Georgia et al. investigate how cell fate transitions are regulated during pancreatic organogenesis. With conditional depletion of Dnmt1 and lineage tracing experiments, they present evidence for a physiological link between DNMT1, p53, and development of the pancreas. These results underscore the context-dependent roles of DNMT1 in tissue-specific differentiation and suggest that Dnmt1 is required to prevent p53-dependent apoptosis during lineage specification in pancreatic progenitor cells.
Keywords: pancreatic development, DNMT1, differentiation, DNA methylation, p53
Abstract
In the developing pancreas, self-renewal of progenitors and patterning of cell fates are coordinated to ensure the correct size and cellular makeup of the organ. How this coordination is achieved, however, is not clear. We report that deletion of DNA methyltransferase 1 (Dnmt1) in pancreatic progenitors results in agenesis of the pancreas due to apoptosis of progenitor cells. We show that DNMT1 is bound to the p53 regulatory region and that loss of Dnmt1 results in derepression of the p53 locus. Haploinsufficiency of p53 rescues progenitor cell survival and cellular makeup of the Dnmt1-deleted pancreas.
The vertebrate pancreas is a complex organ made of functionally different tissue types that arise from a common progenitor cell pool during embryogenesis (Gu et al. 2003; Jensen 2004; Gittes 2009). Lineage tracing analysis indicates that a pool of pancreatic progenitor cells persists throughout organogenesis to give rise to differentiated mature cell types in a spatially and temporally regulated manner (Herrera 2000; Jensen et al. 2000a; Gu et al. 2002). The maintenance of the progenitor cell pool requires that progenitor cell self-renewal is balanced with cell cycle exit to coordinate differentiation of mature cell types of the pancreas (Georgia et al. 2006).
DNA methylation is a potential epigenetic mechanism that may be involved in maintaining the balance between self-renewal and terminal differentiation in the developing pancreas. DNA methylation may serve as a means of repressing pluripotency genes in multipotent progenitor cells or a means of repressing lineage-specific differentiation genes in multipotent progenitor cells (Reik 2007). It is not clear whether pancreatic progenitor DNA methylation patterns must be inherited during self-renewal to maintain a functional progenitor population by repressing differentiation genes or to serve as a basis for establishing lineage-specific methlylation patterns during differentiation.
DNA methyltransferase 1 (Dnmt1) is the enzyme that propagates DNA methylation patterns during cell division. Previous studies on Dnmt1 have yielded interesting yet divergent data about its role in maintenance and differentiation of progenitor cell types. Dnmt1-null mouse embryonic stem cells (mESCs) undergo limited differentiation and apoptosis and are eliminated in competition assays with wild-type mESCs yet have a proliferative capacity comparable with that of wild-type mESCs when maintained in an undifferentiated state (Lei et al. 1996; Panning and Jaenisch 1996). Dnmt1-null embryos expire from growth arrest and severe neural tube distortions by embryonic day 8.5 (E8.5), which indicates that Dnmt1-null mESCs are capable of undergoing more extensive differentiation in vivo than in vitro (Li et al. 1992; Lei et al. 1996). Loss of Dnmt1 in hematopoetic progenitor cells results in defects in self-renewal and niche retention and an inappropriate shift to myeloid progenitor cell cycling and differentiation (Trowbridge et al. 2009). Studies in human adult epidermal stem cells indicate that Dnmt1 maintains progenitor self-renewal by repressing genes that trigger cell cycle exit and premature differentiation (Sen et al. 2010). During neurogenesis, Dnmt1 restricts precocious astrogliogenesis of neural precursor cells, but deletion of Dnmt1 in telencephalic precursors results in cell death and forebrain degeneration (Fan et al. 2005; Hutnick et al. 2009). Taken together, these studies present context-dependent roles for Dnmt1 that may have differential effects in vitro or in vivo.
To investigate the in vivo role of DNMT1 in progenitor maintenance and concomitant lineage specification during organogenesis, we conditionally deleted Dnmt1 in pancreatic progenitor cells. Analysis of progenitor cell self-renewal dynamics revealed that Dnmt1-null pancreatic progenitor cells undergo G2 arrest. Our studies indicate that DNMT1 functions to repress p53. In its absence, p53 expression is derepressed, resulting in apoptosis of pancreatic progenitor cells. We show that p53 haploinsufficiency rescues progenitor cell survival and restores pancreatic organogenesis in the Dnmt1-null embryos. These data suggest that Dnmt1 is required to prevent p53-dependent apoptosis during lineage specification of pancreatic progenitor cells.
Results and Discussion
Loss of Dnmt1 results in a severely atrophic pancreas
We bred mice with a floxed allele of Dnmt1 (DNMT1fl/fl) with mice transgenic for Cre recombinase under the control of the Pdx1 promoter (Jackson-Grusby et al. 2001; Gu et al. 2002). This restricted Dnmt1 excision to the pancreatic epithelium (DNMT1PC). To facilitate lineage tracing studies, we also bred in the stop-floxed-R26RYFP to mark all cells derived from progenitor cells that expressed the Pdx1-Cre transgene (Srinivas et al. 2001). To confirm that deletion of Dnmt1 resulted in hypomethylation of the self-renewing pancreatic progenitor pool, we used immunohistochemistry to detect 5-methyl-cytosine (5mC). Unlike the control epithelium, where 5mC uniformly stained the epithelium and surrounding mesenchyme, 5mC staining was grossly diminished in the epithelium of the DNMT1PC pancreas but maintained in the surrounding mesenchyme (Supplemental Fig. 1A,B). This indicated that deletion of Dnmt1 resulted in hypomethylation of the pancreatic epithelium during organogenesis. Previous reports have shown that loss of Dnmt1 leads to derepression of intracisternal A particle (IAP), a core retroviral element protein (Fan et al. 2001). Consistent with these previous studies, IAP staining was not detected in the pancreatic epithelium of control E13.5 embryos, but high levels of IAP were detected in the DNMT1PC pancreatic epithelium (Supplemental Fig. 1C,D). Some glucagon-positive cells in the DNMT1PC pancreatic epithelium were negative for IAP expression and were likely derived from cells that had escaped Cre-mediated recombination. These data indicated that Pdx1-Cre expression efficiently deleted Dnmt1 from the large majority of pancreatic progenitor cells early in development and led to hypomethylation of the pancreatic epithelial cells.
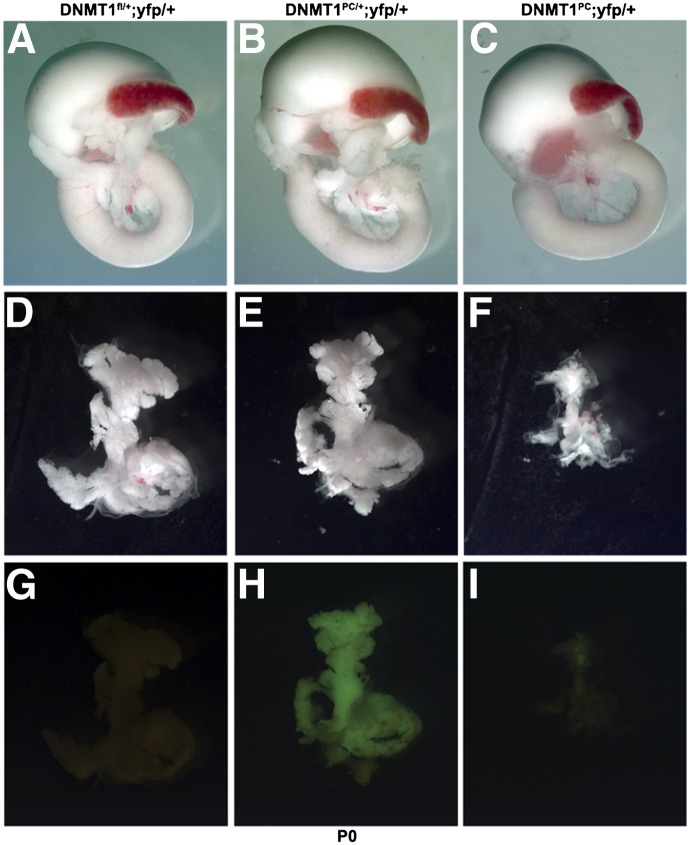
We next examined the effects of Dnmt1 deletion on pancreatic organogenesis by analyzing DNMT1PC litters at birth. DNMT1PC animals were born alive at expected Mendelian ratios. Examination of littermates indicated that the pancreas of pups in which one allele of Dnmt1 was deleted were grossly normal and comparable with wild-type control littermates (Fig. 1 A,B, D,E). YFP expression was absent in the control animals and homogenously distributed throughout the pancreas in heterozygous pups (Fig. 1G,H). Strikingly, gross examination of the DNMT1PC pancreas revealed a severely atrophic pancreas (Fig. 1C,F). The rudimentary DNMT1PC pancreas displayed little YFP expression, indicating that most of the atrophic pancreatic tissue was derived from cells that had escaped recombination (Fig. 1I). These data indicated that Dnmt1 is essential for the formation of the pancreas.
Figure 1.
DNMT1 deletion results in an atrophic pancreas. (A–C) Gastrointestinal tract of control (DNMT1fl/fl) (A), heterozygous (DNMT1PC/+) (B), and conditional knockout (DNMT1PC) (C) pancreas at postnatal day 0 (P0). (D–F) Pancreas dissected from the gastrointestinal tract illustrate the atrophic size of the DNMT1PC pancreas. (G–I) Lineage trace analysis indicates that there is efficient recombination in the heterozygous DNMT1PC/+ pancreas (H), but very few recombined cells persist in the DNMT1PC pancreas (I).
Previous studies in zebrafish indicated that loss of Dnmt1 resulted in degeneration of the acinar pancreas, but ductal and endocrine lineages were spared, leaving open the possibility that the atrophic pancreas that we observed could consist of primarily endocrine and ductal cells (Anderson et al. 2009). To determine whether the absence of Dnmt1 resulted in loss of specific cell lineages, we carried out immunohistological analysis for differentiated cell types in the DNMT1PC pancreas. Antibody staining against exocrine cells expressing amylase and endocrine cells expressing insulin showed that scattered clusters of both endocrine and exocrine cells were present in the DNMT1PC pancreas, but the typical rosette architecture of the acinar tissue and islet clusters of insulin cells was disrupted (Supplemental Fig. S2A–D). Cells that stained for the ductal marker mucin were scattered throughout the DNMT1PC pancreas (data not shown). From this analysis, we concluded that deletion of Dnmt1 from pancreatic epithelial progenitor cells did not impact any particular lineage and instead disrupted the architecture and severely reduced the numbers of differentiated cells of all lineage of the mature pancreas.
Deletion of Dnmt1 in pancreatic progenitors results in the absence of differentiated cells
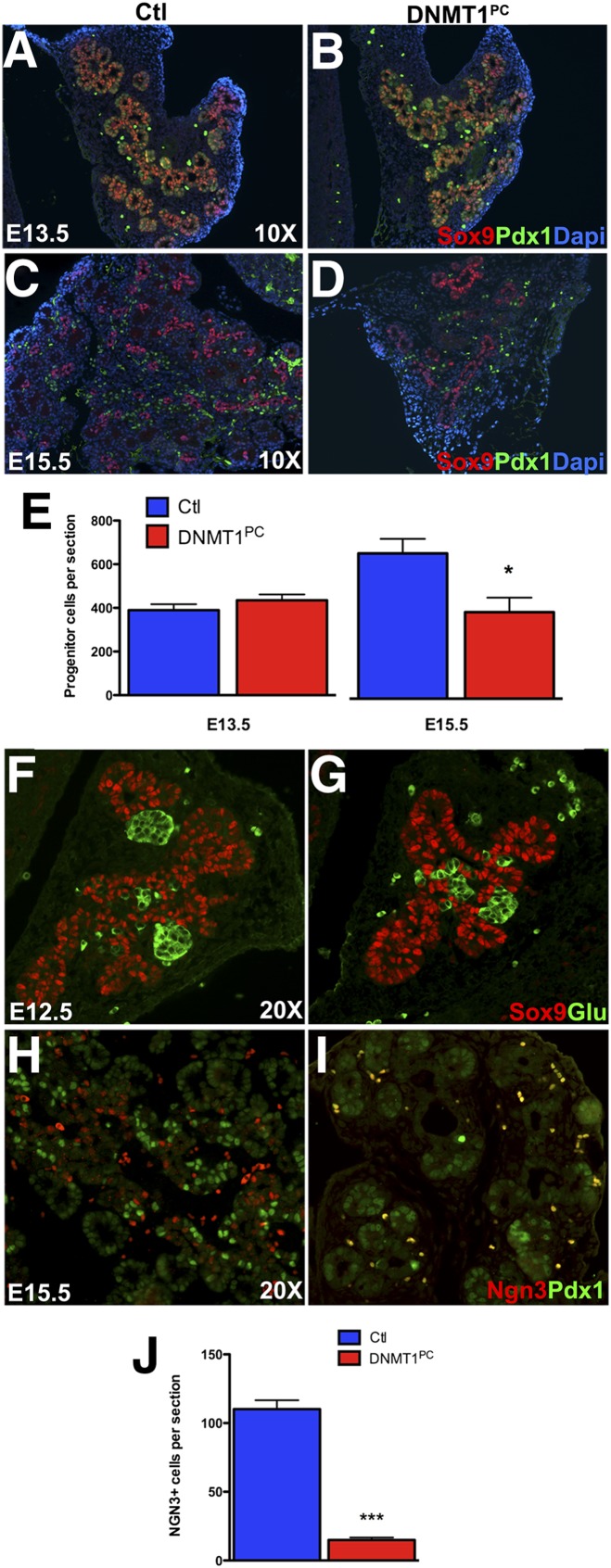
We investigated whether a severely reduced pancreas size could result from depletion of the pancreatic progenitor pool during embryogenesis. We examined DNMT1PC and control littermate pancreas by immunostaining for pancreatic progenitor markers Pdx1 and Sox9. At E13.5, the overlapping Pdx1 and Sox9 populations were comparable in the control and DNMT1PC pancreatic epithelium (P = 0.2568) (Fig. 2A,B,E). In contrast, the number of cells staining for Sox9 at E15.5 was reduced by >40% in the DNMT1PC pancreas compared with the control littermate pancreas (P = 0.0137) (Fig. 2C,D,E). This indicated that the progenitor pool is depleted during pancreatic organogenesis and results in a severely atrophic pancreas. The depletion of pancreatic progenitors could be due to either a defect in self-renewal or precocious differentiation. Precocious differentiation leading to depletion of the pancreatic progenitor pool has been previously described (Jensen et al. 2000b; Bhushan et al. 2001). To assess precocious differentiation, we carried out immunohistochemistry of E12.5 pancreas with antibodies against Sox9 to mark progenitor cells and glucagon to mark differentiated cells. The size of the pancreatic epithelial bud and the number of glucagon cells from E12.5 embryos immunostained with Sox9 and glucagon were comparable in DNMT1PC and control littermates (Fig. 2F–G). To further assess differentiation, we compared global gene expression by microarray analysis in control and DNMT1PC pancreas. There was less than a twofold difference between most transcripts in the control and DNMT1PC pancreas, which suggests that the majority of the cells in the DNMT1PC pancreas had a transcriptome similar to that of the control pancreas (Supplemental Fig. 3A). Gene enrichment analysis indicated that a subset of genes involved in pancreatic differentiation was specifically down-regulated. Neurogenin 3, the gene required for differentiation of all endocrine cell types, was one of the most down-regulated gene in the array analysis, with a >12-fold decrease in expression in the DNMT1PC pancreas (Schwitzgebel et al. 2000). Other transcription factors involved in endocrine and exocrine lineage specification—such as Nkx6.1, Nkx2.2, Rfx6, Rbjk-l, and Ptf1—were specifically down-regulated (Supplemental Fig. 3B). We sought to confirm the gene expression profiles by immunohistochemistry. Immunostaining of the control pancreas at E15.5 showed Ngn3+ cells along the embryonic duct surrounded by pancreatic progenitor cells expressing Pdx1. In striking contrast, the number of Ngn3+ cells in the DNMT1PC pancreas was <15% that of the control littermates (P < 0.005) (Fig. 2H–J). These results indicated that in the absence of Dnmt1, pancreatic epithelial cells were not depleted due to precocious differentiation, and in fact, very few differentiated cells were observed in the DNMT1PC pancreas.
Figure 2.
DNMT1PC pancreatic atrophy is the result of progenitor cell depletion. (A,B) Immunohistochemistry for Sox9 and Pdx1 at E13.5 indicates no notable difference in the size or morphology in control (A) and DNMT1PC (B) pancreatic epithelium. (C,D) Immunohistochemistry of pancreatic epithelium for Sox9 and Pdx1 at E15.5 reveals a decrease in the number progenitor cells in the DNMT1PC pancreatic epithelium. (E) Quantification of progenitor cells (E13.5) and Sox9 cells (E15.5). (F,G) Immunohistochemical comparison of the progenitor and glucagon populations at E12.5 in the control and DNMT1PC suggests that precocious differentiation is not responsible for progenitor depletion. (H,I). By E15.5, DNMT1PC pancreas have little expression of pre-endocrine cell marker Ngn3 (I) in comparison with control littermates (H). (J) Quantification of Ngn3+ cells at E15.5.
Loss of Dnmt1 resulted in accumulation of progenitor cells in the G2 phase
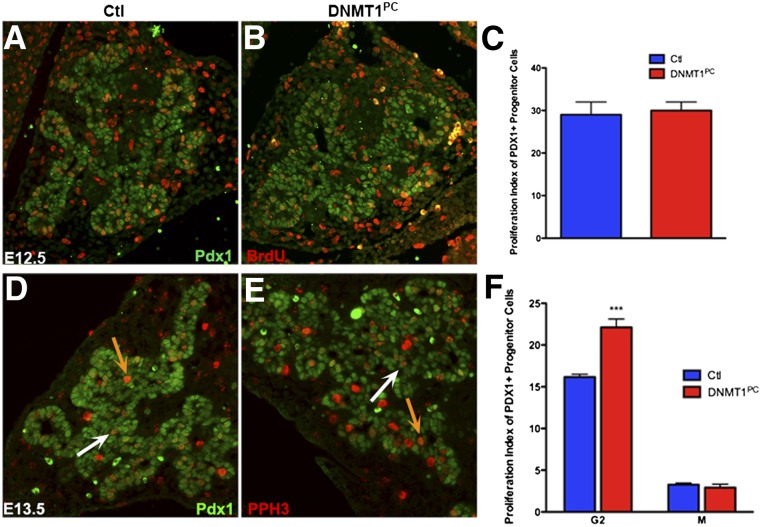
To investigate whether loss of Dnmt1 resulted in depletion of pancreatic progenitor cells due to defects in self-renewal, we analyzed embryos from pregnant dams injected with a short pulse of BrdU. Control and DNMT1PC pancreas were immunostained with antibodies against Pdx1 to mark pancreatic progenitor cells and BrdU to identify cells actively in S phase of the cell cycle (Fig. 3A,B). The Pdx1+BrdU+ cells were counted to determine the proliferation index, calculated as (Pdx1+BrdU+)/Pdx1+. There were no differences in the proliferation index between control and DNMT1PC progenitor cells at E12.5 (Fig. 3C). Because a short BrdU pulse only measures the cells that are actively in S phase, we pursued additional proliferation studies to determine whether the progenitor cells were completing the cell cycle. Control and DNMT1PC pancreas were immunostained with antibodies against phosphorylated histone H3, a marker for G2/M phase, and Pdx1 (Fig. 3D,E). Cells with a speckled PHH3 pattern were categorized as G2 phase of the cell cycle (Fig. 3D,E, white arrows), and cells with more robust staining were categorized as M phase (Fig. 3D,E, orange arrows). Ratios of Pdx1+PHH3+/Pdx1+ were used to calculate the proliferation index. Comparing control and DNMT1PC pancreas revealed that the number of M-phase progenitor cells was not significantly different, but the number of progenitor cells in G2 phase in the DNMT1PC pancreas was increased by 40% (P < 0.005) (Fig 3F). The fact that S-phase and M-phase progenitor cells did not change in the DNMT1PC pancreas indicated that cell cycle entry and mitosis of pancreatic epithelial cells were not perturbed by the deletion of Dnmt1. The accumulation of a subset of pancreatic epithelial cells in G2 arrest suggested that depletion of these cells could be due to apoptosis.
Figure 3.
A subset of DNMT1PC progenitor cells accumulates in G2 arrest. (A–C) E12.5 pancreatic sections immunostained for BrdU and Pdx1 to quantify progenitor proliferation. (D–F) Quantification of pancreatic progenitor cell proliferation at E13.5 as a percentage of phosphorylated histone H3+ Pdx1+ double cells indicates that there is an increase in the number of cells in G2 in the DNMT1PC pancreas. (White arrows) G2-phase cells; (orange arrows) M-phase cells. (***) P < 0.005.
Deletion of Dnmt1 results in p53-dependent apoptosis of pancreatic progenitors
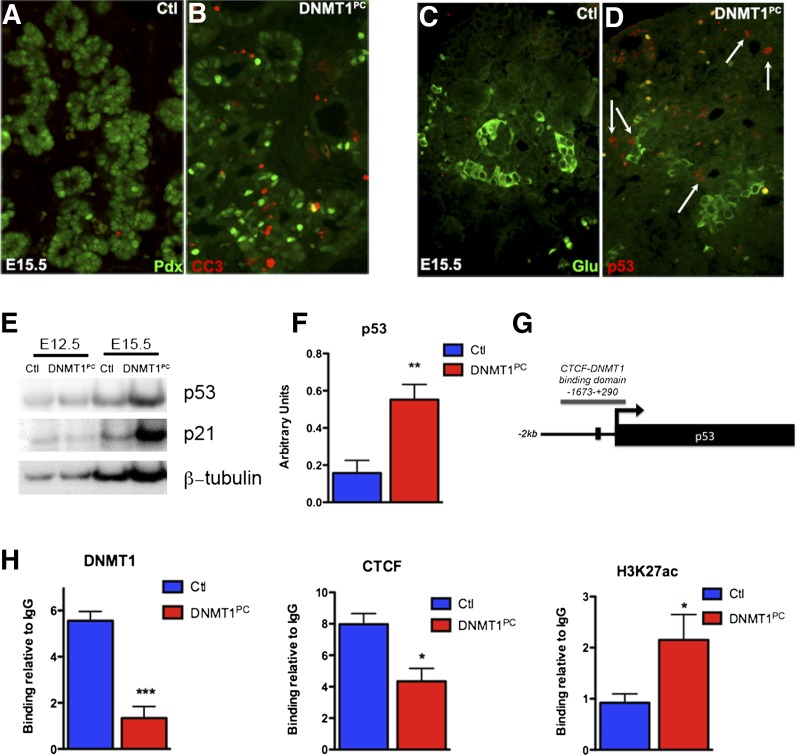
We next investigated whether apoptosis of pancreatic epithelial cells could result in the atrophic phenotype of the DNMT1PC pancreas at birth. Pancreatic sections from E15.5 DNMT1PC and control littermate embryos were stained for cleaved caspase 3, a marker for apoptosis, and Pdx1. Cleaved caspase 3 was not detected in control pancreas; however, DNMT1PC pancreas showed a number of cells within the pancreatic epithelium that were positive for cleaved caspase 3 (Fig. 4A,B).
Figure 4.
Progenitor depletion is a result of apoptosis in the DNMT1PC pancreatic epithelium. (A,B) Immunohistochemistry for cleaved caspase 3 did not reveal any apoptosis in the control pancreas (A), but apoptosis was widespread in the DNMT1PC pancreas (B). (C,D) The presence of apoptosis markers coincides with the appearance of p53 accumulation in the DNMT1PC pancreatic epithelium (D, white arrows) and is absent in control littermates at E15.5 (C). (E) Western blot analysis for p53 protein and its downstream target, p21, at E12.5 and E15. (F) Densitometric quantification of Western blots indicates a sixfold increase in p53 protein at E15.5. (G) Schematic of the ChIP assay binding in the p53 locus. (H) ChIP analysis indicates that DNMT1 binds to the p53 locus in control pancreas but is lost in DNMT1PC pancreas. Chromatin insulator protein CTCF is also bound to the p53 locus in control pancreas but is lost in the DNMT1PC pancreas. The activating histone mark H3K27ac is enriched in the DNMT1PC pancreas.
Several studies in fibroblast and cancer cell lines indicated that loss of Dnmt1 resulted in p53-dependent apoptosis, although the mechanism remains unclear (Jackson-Grusby et al. 2001; Chen et al. 2007; Anderson et al. 2009). To determine whether deletion of Dnmt1 in pancreatic progenitors resulted in changes in p53, we first assessed whether levels of p53 were altered in DNMT1PC pancreas. Immunohistochemical analysis using a p53 antibody revealed that p53 accumulated in the DNMT1PC epithelium, while no p53 staining was observed in control littermate pancreas (Fig. 4C,D, white arrows). To confirm this accumulation of p53 in DNMT1PC pancreas, we measured p53 using Western blot analysis at E12.5, prior to detectable differences in the pancreatic epithelium, and at E15.5, after the onset of p53 expression. Western blot analysis indicated that there was no difference in the accumulation of p53 at E12.5, but there was a significant sixfold increase in p53 in the DNMT1PC pancreas compared with control pancreas from littermates at E15.5 (P = 0.0098) (Fig 4E,F). Western blot analysis indicated that the p53 downstream transcriptional target p21 accumulated in the DNMT1PC pancreas (Fig. 4E). To assess whether the increase in p53 protein correlated with an increase in mRNA levels of p53, we used quantitative PCR to quantify any changes in p53 gene expression. Our analysis revealed a 20-fold increase in p53 mRNA in DNMT1PC pancreas compared with control pancreas from littermates (data not shown). The increase in p53 mRNA indicated that Dnmt1 could potentially regulate p53 at a transcriptional level.
We next assessed how loss of Dnmt1 resulted in transcriptional up-regulation of p53. We hypothesized that DNMT1 could play a role in directly repressing p53 transcription and that loss of Dnmt1 would result in derepression of p53. Supporting our hypothesis, a previous study showed that DNMT1 was bound to the p53 promoter 2 kb upstream of the p53 transcription start site. Interestingly, this upstream regulatory region also contained a binding sequence for the transcriptional repressor CTCF. Binding of CTCF to this region suppressed reporter activity (Su et al. 2009). We reasoned that DNMT1 and CTCF associated at the p53 regulatory region, and binding of CTCF to the p53 regulatory region may be disrupted in the absence of Dnmt1. To test this, E14.5 pancreas from DNMT1PC and control littermates were subjected to chromatin immunoprecipitation (ChIP) analysis (Fig. 4G). In control pancreas, both DNMT1 and CTCF were bound to the p53 regulatory region, confirming that DNMT1 and CTCF binding was associated with p53 repression (P < 0.005) (Fig. 4H). In contrast, there was no enrichment of CTCF in DNMT1PC embryos (P = 0.0268) (Fig. 4H). Moreover, enrichment of H3K27ac within the p53 promoter region was observed in the DNMT1PC embryos, which is consistent with transcriptional activation (P = 0.0423) (Fig. 4H). These results suggested that DNMT1 directly repressed p53 transcription, and loss of Dnmt1 resulted in derepression of p53.
Haploinsufficiency of p53 rescues pancreatic organogenesis
To investigate whether inactivation of Dnmt1 resulted in p53-mediated apoptosis of pancreatic progenitors, we crossed the p53-null animals into the DNMT1PC background. p53-null animals are phenotypically indistinguishable from wild-type littermates at birth, and pancreas formation was not perturbed (Jacks et al. 1994).
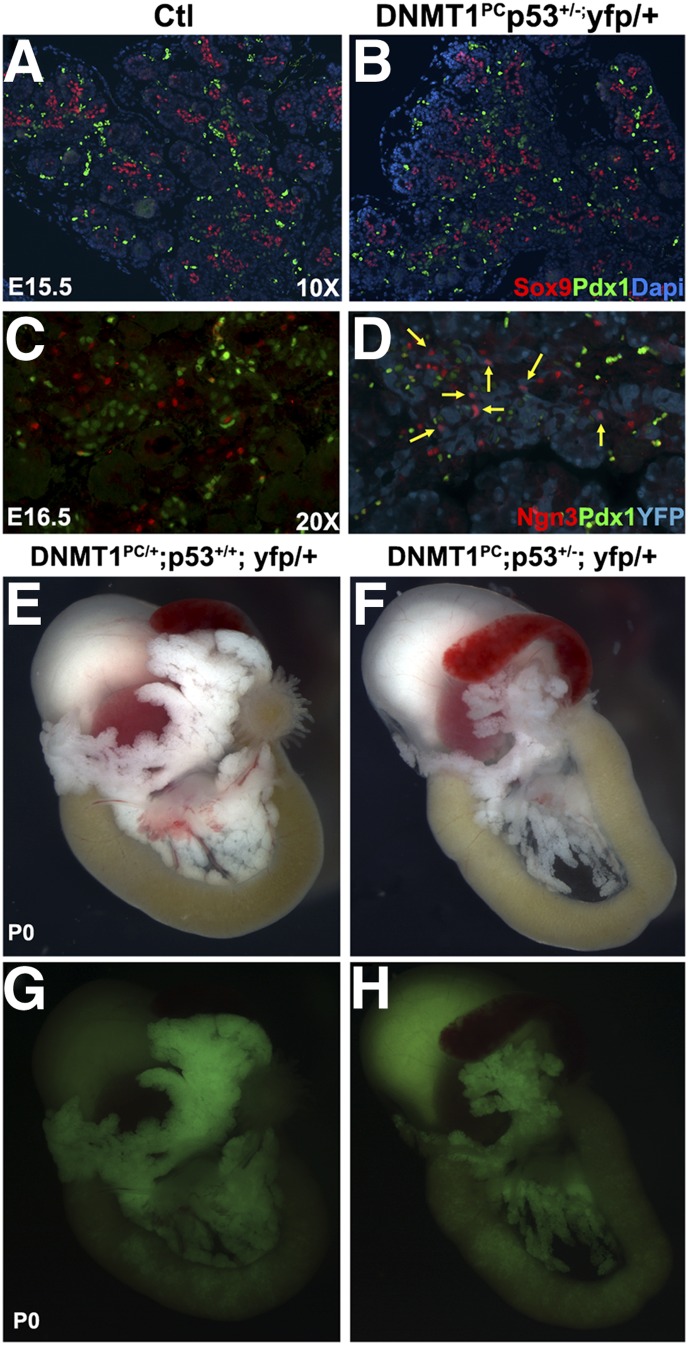
We first assessed whether haploinsufficiency of p53 restored pancreatic progenitor populations at E15.5. Immunohistochemical assessment of Sox9 and Pdx1 revealed no significant differences in the pancreatic epithelium between the control and DNMT1PCp53+/− pancreas (Fig. 5A,B). To evaluate whether p53 haploinsufficiency would restore the expression of ngn3, we examined DNMT1PCp53+/− pancreas for expression of Ngn3. Immunohistochemistry at E16.5 indicated that Ngn3+ and Pdx1+ cell populations persisted through organogenesis with a staining pattern similar to that of control littermates. Lineage tracing indicated that DNMT1PCp53+/− cells expressing Ngn3 had undergone recombination, thus leading us to conclude that the expression of Ngn3 was restored by p53 haploinsufficiency (Fig. 5C,D). At birth, DNMT1PC and DNMT1PCp53+/− animals were present at the expected Mendelian ratios. Gross histological examination of the neonatal pancreas revealed that while the pancreas of DNMT1PC animals were severely atrophic, DNMT1PCp53+/− littermates had a restoration of a full, albeit smaller, pancreas (Fig. 5E,F). Lineage tracing suggested that recombination was extensive, although not complete, in the DNMT1PCp53+/− background (Fig. 5G,H). Insulin and YFP immunostaining indicated normal morphology of the endocrine tissue compartment in the control and DNMT1PCp53+/− littermate pancreas (Supplemental Fig. 4A,B). Immunostaining for amylase and YFP indicated that exocrine cells formed morphologically normal rosette structures (Supplemental Fig. 4C,D). Mucin+/YFP+ ductal structures were present and morphologically normal in both the control and DNMT1PCp53+/− pancreas (Supplemental Fig. 4E,F). Taken together, these data indicated that p53 haploinsufficiency rescued the ability of Dnmt1-null pancreatic progenitor cells to differentiate into a normal mature organ during embryogenesis. The area of DNMT1PCp53+/− islets decreased by postnatal day 15, and blood glucose levels were elevated into the diabetic range. This suggests that while haploinsufficiency of p53 may permit organogenesis in the DNMT1PC pancreas, these cells are not able to support glucose homeostasis in adolescent mice (Supplemental Fig. 5A–E).
Figure 5.
Haploinsufficiency of p53 rescues apoptosis and allows for cell survival during differentiation in DNMT1PCp53+/− pancreas. (A,B) Immunohistochemical staining for Sox9 and Pdx indicates no significant difference in the control (A) and DNMT1PCp53+/− (B) cell populations at E15.5. (C,D) Immunohistochemical analysis of control (C) and DNMT1PCp53+/−yfp+ (D) pancreas revealed a restoration of Ngn3+ expression in the absence of Dnmt1. Overlap with YFP indicates that these cells have arisen from cells that underwent recombination. (E,F) Gross analysis of control and DNMT1PCp53+/−yfp+ indicates that mutant pancreas are comparable with control littermates at P0. (G,H) Lineage tracing by YFP expression indicates that most cells in the DNMT1PCp53+/−yfp+ pancreas have undergone recombination.
Our results suggest that p53 repression mediated by DNMT1 is not required for self-renewal of pancreatic progenitor cells per se but is critical for progenitor cell survival during differentiation. Furthermore, p53 haploinsufficiency prevented apoptosis and rescued the survival of Dnmt1-null progenitor cells; this suggests that p53 is a primary target of DNMT1 action in pancreatic progenitor cells. Recently, the role of Dnmt1 in epidermal progenitor cells suggested that Dnmt1 was required to retain proliferative stamina and suppress differentiation (Sen et al. 2010). Deletion of Dnmt1 in epidermal progenitors resulted in precocious differentiation and eventual tissue loss. Although the phenotype in the pancreatic deletions of Dnmt1 also was the eventual loss of tissue, no evidence of premature differentiation or defects in proliferation were observed. The mechanism by which Dnmt1 maintains progenitor populations may vary in different tissue depending on the cellular context. This suggests that at a molecular level, DNMT1 may associate with different effectors to carry out repressive functions depending on the cellular context.
We attribute the inability of the DNMT1PC pancreatic progenitor population to undergo differentiation to the accumulation of p53. Part of p53 accumulation may be due to the derepression of the p53 locus when DNMT1 is not present on the promoter. It is to be noted that there is no CpG methylation around the DNMT1-CTCF-binding area (Su et al. 2009; our unpublished results), suggesting that DNMT1 is working as part of a repressive complex independent of its DNA methylation activity. Methylation-independent functions of DNMT1 have been shown to repress target genes as part of complexes with E2F1 and histone deacetylases (Valdez et al. 2011; Clements et al. 2012). The inability of DNMT1PC cells to differentiate because of derepression of p53 is similar to the mdm2−/− mouse phenotype (Jones et al. 1995; Montes de Oca Luna et al. 1995). While mdm2−/− are embryonic-lethal at E6.5, mdm2−/−p53−/− compound mutants are normal and viable. Analysis of compound mdm2−/−p53515c-515c, which express a mutated p53 that can mediate growth arrest but not apoptosis, revealed hematapoetic and lymphoid deficiencies because of progenitor cell cycle and differentiation defects (Liu et al. 2007). Thus, it is possible that repression of p53 serves to facilitate pancreatic progenitor cell differentiation during normal organogenesis and that the absence of Dnmt1 results in transcriptional derepression and subsequent accumulation of p53 that initiates cell cycle arrest and apoptosis during differentiation. The ability of DNMT1PCp53+/− pancreatic progenitor cells to persist through differentiation and form a pancreas at birth suggests that Dnmt1 is not required to maintain progenitor cell self-renewal or differentiation but is required to attenuate the role of p53 as a checkpoint during differentiation.
Materials and methods
Information on mouse strains is in the Supplemental Material. All animal experiments were performed in accordance with NIH policies on the use of laboratory animals and approved by the Animal Research Committee of the Office for the Protection of Research Subjects at University of California at Los Angeles. For immunohistochemical experiments, tissues were prepared, oriented, and stained, and images were captured as previously described (Georgia et al. 2006). Calculations of the proliferative index were done as previously described using at least three embryos from each genotype (Zhong et al. 2007). ChIP experiments with the E14.5 dorsal pancreas cells were carried out using the micro-ChIP protocol (Dahl and Collas 2008). Statistical significance was determined using Student's t test. Information on mouse strains, antibodies, and primers is in the Supplemental Material.
Acknowledgments
We thank Guoping Fan (University of California at Los Angeles) for providing the Dnmt1lox/lox mice and helpful comments. We are grateful to Lendy Le for technical assistance. S.G. is supported by an National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) K01 (DK088995). This work was supported by grants from NIDDK (DK080996 and DK068763), JDRF, and the Helmsley Trust to A.B. S.G. and M.K. performed the experiments. A.B. and S.G. conceived and planned the experiments and interpreted data. A.B. and S.G. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.207001.112.
References
- Anderson RM, Bosch JA, Goll MG, Hesselson D, Dong PDS, Shin D, Chi NC, Shin CH, Schlegel A, Halpern M, et al. 2009. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev Biol 334: 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan A, Itoh N, Kato S, Thiery JP, Czernichow P, Bellusci S, Scharfmann R 2001. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development 128: 5109–5117 [DOI] [PubMed] [Google Scholar]
- Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E 2007. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet 39: 391–396 [DOI] [PubMed] [Google Scholar]
- Clements EG, Mohammad HP, Leadem BR, Easwaran H, Cai Y, Van Neste L, Baylin SB 2012. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res 40: 4334–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Collas P 2008. A rapid micro chromatin immunoprecipitation assay (microChIP). Nat Protoc 3: 1032–1045 [DOI] [PubMed] [Google Scholar]
- Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, Biniszkiewicz D, Bates B, Lee PP, Kuhn R, et al. 2001. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci 21: 788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H, et al. 2005. DNA methylation controls the timing of astrogliogenesis through regulation of JAK–STAT signaling. Development 132: 3345–3356 [DOI] [PubMed] [Google Scholar]
- Georgia S, Soliz R, Li M, Zhang P, Bhushan A 2006. p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. Dev Biol 298: 22–31 [DOI] [PubMed] [Google Scholar]
- Gittes GK 2009. Developmental biology of the pancreas: A comprehensive review. Dev Biol 326: 4–35 [DOI] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA 2002. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129: 2447–2457 [DOI] [PubMed] [Google Scholar]
- Gu G, Brown JR, Melton DA 2003. Direct lineage tracing reveals the ontogeny of pancreatic cell fates during mouse embryogenesis. Mech Dev 120: 35–43 [DOI] [PubMed] [Google Scholar]
- Herrera PL 2000. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 127: 2317–2322 [DOI] [PubMed] [Google Scholar]
- Hutnick LK, Golshani P, Namihira M, Xue Z, Matynia A, Yang XW, Silva AJ, Schweizer FE, Fan G 2009. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum Mol Genet 18: 2875–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA 1994. Tumor spectrum analysis in p53-mutant mice. Curr Biol 4: 1–7 [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet 27: 31–39 [DOI] [PubMed] [Google Scholar]
- Jensen J 2004. Gene regulatory factors in pancreatic development. Dev Dyn 229: 176–200 [DOI] [PubMed] [Google Scholar]
- Jensen J, Heller RS, Funder-Nielsen T, Pedersen EE, Lindsell C, Weinmaster G, Madsen OD, Serup P 2000a. Independent development of pancreatic α- and β-cells from neurogenin3-expressing precursors: A role for the notch pathway in repression of premature differentiation. Diabetes 49: 163–176 [DOI] [PubMed] [Google Scholar]
- Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD 2000b. Control of endodermal endocrine development by Hes-1. Nat Genet 24: 36–44 [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378: 206–208 [DOI] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122: 3195–3205 [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Liu G, Terzian T, Xiong S, Van Pelt CS, Audiffred A, Box NF, Lozano G 2007. The p53–Mdm2 network in progenitor cell expansion during mouse postnatal development. J Pathol 213: 360–368 [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206 [DOI] [PubMed] [Google Scholar]
- Panning B, Jaenisch R 1996. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev 10: 1991–2002 [DOI] [PubMed] [Google Scholar]
- Reik, W 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Schwitzgebel VM, Scheel DW, Conners JR, Kalamaras J, Lee JE, Anderson DJ, Sussel L, Johnson JD, German MS 2000. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development 127: 3533–3542 [DOI] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA 2010. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463: 563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CH, Shann YJ, Hsu MT 2009. p53 chromatin epigenetic domain organization and p53 transcription. Mol Cell Biol 29: 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge JJ, Snow JW, Kim J, Orkin SH 2009. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 5: 442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschen S-I, Dhawan S, Gurlo T, Bhushan A 2009. Age-dependent decline in β-cell proliferation restricts the capacity of β-cell regeneration in mice. Diabetes 58: 1312–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez CD, Davis JN, Odeh HM, Layfield TL, Cousineau CS, Berton TR, Johnson DG, Wojno KJ, Day ML 2011. Repression of androgen receptor transcription through the E2F1/DNMT1 axis. PLoS ONE 6: e25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Georgia S, Tschen SI, Nakayama K, Bhushan A 2007. Essential role of Skp2-mediated p27 degradation in growth and adaptive expansion of pancreatic β cells. J Clin Invest 117: 2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]