, Paszkowski J 1999. An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J 18: 4505–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhans A, Vartanian JP, Wain-Hobson S 1990. DNA recombination during PCR. Nucleic Acids Res 18: 1687–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orel N, Kyryk A, Puchta H 2003. Different pathways of homologous recombination are used for the repair of double-strand breaks within tandemly arranged sequences in the plant genome. Plant J 35: 604–612 [DOI] [PubMed] [Google Scholar]
- Osakabe K, Osakabe Y, Toki S 2010. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc Natl Acad Sci 107: 12034–12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Duchateau P 2007. Meganucleases and DNA double-strand break-induced recombination: Perspectives for gene therapy. Curr Gene Ther 7: 49–66 [DOI] [PubMed] [Google Scholar]
- Pastwa E, Blasiak J 2003. Non-homologous DNA end joining. Acta Biochim Pol 50: 891–908 [PubMed] [Google Scholar]
- Potts PR 2009. The Yin and Yang of the MMS21-SMC5/6 SUMO ligase complex in homologous recombination. DNA Repair (Amst) 8: 499–506 [DOI] [PubMed] [Google Scholar]
- Potts PR, Porteus MH, Yu H 2006. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J 25: 3377–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H 1998. Repair of genomic double-strand breaks in somatic plant cells by one-sided invasion of homologous sequences. Plant J 13: 331–339 [Google Scholar]
- Puchta H 1999. Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics 152: 1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H 2005. The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J Exp Bot 56: 1–14 [DOI] [PubMed] [Google Scholar]
- Puchta H, Dujon B, Hohn B 1996. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci 93: 5055–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss B, Schubert I, Kopchen K, Wendeler E, Schell J, Puchta H 2000. RecA stimulates sister chromatid exchange and the fidelity of double-strand break repair, but not gene targeting, in plants transformed by Agrobacterium. Proc Natl Acad Sci 97: 3358–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risseeuw E, Offringa R, Franke-van Dijk ME, Hooykaas PJ 1995. Targeted recombination in plants using Agrobacterium coincides with additional rearrangements at the target locus. Plant J 7: 109–119 [DOI] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. 2010. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods 8: 67–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H, Melamed-Bessudo C, Levy AA 2005. High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc Natl Acad Sci 102: 12265–12269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev G, Levy AA 1997. The maize transposable element Ac induces recombination between the donor site and an homologous ectopic sequence. Genetics 146: 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. 2006. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res 34: e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CN Jr, Via LE 1993. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques 14: 748–750 [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF 2009. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459: 442–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF 2013. Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol 64 (in press). [DOI] [PubMed] [Google Scholar]
- Watanabe K, Pacher M, Dukowic S, Schubert V, Puchta H, Schubert I 2009. The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21: 2688–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DA, Townsend JA, Winfrey RJ Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF 2005. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J 44: 693–705 [DOI] [PubMed] [Google Scholar]
- Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, et al. 2010. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci 107: 12028–12033 [DOI] [PMC free article] [PubMed] [Google Scholar]