Abstract
Herpesvirus-associated ubiquitin specific protease (HAUSP) regulates the stability of p53 and MDM2, implicating HAUSP as a therapeutic target for tuning p53-mediated anti-tumor activity. Here, we report the structural analysis of HAUSP with Kaposi’s sarcoma-associated herpesvirus vIRF4 and the discovery of two vIRF4-derived peptides, vif1 and vif2, as potent and selective HAUSP antagonists. This analysis reveals a bilateral belt-type interaction resulting in inhibition of HAUSP. The vif1 peptide binds the HAUSP TRAF domain, competitively blocking substrate binding, while the vif2 peptide binds both the HAUSP TRAF and catalytic domains, robustly suppressing its deubiquitination activity. Consequently, peptide treatments comprehensively blocked HAUSP, leading to p53-dependent cell cycle arrest and apoptosis in culture and tumor regression in xenograft mouse model. Thus, the virus has developed a unique molecular strategy to target the HAUSP-MDM2-p53 pathway, and these virus-derived short peptides represent biologically active HAUSP antagonists.
The p53 pathway is partially abrogated in more than a half of tumors through the inactivation of various signaling or effector components. Several recent studies have shown that the restoration of p53 activity alone can robustly induce tumor regression1,2, asserting the p53 pathway as a prime target for new cancer drug development. Accordingly, a number of strategies for targeting wild-type p53 have been designed. These include restoring p53 function by antagonizing its negative regulator, the E3 ubiquitin ligase MDM2, via peptides and small molecules, which have been shown to induce p53-dependent suppression of tumor cell growth in vitro and in vivo3–5. However, MDM2 inhibition alone may not be sufficient to potently activate p53 within the in vivo tumor microenvironment6.
It has been fairly well established that the level of p53 protein is capricious and is subtly regulated by ubiquitination and deubiquitination systems. The herpesvirus-associated ubiquitin specific protease (HAUSP) is a well-characterized deubiquitinase enzyme that has the ability to remove ubiquitin moieties from ubiquitinated substrates. It was initially identified as a binding partner of herpes simplex virus (HSV) ICP0 to cooperatively facilitate viral replication7. Since then, further studies have shown that HAUSP can bind to various other substrates and is involved in the stress response pathway, epigenetic silencing, neurodegenerative disorders, and progression of infections by DNA viruses8. Additionally, HAUSP functions as a pivotal component of the p53-MDM2-MDMX signaling pathway, thus participating in the delicate balance that maintains p53 protein levels critical for normal cellular homeostasis and diverse stress responses9–11. Intriguingly, the reduction or ablation of HAUSP leads to DNA-damage-induced MDMX degradation and MDM2 instability, both of which robustly stabilize p5312.
Structural analyses reveal that both p53 and MDM2 specifically recognize the amino-terminal tumor necrosis factor–receptor associated factor (TRAF)–like domain of HAUSP, and HAUSP-binding elements were mapped to a peptide fragment in the carboxyl-terminus of p53 and to a short-peptide region preceding the acidic domain of MDM213,14. The co-crystal structures of the HAUSP TRAF-like domain in complex with these p53 and MDM2 short peptides demonstrate that the MDM2 peptide recognizes the same surface groove of HAUSP as that recognized by p53. However, the MDM2 peptide mediates more extensive interactions, indicating that MDM2, rather than p53, is the primary substrate of HAUSP under normal physiological conditions13,14. An Epstein-Barr virus (EBV) viral protein, Epstein-Barr nuclear antigen 1 (EBNA1), also binds the same region of HAUSP as that bound by the MDM2 and p53 peptides, competitively blocking these cellular interactions15,16. Thus, HAUSP appears to play multiple roles in regulating the p53-MDM2 pathway as well as in EBV-induced tumor cell survival, and is hence a potential chemotherapeutic target for p53-mediated suppression of tumor cell growth. In this report, we set out to answer the question of how viruses regulate HAUSP activity to escape host’s p53-mediated growth control. Our study describes the structural analysis of Kaposi’s sarcoma-associated herpesvirus (KSHV) vIRF4-HAUSP complex, and the discovery of two short viral peptides, vif1 and vif2 as potent, selective HAUSP antagonists.
RESULTS
Interaction between HAUSP and vIRF4, and its complex structure
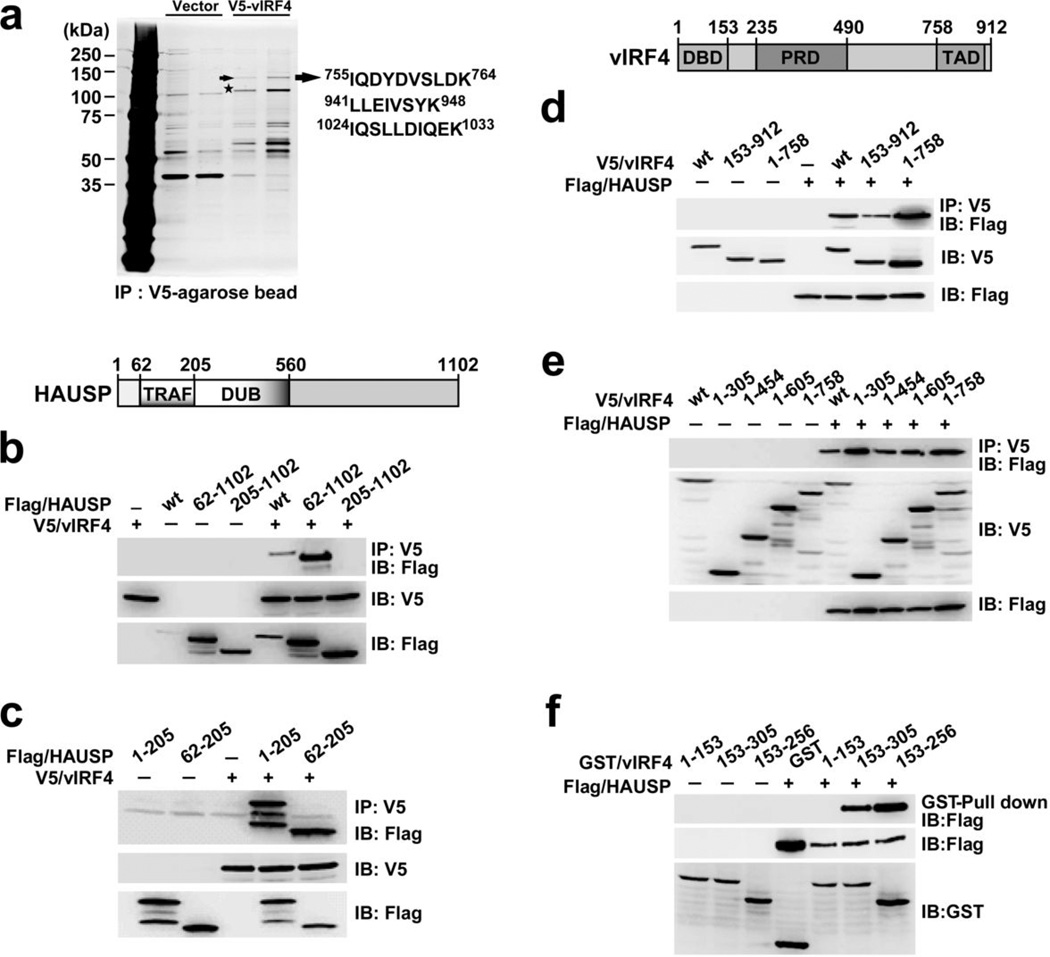
To study the HAUSP interaction network in KSHV (γ-2 herpesvirus), we performed a yeast-two hybrid screen with a KSHV library17 and mass spectrometry analysis. Both studies independently discovered a novel interaction between HAUSP and vIRF4 (Fig. 1a). Detailed binding assays indicate that the HAUSP TRAF domain (HAUSP62–205) specifically interacts with vIRF4153–256 (Fig. 1b–f). An isothermal titration calorimetry (ITC) assay revealed a robust interaction between HAUSP62–205 and vIRF4153–256, with a dissociation constant (Kd) of 76 nM; this Kd value is markedly higher than those reported for other HAUSP TRAF domain binding substrates (Kd = 0.5–15 µM)13–15,18 (Supplementary Fig. 1a,b).
Figure 1. vIRF4 interacts with HAUSP.
(a) Silver-stained purified V5-vIRF4 complexes 48 h post-transfection with V5-vIRF4. Arrow, HAUSP; asterisk, V5-vIRF4. (b) The HAUSP TRAF domain is sufficient to interact with vIRF4 (wt). Schematic representation of HAUSP: TRAF, TRAF-like domain; DUB, deubiquitinase domain. 293T cells were transiently transfected and coimmunoprecipitated (Co-IP) with an anti-V5 antibody and immunoblotted (IB) with an anti-Flag antibody. (c) 293T cells transfected with the indicated constructs were subjected to Co-IP and IB. (d) The central region of vIRF4 is required for HAUSP interaction. Schematic representation of vIRF4: DBD, DNA-binding domain; PRD, proline-rich domain; and TAD, transactivation domain. 293T cells were transfected with the indicated constructs, followed by Co-IP and IB. (e) Co-IP of HAUSP with wt or several vIRF4 mutants. 293T cells transfected with the indicated vIRF4 constructs along with HAUSP subjected to IP and IB as in (b). (f) vIRF4153–256 is sufficient to bind to HAUSP. Cells were transfected with the indicated vIRF4 mutants in combination with empty vector or HAUSP, and subjected to GST pull-down and IB with an anti-Flag antibody.
To gain further insight into the molecular basis of the HAUSP-vIRF4 interaction, the HAUSP62–205-vIRF4153–256 complex was crystallized using an in situ proteolysis technique19. The three-dimensional structure of this crystallized complex was determined by the molecular replacement method using the HAUSP TRAF domain structure (PDB accession code 2F1W) as a search model, and refined to 1.6 Å resolution (Table 1 and Fig. 2a). All residues of HAUSP62–205 except Asp62 are included in the final model, whereas only 15 residues (Ser202 to Met216) of vIRF4 are visible in the electron density map (Supplementary Fig. 1c). The overall structure of the HAUSP TRAF domain comprises a typical eight-stranded anti-parallel β-sandwich fold (Fig. 2a) that forms a shallow groove in its surface structure (Fig. 2b, displayed in the middle). No significant conformational changes were observed between the peptide-free (PDB accession code 2F1W) and vIRF4-bound TRAF domains, except that the C-terminal region of the TRAF domain was less extended upon vIRF4-binding (data not shown).
Table 1.
X-ray data collection and refinement statistics
| HAUSP62–205-vIRF4153–256 complex | |
|---|---|
| Data collection | |
| Space group | P3221 |
| Cell dimensions a, b, c (Å) |
72.46, 72.46, 53.84 |
| Resolution (Å) | 1.60(1.66–1.60) * |
| Rsym | 0.061(0.266) |
| I / σI | 65.2(11.9) |
| Completeness (%) | 100(100) |
| Redundancy | 21.7(21.8) |
| Refinement | |
| Resolution (Å) | 30.0–1.60 |
| No. reflections | 20,747 |
| Rwork / Rfree | 0.158/0.174 |
| No. atoms | 1,560 |
| Protein (HAUSP/vIRF4) | 1,216/106 |
| Water | 106 |
| B-factors | 14.81 |
| Protein (chain A/B) | 12.85/14.76 |
| Water | 24.85 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.131 |
Values in parentheses are for highest-resolution shell.
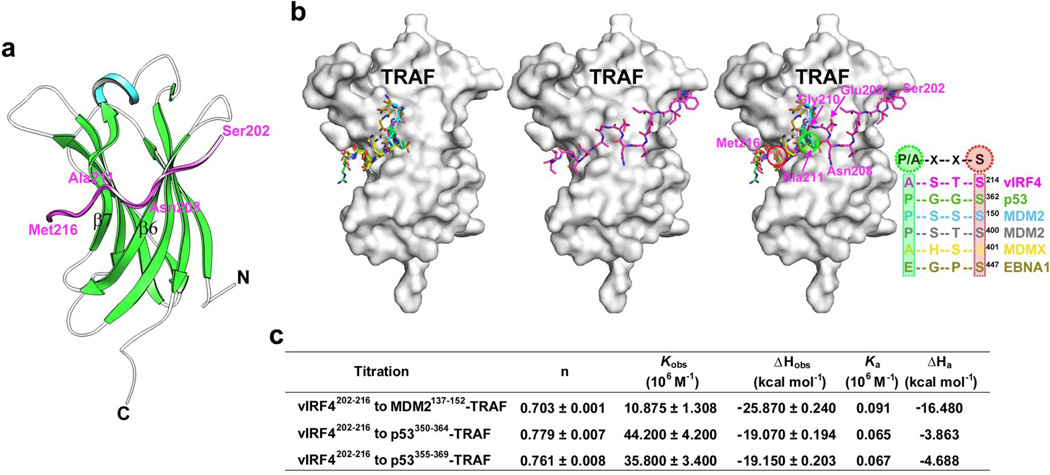
Figure 2. Structural basis for the interaction between HAUSP and vIRF4.
(a) Ribbon representation of the vIRF4-HAUSP TRAF domain complex. Alpha-helix and β-sheets of the TRAF domain are shown in cyan and green, respectively. The β6 and β7 strands are indicated. The viral peptide (Ser202 to Met216) bound to the TRAF domain is represented in magenta. (b) Distinctiveness of HAUSP TRAF-vIRF4 interaction. Cellular substrates and EBV EBNA1 utilize a similar strategy in the interaction with HAUSP TRAF (left) whereas vIRF4 employs an unusual strategy for binding to the TRAF (middle). All target binding peptides are superimposed onto the HAUSP TRAF domain (right). The peptides include vIRF4 (magenta), p53 (green), MDM2 (cyan), MDM2 (gray), MDMX (yellow), and EBNA1 (olive). Residues in vIRF4 are labeled in magenta. The consensus sequence motif for each peptide is shown, and the most conserved residues are circled. (c) Thermodynamic parameters of competitive binding of vIRF4 with the TRAF domain against cellular substrates. Each cellular substrate peptide was first titrated into the TRAF domain, and the competitor vIRF4202–216 was then titrated against each peptide.
Detailed characterization of the HAUSP TRAF-vIRF4 interaction
Unlike previous studies that used synthetic or chimerically fused peptides of 4–10 amino acids in length in complex with the HAUSP TRAF domain13–15,18, an in situ proteolysis treatment of the HAUSP62–205-vIRF4153–256 protein complex yielded a crystal structure with a markedly longer 15-residue vIRF4 peptide. The peptide consists of an upstream (Ser202 to Asn208) and a downstream (Ala211 to Met216) region, which are linked by the Glu209 and Gly210 residues (Fig. 2a,b and Supplementary Fig. 1d). The long peptide is positioned on the TRAF domain surface groove in a novel belt-type arrangement that threads through the left and right armholes (Fig. 2b, displayed in the middle). This binding pattern is significantly different from other HAUSP substrates, which surround the left armhole of the TRAF domain (Fig. 2b, displayed in the left).
The downstream region (Ala211 to Met216) of vIRF4 corresponds to the peptides previously reported for p53, MDM2, MDMX, and EBNA1 to complex with HAUSP TRAF (Fig. 2b, displayed in the right). This region contains the well-conserved 4-residue consensus sequence P/A××S binding motif (Fig. 2b). The equivalent motif of vIRF4 consists of Ala211, Ser212, Thr213, and Ser214, and engages in extensive polar and nonpolar interactions (Supplementary Fig. 1d) with one side of the TRAF β-sheet, particularly the β7 strand (Fig. 2a). Overall, the interaction pattern between HAUSP TRAF and the downstream region of vIRF4 is similar to those previously reported for other peptides13–15,18 (Fig. 2b). The upstream region (Ser202 to Asn208) of vIRF4 binds to the HAUSP TRAF domain in a novel extended conformation. This region participates in extensive interactions mainly with the other side of the β-sheet of the TRAF domain, especially the β6 strand (Fig. 2a,b). TRAF Arg153 appears to play a decisive and unique role in TRAF’s interaction with the upstream region of the vIRF4 peptide (Supplementary Fig. 1d). On the other hand, the vIRF4 Glu209 and Gly210 residues grasp the TRAF β6 and β7 strands (Fig. 2a,b and Supplementary Fig. 1d). These indicate that the distinctive upstream region of the vIRF4 peptide may be vital for stabilizing the interaction of the downstream consensus region with the HAUSP TRAF domain. Indeed, ITC analysis demonstrated that the deletion of this upstream region (vIRF4209–216) results in a 25-fold decrease in TRAF binding affinity compared with vIRF4202–216 (Supplementary Fig. 1a,b), suggesting that vIRF4 uses a different interaction strategy from other HAUSP TRAF-binding cellular substrates as well as EBV EBNA1.
HAUSP binding to vIRF4202–216 vs other known substrates
ITC analysis with the peptides containing the HAUSP-binding consensus sequence (P/A××S) or the vIRF4 upstream region (vIRF4202–216) showed that the vIRF4202–216 peptide exhibited 28- to 40-fold tighter binding (Kd of 0.39 µM) to the TRAF domain than peptides derived from MDM2 and p53 (Supplementary Fig. 1a,b). To evaluate whether vIRF4 competes with cellular substrates for binding to the HAUSP TRAF domain, each peptide (MDM2137–152, p53350–364, or p53355–369) was titrated into the HAUSP TRAF domain, resulting in association constants (Ka) of 9.1×104 M−1, 6.5×104 M−1, and 6.7×104 M−1, respectively (Fig. 2c). When vIRF4202–216 was subsequently titrated against HAUSP cellular substrates as a competitor, the observed association constant (Kobs) of each titration was markedly increased to 10.9×106 M−1, 44.2×106 M−1, and 35.8×106 M−1, respectively. These results indicate a considerably tighter interaction between the HAUSP TRAF domain and vIRF4202–216 compared to its cellular substrates, MDM2 and p53 (Fig. 2c and Supplementary Fig. 2a).
To further gauge the competitive nature of vIRF4 binding, the TRAF domain was incubated with an equal molar amount of vIRF4153–216 in the presence of a 5-fold molar excess of MDM2137–152, followed by size exclusion chromatography analysis. This showed that the vIRF4202–216 peptide was still able to form a stable complex with TRAF even in the presence of a 5-fold molar excess of MDM2 peptide (Supplementary Fig. 2b). This competitive nature of the vIRF4202–216 peptide is likely due to its upstream region since the HAUSP binding affinity of the upstream-region-deleted vIRF4209–216 peptide (Kd of 9.57 µM) was comparable to that of MDM2137–152 (Kd of 11.06 µM), p53350–364 (Kd of 15.46 µM), and p53355–369 (Kd of 15.46 µM) (Supplementary Fig. 1b).
NMR analysis of the HAUSP-vIRF4 interaction
The ITC analysis results revealed that while the TRAF binding affinity (Kd = 0.54 µM) of vIRF4153–216 was comparable to that of vIRF4202–216, the C-terminally extended vIRF4153–256 exhibited a 7-fold higher affinity (Kd = 0.076 µM). vIRF4153–256/Δ202–216 (Kd = 3.45 µM) led to a significant reduction of TRAF binding affinity, whereas vIRF4153–256/Δ202–216/Δ237–256 (Kd = 4.03 µM) had no further effect (Supplementary Fig. 1b). These results collectively indicate that, in addition to the vIRF4202–216 residues, the vIRF4217–236 sequence plays an important role in TRAF binding.
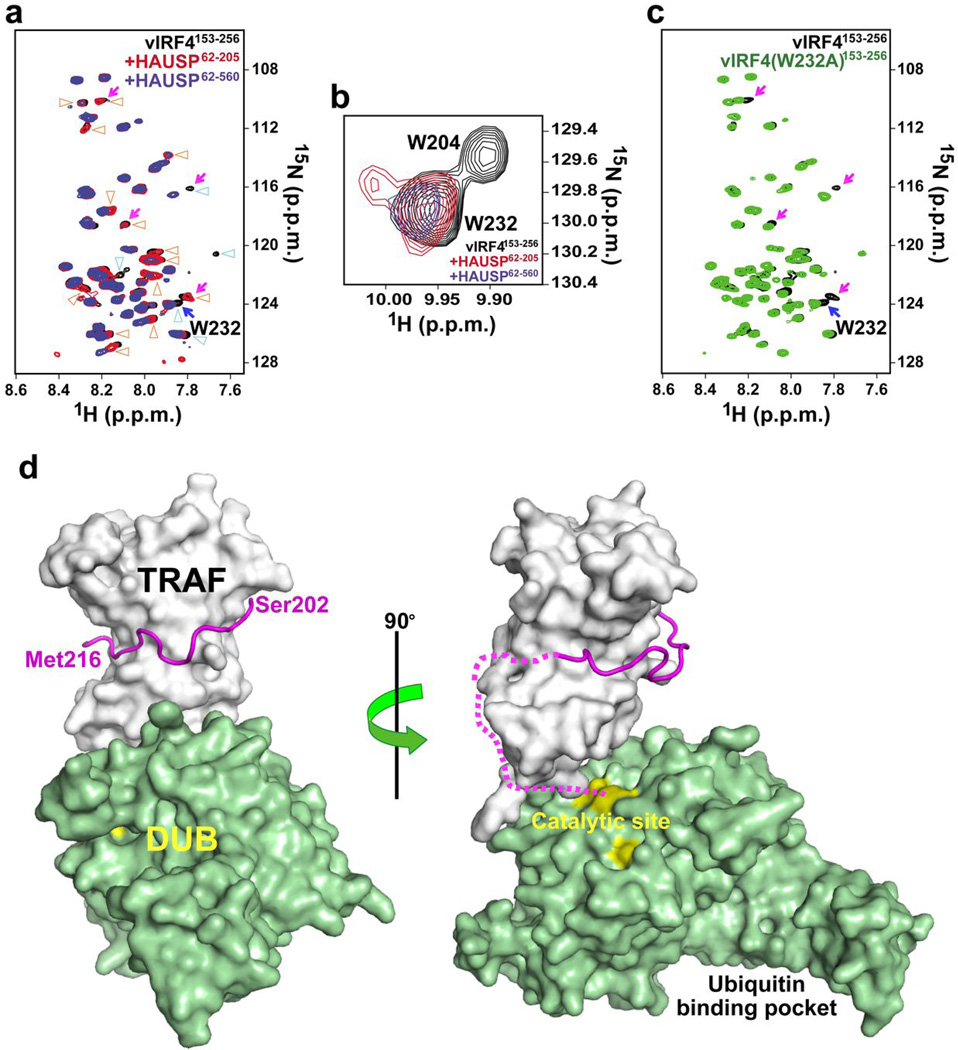
To further dissect these ITC results, we analyzed NMR chemical shift perturbations of vIRF4153–256 in the presence of HAUSP62–205 (TRAF domain) or HAUSP62–560 (TRAF-deubiquitinase (DUB)-domain). Figure 3a shows the superposition of the 2D 1H-15N correlation spectra of vIRF4153–256 in the absence and presence of each HAUSP fragment. Signal changes (denoted by blue) of vIRF4153–256 were observed upon the binding of HAUSP62–205 (red) compared to free vIRF4153–256 (black), and additional changes (orange) were detected upon the binding of HAUSP62–560 (blue). The 2D 1H-15N correlation spectra and mutational analysis revealed that the ε-NH proton of vIRF4 Trp204 changes dramatically upon binding of TRAF-containing HAUSP62–205 (Fig. 3b, red and Supplementary Fig. 3a), consistent with crystal structure data showing that Trp204 is located in the TRAF binding region of vIRF4202–216. On the other hand, the ε-NH signal of Trp232 was perturbed by binding of the TRAF-DUB-containing HAUSP62–560, but not by binding of the TRAF-containing HAUSP62–205 (Fig. 3b, blue). This latter suggests that Trp232 is involved in the interaction of vIRF4 with the HAUSP DUB domain.
Figure 3. Bilateral interaction of vIRF4 with HAUSP.
(a) NMR analysis of the interaction between the vIRF4 peptide and HAUSP TRAF-DUB domain. Shown is the backbone amide region of the 2D 1H-15N correlation spectra of vIRF4153–256 in the presence of an equimolar amount of HAUSP62–205 (red) or HAUSP62–560 (blue). The 1H-15N spectrum of free vIRF4153–256 is represented in black. Blue triangles, signal changes of vIRF4153–256 observed upon binding of HAUSP62–205. Orange triangles, additional changes detected upon binding of HAUSP62–560. Magenta arrows, residues close to vIRF4 Trp232. (b) Signal changes of the tryptophan ε-NH protons upon interaction with HAUSP62–205 (red) or HAUSP62–560 (blue). Free vIRF4153–256 is represented in black contours. (See also Supplementary Fig. 3a). (c) The Trp232 backbone assignment. Superposition of the 1H-15N correlation spectra of free vIRF4153–256 (black) and vIRF4 (W232A)153–256 (green). Residues located close to Trp232 were identified by comparing the two spectra (magenta arrows). Blue arrow, the assigned Trp232 backbone. (See also Supplementary Fig. 3b). (d) Proposed molecular interaction scheme between HAUSP and two different vIRF4-derived peptides. This model is based on the vIRF4-TRAF complex structure from the present study and the HAUSP structure containing the TRAF and DUB domains (PDB accession code 2F1Z). The vIRF4202–216 peptide is displayed as a magenta loop, while the vIRF4217–236 peptide is depicted as magenta short-dashed line. Catalytic triad (yellow) is highlighted in the catalytic site. The ubiquitin binding pocket is indicated.
Selective isotope (15N) labeling of Trp232 was performed to further identify backbone amide signals derived from residues located near Trp232, by comparing vIRF4 and W232A mutant (Fig. 3c, denoted by pink arrows and Supplementary Fig. 3b). Results indicate that the residues near Trp232 are involved in binding to the HAUSP catalytic DUB domain (Fig. 3a). However, while vIRF4 and its peptide fragments (vIRF4202–216 and vIRF4220–236) were individually capable of interacting with HAUSP, neither peptide was able to bind to the HAUSP DUB domain alone (Fig. 1b and Supplementary Fig. 4a, data not shown), suggesting that a tight interaction with the N-terminal TRAF domain of HAUSP may be required for the binding of vIRF4 to the central DUB domain of HAUSP. These data suggest a bilateral mode of interaction between vIRF4 and HAUSP (Fig. 3c) wherein vIRF4202–216 interacts with the HAUSP TRAF domain (primarily β6 and β7) with an unusually high binding affinity, while vIRF4217–236 contacts the HAUSP DUB domain.
vIRF4 independently targets HAUSP and MDM2 to downregulate p53
As previously shown20, vIRF4 expression considerably increases MDM2 levels and consequently decreases p53 levels in TREx/BCBL-1 vIRF4 cells expressing AU-vIRF4 in a tetracycline-inducible manner. Unlike vIRF4, vIRF41–605 mutant that no longer interacts with MDM2 did not induce MDM2 stabilization and p53 degradation, whereas vIRF4Δ202–256 mutant that no longer interacts with HAUSP increased MDM2 amount and marginally decreased p53 amount (Supplementary Fig. 4b,c). Consistent with these endogenous p53 amounts, vIRF4 markedly induced p53 ubiquitination, vIRF4Δ202–256 mutant did so at reduced levels and vIRF41–605 mutant did not do so (Supplementary Fig. 4c). Thus, while vIRF4 interacts with HASUP and MDM2 to control p53 levels, the vIRF4-MDM2 interaction contributes more significantly to p53 downregulation than the vIRF4-HAUSP interaction.
Comprehensive inhibition of HAUSP deubiquitinase activity by vif1 and vif2 peptides
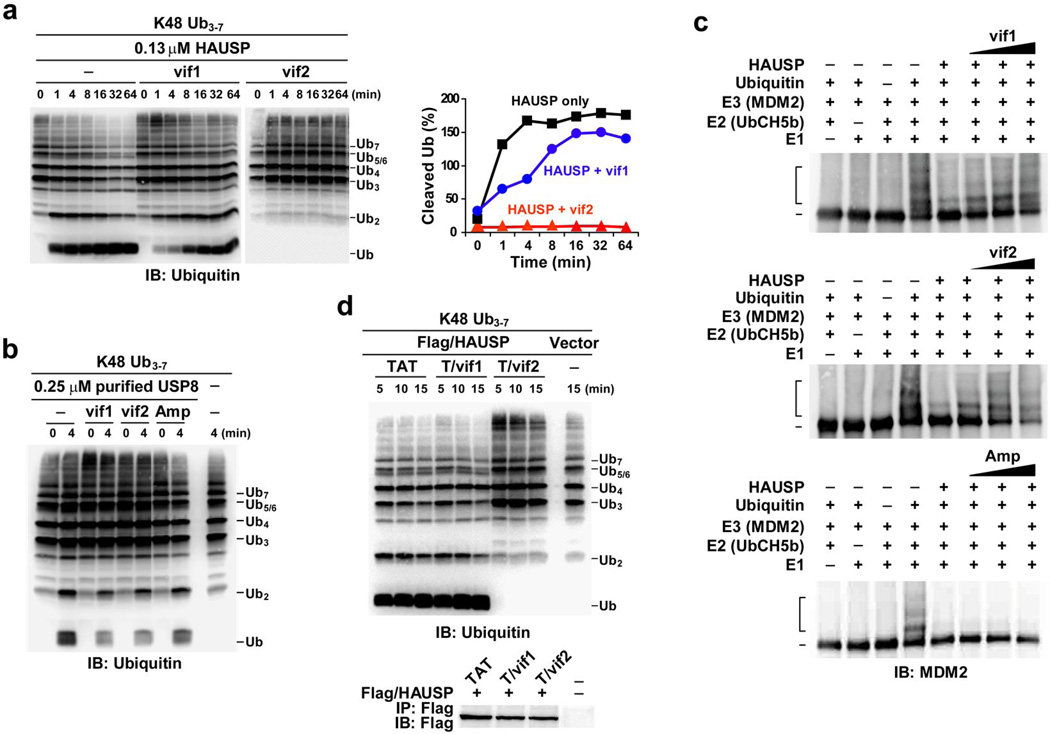
An in vitro DUB assay of immunopurified Flag-HAUSP complexes with K48-linked 3–7 polyubiquitin chains showed that vIRF4 effectively suppressed HAUSP DUB activity in a binding-dependent manner (Supplementary Fig. 5a). To further assess the effects of these vIRF4 short sequences on HAUSP enzymatic activity, the vIRF4 peptides corresponding to residues 202–216 (hereafter referred to as “vif1”) and 220–236 (hereafter referred to as “vif2”) were mixed with purified HAUSP and then subjected to an in vitro DUB assay with K48- or K63-linked 3–7 polyubiquitin chains. An “Amp” peptide derived from the amphipathic helix sequence of the herpesvirus saimiri Tip protein21 was included as a negative control. These results showed that the vif2 peptide markedly suppressed HAUSP DUB activity, the vif1 peptide minimally inhibited activity, and Amp peptide had no effect on HAUSP DUB activity (Supplementary Fig. 5b). Comparative kinetic analysis showed that while the vif1 peptide weakly attenuated HAUSP DUB activity, the vif2 peptide completely suppressed HAUSP DUB activity (Fig. 4a, right panel). These results strongly support that the vif2 peptide (residues 220–236) may directly contact the catalytic domain of HAUSP and thereby inhibit its DUB activity. To confirm the specificity of the vif1 and vif2 peptides toward HAUSP, an in vitro DUB assay was conducted using USP8, which is in the same family as HAUSP22. By contrast, neither the vif1 peptide nor the vif2 peptide was capable of inhibiting USP8 DUB activity (Fig. 4b).
Figure 4. Effect of vIRF4 peptides on HAUSP DUB enzymatic activity.
(a) Effect of vif1 and vif2 peptides on HAUSP DUB activity toward ubiquitin chains. Right: time-course measuring the appearance of cleaved mono- and diubiquitin reaction products was determined by semiquantification of IB, shown on the left. (b) The vif1 and vif2 peptides cannot inhibit USP8 deubiquitinase enzymatic activity in vitro. Purified USP8 was premixed with the vif1, vif2, or Amp (nonspecific) peptide for 5 min and then subjected to an in vitro DUB assay with the K48-Ub3–7 chain. (c) Effect of vif1 and vif2 peptides on HAUSP DUB activity toward ubiquitinated MDM2. Human recombinant purified MDM2 was incubated with purified E1, E2, and ubiquitin prior to the DUB assay. HAUSP preincubated with increasing concentrations of each peptide or HAUSP alone was incubated with ubiquitinated MDM2. (d) Ex vivo effect of TAT-vif1 and TAT-vif2 peptides on HAUSP DUB activity. At 24 h posttransfection with vector or Flag-tagged HAUSP, 293T cells were treated with 100 µM of each peptide for an additional 12 h, followed by IP with anti-Flag agarose beads and elution with Flag peptide. Purified HAUSP complexes were incubated with K48-Ub3–7 chains for the indicated intervals. 1% of the IP complex was used as the input.
We then investigated whether the vif1 and vif2 peptides can inhibit HAUSP DUB activity against ubiquitinated substrates through substrate binding competition. To this end, we first generated ubiquitinated MDM2 using purified E1 (UBE1), E2 (UbcH5b), and E3 (MDM2) proteins, and then performed an in vitro DUB assay employing purified HAUSP alone or HAUSP preincubated with increasing amounts of each peptide (Fig. 4c and Supplementary Fig. 5c). The vif2 peptide efficiently blocked HAUSP enzymatic activity against both K48-linked polyubiquitin chains and ubiquitinated MDM2 (Fig. 4a,c). In striking contrast to its ineffectiveness against K48-linked polyubiquitin chains, the vif1 peptide effectively blocked HAUSP DUB activity when ubiquitinated MDM2 was used as a substrate (Fig. 4c). Finally, incubation with the Amp peptide showed no effect on HAUSP DUB activity under the same conditions (Fig. 4c).
To further delineate the vIRF4 peptides’ action in vivo, the vif1 and vif2 peptides were fused with the HIV-1 TAT protein transduction domain for intracellular delivery23,24 and tested for their potential effects on in vivo HAUSP DUB activity. Consistent with the previous in vitro DUB assay, the TAT-vif2 peptide showed the strongest inhibitory activity toward ex vivo HAUSP enzymatic activity, but the TAT-vif1 peptide showed no effect under the same conditions (Fig. 4d). These results thus further corroborate the notion that vif1 interferes with HAUSP substrate binding, while vif2 inhibits HAUSP DUB activity.
Anti-tumor activity of the TAT-vif1 and TAT-vif2 peptides
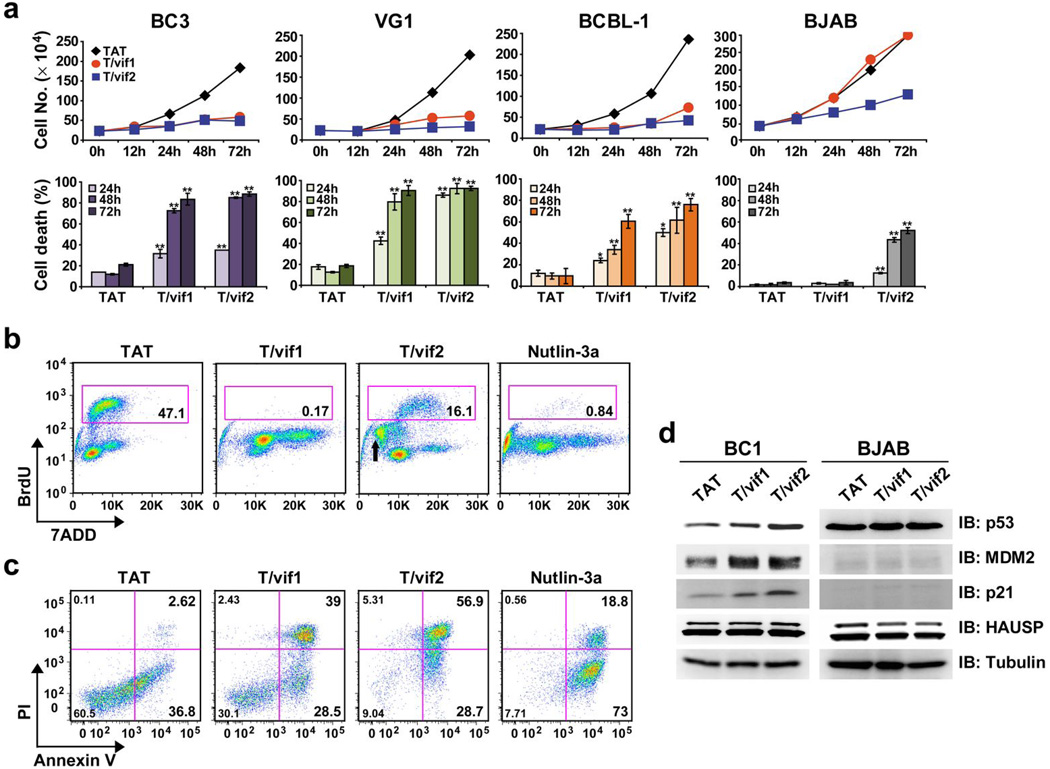
Since HAUSP plays a pivotal role in the regulation of the p53 pathway10,11, we accordingly investigated the potential effect of each peptide on KSHV-induced primary effusion lymphoma (PEL) tumor cell lines harboring p53(wt/wt)25,26. Cell lines with mutated, non-functional p53 were included as controls. These results showed that time-dependent anti-proliferative and cytotoxic activities differed depending on p53 status (Fig. 5a). In contrast to treatment with the HIV-1 TAT peptide, which showed no effect on cell proliferation and cell death, incubation of PELs with various concentrations (25, 50, or 100 µM) of the TAT-vif2 peptide not only robustly suppressed cell proliferation, but also induced profound cell death. The TAT-vif1 peptide did the same but with a much weaker activity than the TAT-vif2 peptide (Supplementary Fig. 6a). Significantly, BJAB Burkitt lymphoma tumor cells carrying mutant p5327 continued to proliferate in the presence of the TAT-vif1 peptide and showed only minor growth retardation and cell death in the presence of the TAT-vif2 peptide (Fig. 5a). Prostate cancer cells, LnCap (p53wt/wt), PC3 (p53−/−), and DU145 (p53m/m)28, which carry different functional p53 genotypes, showed the p53-dependence of TAT-vif1- and TAT-vif2-mediated cell death: LnCap cells, but not PC3 and DU145 cells, were highly susceptible to TAT-vif1- and TAT-vif2-mediated cell growth inhibition (Supplementary Fig. 6b). These data collectively demonstrate that both the vif1 and vif2 peptides have vigorous cell killing activities against p53(wt/wt)-containing tumor cells.
Figure 5. Cytotoxic effect of TAT-vif1 or TAT-vif2 peptide on PEL cells.
(a) Growth inhibition of PEL cells induced by TAT-vif1 and TAT-vif2 peptides. BC3 (p53wt/wt), VG1 (p53wt/wt), BCBL-1 (p53wt/mt), and BJAB (p53mt/mt) cells were treated with 100 µM of individual peptide for the indicated periods of time. *P < 0.01 and **P < 0.001. (b) TAT-vif1 and TAT-vif2 peptides induce cell cycle arrest of PEL cells. Asynchronously growing BC-1 (p53wt/wt) cells were treated with 100 µM of each peptide or 10 µM of Nutlin-3a for 48 h. Cells were pulse-labeled with BrdU and analyzed for DNA content by flow cytometry. BrdU incorporation during the S phase is quantified as the percentage of stained cells. The sub-G1 population in TAT-vif2 peptide–treated BC-1 cells is denoted by an arrow. (c) TAT-vif1- and TAT-vif2 induced cell death of PELs. Apoptosis in BC-1 cells was assessed at 48 h after treatment with 10 µM of Nutlin-3a or 100 µM of each peptide by Annexin V/PI staining. Apoptosis was measured by flow cytometry analysis. Numbers indicate the percentage of cells in each quadrant. (d) Effect of TAT-vif1 and TAT-vif2 peptides on p53 and its transcription target protein levels. VG1 and BJAB cells were treated with the same dose as that used in (a–c) for 6 h. Aliquots of cell lysates containing 10 mg of protein were analyzed by IB with the indicated antibody.
One of the main cellular consequences of p53 activation in proliferating cells is cell cycle arrest through transcriptional upregulation of the cyclin-dependent kinase inhibitor p21, which causes G1-S or G2-M cell cycle arrest29,30. Indeed, treatment of PEL cells with the TAT-vif1 or TAT-vif2 peptide markedly increased the G1 and G2-M phase fraction and nearly completely depleted S-phase cells (Fig. 5b and Supplementary Fig. 7). Interestingly, significant sub-G1 accumulation (reflecting cell death) was observed in TAT-vif2 treated cells compared to TAT or TAT-vif1 treated cells (Fig. 5b and Supplementary Fig. 7 arrows). Annexin V and propidium iodide (PI) staining assays showed that while TAT-vif1 and TAT-vif2 peptide treatment effectively induced apoptotic cell death in PEL cells carrying p53wt/wt compared to TAT treatment, the TAT-vif2 peptide more rapidly and dramatically induced apoptotic cell death than the TAT-vif1 peptide (Fig. 5c and Supplementary Fig. 8). Treatment of Nutlin-3a, which blocks the interaction between MDM2 and p53 and thus induces extensive apoptosis, also led to considerable cell death, comparable to either peptide treatment.
As the inhibition of HAUSP enzymatic activity stabilizes and activates p53, we examined the effect of each peptide on intracellular levels of p53 and its transcriptional targets, p21, MDM2, NOXA, and PUMA. Incubation of exponentially growing PEL, LnCap, and DU145 tumor cells with either peptide for 6 h led to increased p53, p21, MDM2, PUMA, and NOXA at different levels (Fig. 5d and Supplementary Fig. 9). By contrast, BJAB cells exposed to the same conditions showed no detectable changes in p53, MDM2, and p21 levels. Neither the vif1 nor vif2 peptide treatment altered HAUSP levels of various PEL cells as well as BJAB cells (Fig. 5d and Supplementary Fig. 9). These results demonstrate that the TAT-vif1 and TAT-vif2 peptides differentially affect the p53 pathway in cancer cells with functional p53(wt/wt).
In vivo tumor regression induced by the TAT-vif1 and TAT-vif2 peptides
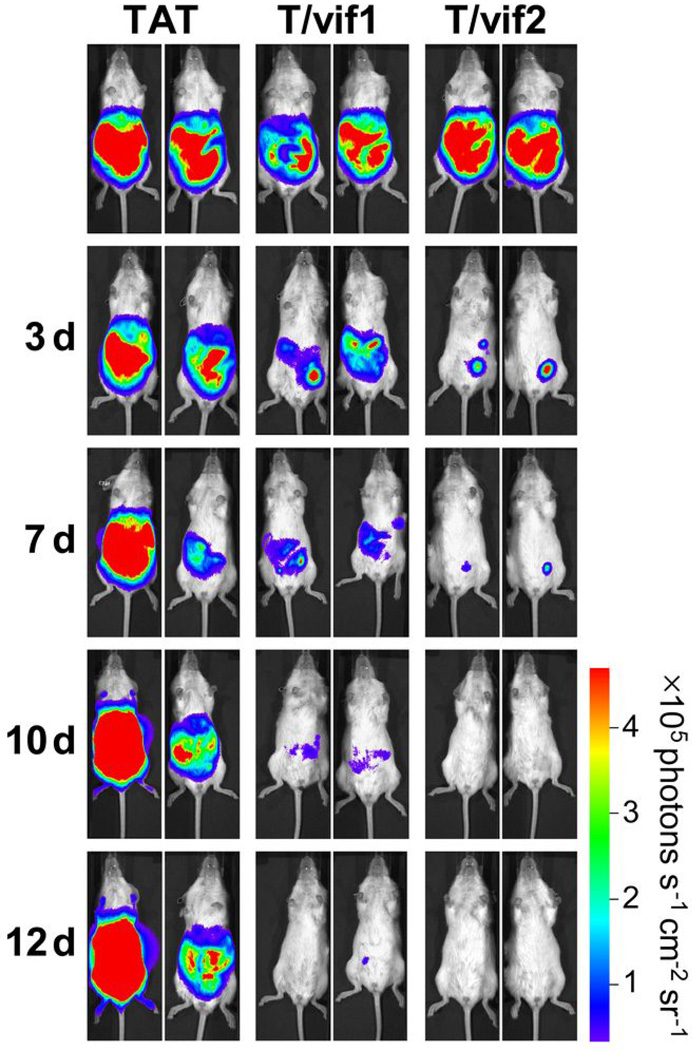
To evaluate the in vivo anti-tumor activity of the vif1 and vif2 peptides, we utilized NOD/SCID xenograft mice intraperitoneally injected with BCBL-1 luciferase cells as previously shown31–33. After being injected with the tumor cells, all of the mice developed PEL, with evident distention and ascites in the peritoneal cavity as well as markedly increased luminescence (data not shown). Mice with advanced PEL were challenged with 1 mg (equivalent to ~100 µM) of each peptide on days 3, 5, 7, and twice weekly for subsequent weeks by intraperitoneal injection. Treatment with the TAT-vif1 or TAT-vif2 peptide led to little or no traceable luminescence with marked tumor regression (Fig. 6 and Supplementary Fig. 10). The TAT-vif2 peptide caused particularly efficient and powerful tumor regression. By contrast, tumors continuously advanced in mice that received TAT peptide injections (Fig. 6 and Supplementary Fig. 10). The mice treated with the TAT-vif1 or TAT-vif2 peptide showed neither significant weight-loss nor any gross abnormalities upon necropsy at the end of the treatment. These results collectively demonstrate the in vivo anti-tumor activity of the TAT-vif1 and TAT-vif2 peptides.
Figure 6. TAT-vif1 and TAT-vif2 induce tumor suppression in vivo.
Time course of bioluminescent images of tumors formed from BCBL-1-Luc cells in NOD/SCID in response to intraperitoneal injection with 1 mg of TAT, TAT-vif1, or TAT-vif2 peptide for two weeks (See also Supplementary Fig. 10).
Significant increase of anti-tumor activity by the combined treatment of the TAT-vif1 and TAT-vif2 peptides
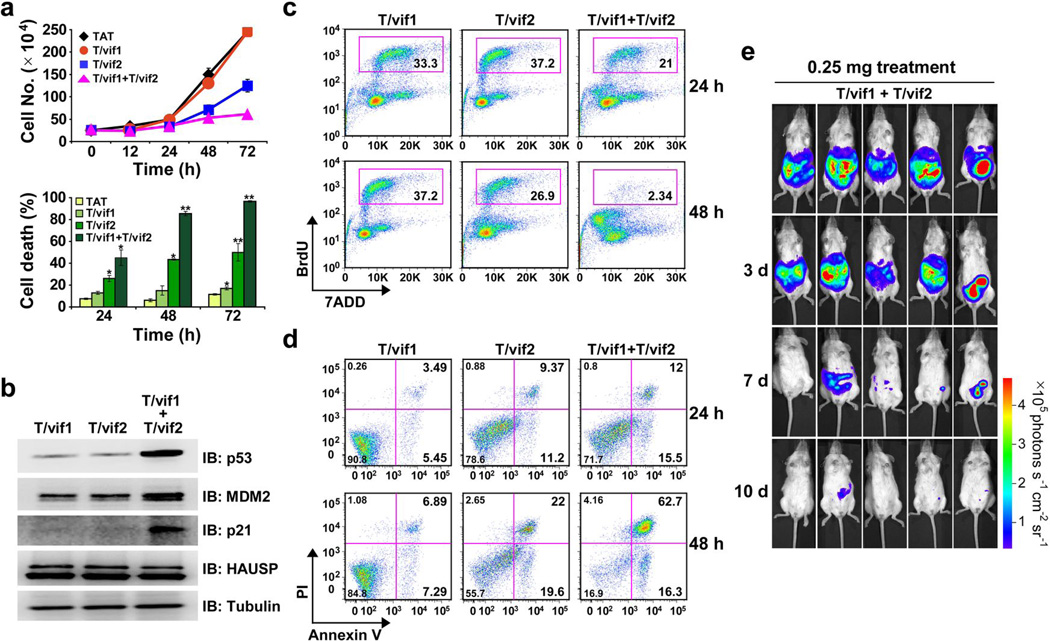
Due to their bilateral inhibition of HAUSP activity, combined treatment of the TAT-vif1 and TAT-vif2 peptides exhibited significant increases in anti-tumor activity: treatment with 25 µM of the TAT-vif1 and TAT-vif2 peptides was highly effective in inducing p53-dependent cell growth suppression, cell cycle arrest, as well as cell death, concordant with that of 100 µM treatment of the TAT-vif1 or TAT-vif2 peptide individually (Fig. 7a,c, and d and Supplementary Fig. 11a,b). Furthermore, incubation of exponentially growing PELs cells with the combination of the TAT-vif1 and TAT-vif2 peptides (25 µM of each) for 6 h robustly increased the levels of p53, p21, and MDM2 (Fig. 7b and Supplementary Fig. 11c). Marked tumor regression was observed when NOD/SCID mice with advanced PEL were challenged with TAT-vif1 and TAT-vif2 peptide combination (each at a dose of 0.25 mg, equivalent to ~25 µM) (Fig. 7e). These results demonstrate the increased anti-tumor activity of the combination of the TAT-vif1 and TAT-vif2 peptides.
Figure 7. Combination therapeutic effects of low dose of TAT-vif1 and TAT-vif2 peptides.
(a) BCBL-1 cells were treated with 25 µM of each peptide, alone or in combination, for the indicated time periods. Cells were examined by trypan blue staining for cell death analysis or cell number counting for cell growth. *P < 0.05 and ** P < 0.01. (b) BCBL-1 cells were incubated for 6 h with 25 µM of TAT-vif1, TAT-vif2, or both. WCL were subjected to SDS-PAGE followed by IB and were analyzed for p53, MDM2, p21, and HAUSP expression. (c) Asynchronously growing BCBL-1 cells were treated with 25 µM of the designated peptide or combined for the indicated time periods. Cells were pulse-labeled with BrdU and analyzed for DNA content by flow cytometry. BrdU incorporation during the S phase is indicated as the percentage of stained cells. (d) Scatter plot of Annexin V-FITC/PI flow cytometry of BCBL-1 cells after exposure to 25 µM peptide treatments for different time periods. Data are representative of three independent experiments. (e) After tumors were established in the NOD/SCID mice, 0.25 mg each of TAT-vif1 and TAT-vif2 peptide were injected together for 2 weeks. The tumors were measured by in vivo bioluminescence imaging.
DISCUSSION
In order to bypass interferon (IFN)- and p53-mediated irreversible cell cycle arrest and apoptosis and complete its life cycle, KSHV has developed a unique mechanism for antagonizing cellular IFN- and p53-mediated host innate immune responses by incorporating viral homologues of cellular IFN regulatory factors (IRFs), called viral IFN regulatory factors (vIRFs). KSHV encodes four vIRFs in a cluster region that have two common biological functions that contribute to host immune evasion: the inhibition of host IFN-mediated innate immunity and the deregulation of p53-mediated tumor suppressive activity34,35. We previously reported that KSHV vIRF4 interacts with MDM2 through its C-terminal region, leading to the inhibition of MDM2 auto-ubiquitination and stabilization of its protein levels, thus enhancing MDM2-mediated p53 degradation20. Here, we show that vIRF4’s actions with HAUSP and MDM2 are functionally and genetically separable, and that the vIRF4-MDM2 interaction has a more significant role in downregulating p53 than the vIRF4-HAUSP interaction. Thus, KSHV vIRF4 has evolved to target two components, MDM2 and HAUSP, of the p53 pathway to comprehensively suppress p53 function. This suggests that vIRF4 affects p53 tumor suppressor-mediated surveillance in a similar but distinct way from other viral proteins such as the EBV EBNA1. Finally, it should be noted that vIRF4 circumvents host growth surveillance to enhance viral replication in infected cells in a manner reminiscent of HSV ICP0 and EBV EBNA1, which also direct HAUSP to promote viral replication and oncogenic processes, respectively7,15. Thus, HAUSP appears to be a common host target for human herpesviruses to support their lifecycle and pathogenesis, and is hence considered to be a potential target for anti-viral therapy.
Using crystal structure analysis facilitated by in situ proteolysis, together with ITC assessment, we demonstrate that while KSHV vIRF4 (residues ~210–216) binds to the same surface groove of the HAUSP TRAF domain as that recognized by MDM2 and p53, it employs a different HAUSP interaction strategy from MDM2 and p53. Besides residues 210–216, vIRF4 utilizes an additional, short upstream region (residues 202–208) to interact extensively with the other side of the TRAF domain β-sheet, especially the β6 strand (Fig. 2a,b). It is this extra interaction property of vIRF4 that results in a much higher binding affinity to the HAUSP TRAF domain than MDM2 and p53.
Interestingly, in a study using a comparably designed peptide EBNA1435–449 containing the P/A××S motif and sequences equivalent to the vIRF4 upstream region, EBV EBNA1435–449 showed a very similar HAUSP TRAF binding affinity (Kd of 0.48 µM) to that of the vIRF4202–216 peptide (Kd of 0.39 µM) (Supplementary Fig. 1b). However, NMR chemical shift mapping analysis demonstrated that, unlike the vIRF4 peptide, the EBNA1 sequences corresponding to the upstream region of the vIRF4 peptide did not affect its interaction with the HAUSP TRAF domain15. This result suggests that KSHV vIRF4 and EBV EBNA1 target HAUSP in similar but distinct manners. It is worth noting that EBNA1 is the only HAUSP TRAF binding substrate that has a charged side chain residue Glu444 at the position of “P/A” in the motif P/A××S motif (Supplementary Fig. 1b and Fig. 2b). This residue seems to be critical for how EBNA1 attains a remarkably higher HAUSP TRAF binding affinity than cellular substrates as the Glu444 side chain interacts with the backbone amide and carbonyl group of Ser155 and Arg153 on the β6 strand, respectively, through bridging water molecules, as well as with the indole nitrogen of TRAF Trp165 (Supplementary Fig. 1e). Moreover, EBNA1 employs four backbone-backbone interactions with the Trp165-Asn169 residues on the TRAF β7 strand (Supplementary Fig. 1e) while cellular substrates utilize only two main chains to interact with the same TRAF strand (data not shown). As a consequence, the intelligently evolved EBNA1 peptide may compete with cellular substrates although they employ similar HAUSP TRAF binding patterns (Fig. 2b). These competences of EBNA1 may collectively result in a comparable effect as that seen with vIRF4202–216 to manage HAUSP interactions with cellular substrates. Furthermore, independently of its inhibition of p53-mediated apoptosis, the EBNA1-HAUSP interaction is also involved in PML disruption to promote the survival of cells with DNA damages36. Thus, two human gamma herpesviruses have evolved to target and deregulate HAUSP function and its related pathways, but the consequences of their action may be significantly different.
Along with the structure of the HAUSP TRAF-DUB domain13, our ITC, NMR, and deubiquitinase enzyme analyses suggest that a peptide, approximately 17-amino-acids (residues 216 to 232) in length, may be sufficiently long enough to stretch from the TRAF domain to the DUB domain of HAUSP, while additional sequences around residues 232–236 may directly contact the catalytic core site of HAUSP, resulting in the robust inhibition of its enzymatic activity. Due to these exceptional properties of vIRF4, the vif1 and vif2 peptides clearly posses two provocative and effective strategies that make them specific and robust HAUSP antagonists: the vif1 peptide binds to the HAUSP TRAF domain with the highest affinity among all reported substrates, blocking the TRAF binding by cellular substrates, whereas the vif2 peptide appears to loosely bind the TRAF domain and the active catalytic site of the DUB domain of HAUSP, suppressing its DUB enzymatic activity. Consequently, vif1 and vif2 peptides comprehensively suppress HAUSP activity, effectively restore p53-dependent apoptosis in wild-type p53-carrying cancer cells, and suppress tumor growth in mouse xenograft models. Our study provides unquestionable evidence that the vif1 and vif2 peptides are not only ideal for abolishing HAUSP activity, but also demonstrates that they have the potential to be therapeutically beneficial reagents against p53 wild-type tumors. However, we should stress that the vIRF4 peptides studied here do not reflect the authentic biology of full-length vIRF4, which targets both HAUSP and MDM2 to comprehensively inactivate host p53-mediated surveillance. Thus, the therapeutic exploitation of the vIRF4 peptides as HAUSP inhibitors should be uncoupled from the biological function of vIRF4 as a survival factor against host’s immune surveillance programs.
Of importance, since HAUSP contains a well-conserved catalytic core domain, it should therefore be a more tractable drug target than blocking the p53-MDM2-MDMX interaction. This is underscored by recent findings showing that HAUSP can be used to therapeutically target p53-independent apoptosis responses in certain tumors. For instance, the loss of HAUSP in colorectal cancer cells potentiates apoptotic cell death by inducing proteasomal degradation of DNMT1, further compounding the potential of HAUSP inhibition as an effective anti-cancer therapy37–40. Accordingly, because we cannot exclude the possibility that HAUSP regulates the function of other known or yet unknown substrates involved in p53-independent apoptosis, our peptides may have additional off-target, p53-independent pro-apoptotic effects.
In summary, our study shows that vIRF4-derived short vif1 and vif2 peptides comprehensively suppress HAUSP activity, effectively restoring p53-dependent apoptosis in wild-type p53-carrying cancer cells and suppressing tumor growth in a mouse xenograft model. Of especial importance, we herein report that the vif1 and vif2 peptides represent potential novel chemotherapeutic molecules for anti-cancer therapies.
METHODS
Crystallization, data collection, and structure determination
Crystallization trials were carried out using an in situ proteolysis technique19. The purified HAUSP62–205-vIRF4153–256 complex (50 mg ml−1) was mixed with trypsin (1 mg ml−1) in a 100:1 (v/v) ratio. The protease-treated protein complexes were used immediately for crystallization trials. Crystals were grown for 1 week under conditions of 5% PEG 3350 and 0.2 M magnesium formate (pH 5.9) in an alternate reservoir containing a 1.5 M NaCl solution at 21 °C. Crystals were transferred to a cryoprotectant solution containing 30% PEG 3350 and 0.2 M magnesium formate (pH 5.9), incubated for 2 h, and then retrieved and placed immediately in a −173 °C nitrogen gas stream. X-ray diffraction data were collected at 1.6 Å resolution on beamline 4A at the Pohang Accelerator Laboratory (Pohang, Korea). All data were processed using the HKL2000 program suite41. The crystal of the protein complex belongs to space group P3221. There is one complex in the asymmetric unit, with a packing density of ~2.26 Å3/Da, corresponding to an estimated solvent content of approximately 45.72%. The crystal structure was determined by molecular replacement using the MOLREP program42. The HAUSP TRAF domain structure (PDB accession code 2F1W) was used as a search model. The initial model was used as a guide to build the remainder of the protein manually into electron density maps with the program COOT43. Refinement was performed with REFMAC544 and included the translation-libration-screw procedure. The final refined model resulted in Rfree and Rcryst values of 0.174 and 0.158, respectively. The model contains 143 amino acids of the HAUSP TRAF domain, 15 residues of vIRF4, and 238 water molecules, and satisfies the quality criteria limits of the program PROCHECK45. The crystallographic data statistics are summarized in Table 1.
Additional methods
Information on cell culture, reagents plasmid constructions, yeast-two hybrid screen, protein purification, immunoblotting and immunoprecipitation, isothermal titiration calorimetry, NMR spectroscopy, in vitro ubiquitination assay, in vitro deubiquitination (DUB) assay, cell proliferation and viability, cell cycle and apoptosis, in vivo bioimaging is available in the Supplementary methods.
ACKNOWLEDGEMENTS
This work was partly supported by CA82057, CA31363, CA115284, CA147868, CA148616, DE019085, Hastings Foundation, Fletcher Jones Foundation, WLBH Foundation, and the GRL Program (K20815000001) from National Research Foundation of Korea (JUJ); and the 21C Frontier Microbial Genomics and Applications Center Program and the National Research Foundation of Korea Grant (NRF-M1AXA002-2010-0029767) funded by the Ministry of Education, Science and Technology, Korea (MHK). We thank the staff at the 4A and 6B beamlines, Pohang Accelerator Laboratory, Korea, for help with the data collection. We thank Drs. Lombardi, Jeong, and Lee for their help. Finally, we thank all lab members for their support and discussions.
Footnotes
Accession Codes
The atomic coordinates and structure factor amplitudes of the protein complex have been deposited in the Protein Data Bank (PDB)46 under accession code 2XXN.
CONTRIBUIONS
H.-R.L. performed all aspect of this study; W.-C.C., J.H. and M.H.K. did X-ray crystallographic and biochemical studies; S.L. and Z.T. assited with the experimental design and collected the data; E.H. and Y.H.J. carried out NMR studies; K.G. did protein purification; J.H. provided KSHV library; H.-R.L., W.-C.C., O.-K.O., M.H.K. and J.J. organized this study and wrote the manuscript. All authors discussed the results and commented on the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests
REFERENCES
- 1.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 3.Brown CJ, Cheok CF, Verma CS, Lane DP. Reactivation of p53: from peptides to small molecules. Trends Pharmacol Sci. 2011;32:53–62. doi: 10.1016/j.tips.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Lain S, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13:454–463. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–559. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 6.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett RD, et al. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks CL, Li M, Hu M, Shi Y, Gu W. The p53--Mdm2--HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene. 2007;26:7262–7266. doi: 10.1038/sj.onc.1210531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummins JM, Vogelstein B. HAUSP is required for p53 destabilization. Cell Cycle. 2004;3:689–692. [PubMed] [Google Scholar]
- 11.Cummins JM, et al. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1. doi: 10.1038/nature02501. p following 486. [DOI] [PubMed] [Google Scholar]
- 12.Meulmeester E, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 13.Hu M, et al. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheng Y, et al. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- 15.Saridakis V, et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol Cell. 2005;18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 16.Holowaty MN, et al. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J Biol Chem. 2003;278:29987–29994. doi: 10.1074/jbc.M303977200. [DOI] [PubMed] [Google Scholar]
- 17.Uetz P, et al. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 18.Sarkari F, et al. Further insight into substrate recognition by USP7: structural and biochemical analysis of the HdmX and Hdm2 interactions with USP7. J Mol Biol. 2010;402:825–837. doi: 10.1016/j.jmb.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Dong A, et al. In situ proteolysis for protein crystallization and structure determination. Nat Methods. 2007;4:1019–1021. doi: 10.1038/nmeth1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HR, et al. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 4 targets MDM2 to deregulate the p53 tumor suppressor pathway. J Virol. 2009;83:6739–6747. doi: 10.1128/JVI.02353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min CK, et al. Role of amphipathic helix of a herpesviral protein in membrane deformation and T cell receptor downregulation. PLoS Pathog. 2008;4:e1000209. doi: 10.1371/journal.ppat.1000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 23.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 24.Gump JM, Dowdy SF. TAT transduction: the molecular mechanism and therapeutic prospects. Trends Mol Med. 2007;13:443–448. doi: 10.1016/j.molmed.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Petre CE, Sin SH, Dittmer DP. Functional p53 signaling in Kaposi's sarcoma-associated herpesvirus lymphomas: implications for therapy. J Virol. 2007;81:1912–1922. doi: 10.1128/JVI.01757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katano H, Sato Y, Sata T. Expression of p53 and human herpesvirus-8 (HHV-8)-encoded latency-associated nuclear antigen with inhibition of apoptosis in HHV-8-associated malignancies. Cancer. 2001;92:3076–3084. doi: 10.1002/1097-0142(20011215)92:12<3076::aid-cncr10117>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 27.Bhatia K, et al. Hemi- or homozygosity: a requirement for some but not other p53 mutant proteins to accumulate and exert a pathogenetic effect. FASEB J. 1993;7:951–956. doi: 10.1096/fasebj.7.10.8344493. [DOI] [PubMed] [Google Scholar]
- 28.Zhang R, Wang H, Agrawal S. Novel antisense anti-MDM2 mixed-backbone oligonucleotides: proof of principle, in vitro and in vivo activities, and mechanisms. Curr Cancer Drug Targets. 2005;5:43–49. doi: 10.2174/1568009053332663. [DOI] [PubMed] [Google Scholar]
- 29.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 31.Keller SA, et al. NF-kappaB is essential for the progression of KSHV- and EBV-infected lymphomas in vivo. Blood. 2006;107:3295–3302. doi: 10.1182/blood-2005-07-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JS, et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol. 2009;11:1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu W, Rochford R, Toomey L, Harrington W, Jr, Feuer G. Inhibition of HHV-8/KSHV infected primary effusion lymphomas in NOD/SCID mice by azidothymidine and interferon-alpha. Leuk Res. 2005;29:545–555. doi: 10.1016/j.leukres.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Lee HR, Lee S, Chaudhary PM, Gill P, Jung JU. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Future Microbiol. 2010;5:1349–1365. doi: 10.2217/fmb.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HR, Kim MH, Lee JS, Liang C, Jung JU. Viral interferon regulatory factors. J Interferon Cytokine Res. 2009;29:621–627. doi: 10.1089/jir.2009.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sivachandran N, Sarkari F, Frappier L. Epstein-Barr nuclear antigen 1 contributes to nasopharyngeal carcinoma through disruption of PML nuclear bodies. PLoS Pathog. 2008;4:e1000170. doi: 10.1371/journal.ppat.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du Z, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Marzo AM, et al. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res. 1999;59:3855–3860. [PubMed] [Google Scholar]
- 39.Agoston AT, et al. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem. 2005;280:18302–18310. doi: 10.1074/jbc.M501675200. [DOI] [PubMed] [Google Scholar]
- 40.Bronner C. Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. Sci Signal. 2011;4:pe3. doi: 10.1126/scisignal.2001764. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski ZMW. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Vagin AaTA. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 46.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]