Little is known about the function of nuclear Piwi proteins. Le Thomas et al. investigate the genomic targets of Piwi and find that Piwi is recruited to chromatin where it colocalizes with RNA polymerase II. Knockdown of Piwi in the germline increases expression of transposable elements targeted by piRNAs. Derepression of transposons with Piwi depletion correlated with increased occupancy of RNA polymerase II. These results show that piRNAs are essential guides for Piwi to recognize targets for transcriptional repression. A related study by Rozhkov et al. appears in this issue.
Keywords: piRNA, Piwi, chromatin, RNA polymerase II, transcription, transposon
Abstract
In the metazoan germline, piwi proteins and associated piwi-interacting RNAs (piRNAs) provide a defense system against the expression of transposable elements. In the cytoplasm, piRNA sequences guide piwi complexes to destroy complementary transposon transcripts by endonucleolytic cleavage. However, some piwi family members are nuclear, raising the possibility of alternative pathways for piRNA-mediated regulation of gene expression. We found that Drosophila Piwi is recruited to chromatin, colocalizing with RNA polymerase II (Pol II) on polytene chromosomes. Knockdown of Piwi in the germline increases expression of transposable elements that are targeted by piRNAs, whereas protein-coding genes remain largely unaffected. Derepression of transposons upon Piwi depletion correlates with increased occupancy of Pol II on their promoters. Expression of piRNAs that target a reporter construct results in a decrease in Pol II occupancy and an increase in repressive H3K9me3 marks and heterochromatin protein 1 (HP1) on the reporter locus. Our results indicate that Piwi identifies targets complementary to the associated piRNA and induces transcriptional repression by establishing a repressive chromatin state when correct targets are found.
Diverse small RNA pathways function in all kingdoms of life, from bacteria to higher eukaryotes. In eukaryotes, several classes of small RNA associate with members of the Argonaute protein family, forming effector complexes in which the RNA provides target recognition by sequence complementarity, and the Argonaute provides the repressive function. Argonaute–small RNA complexes have been shown to regulate gene expression both transcriptionally and post-transcriptionally. Post-transcriptional repression involves cleavage of target RNA through either the endonucleolytic activity of Argonautes or sequestering targets into cytoplasmic ribonucleoprotein (RNP) granules (Hutvagner and Simard 2008).
The mechanism of transcriptional repression by small RNAs has been extensively studied in fission yeast and plants. Several studies showed that Argonaute–small RNA complexes induce transcriptional repression by tethering chromatin modifiers to target loci. In fission yeast, the effector complex containing the Argonaute and the bound siRNA associates with the histone H3 Lys 9 (H3K9) methyltransferase Clr4 to install repressive H3K9-dimethyl marks at target sites (Nakayama et al. 2001; Maison and Almouzni 2004; Sugiyama et al. 2005; Grewal and Jia 2007). Methylation of histone H3K9 leads to recruitment of the heterochromatin protein 1 (HP1) homolog Swi6, enhancing silencing and further promoting interaction with the Argonaute complex. The initial association of Ago with chromatin, however, requires active transcription (Ameyar-Zazoua et al. 2012; Keller et al. 2012). Plants also use siRNAs to establish repressive chromatin at repetitive regions. Contrary to yeast, heterochromatin in plants is marked by DNA methylation, although repression also depends on histone methylation by a Clr4 homolog (Soppe et al. 2002; Onodera et al. 2005). Although siRNA-mediated gene silencing is predominant on repetitive sequences, it is not limited to these sites. Constitutive expression of dsRNA mapping to promoter regions results in production of corresponding siRNAs, de novo DNA methylation, and gene silencing (Mette et al. 2000; Matzke et al. 2004).
In metazoans, small RNA pathways are predominantly associated with post-transcriptional silencing. One class of small RNA, microRNA, regulates expression of a large fraction of protein-coding genes (Friedman et al. 2009). In Drosophila, siRNAs silence expression of transposable elements (TEs) in somatic cells (Chung et al. 2008; Ghildiyal et al. 2008) and target viral genes upon infection (Galiana-Arnoux et al. 2006; Wang et al. 2006; Zambon et al. 2006). Another class of small RNAs, Piwi-interacting RNAs (piRNAs), associates with the Piwi clade of Argonautes and acts to repress mobile genetic elements in the germline of both Drosophila and mammals (Siomi et al. 2011). Analysis of piRNA sequences in Drosophila revealed a very diverse population of small RNAs that primarily maps to transposon sequences and is derived from a number of heterochromatic loci called piRNA clusters, which serve as master regulators of transposon repression (Brennecke et al. 2007). Additionally, a small fraction of piRNAs seems to be processed from the mRNA of several host protein-coding genes (Robine et al. 2009; Saito et al. 2009). The Drosophila genome encodes three piwi proteins: Piwi, Aubergine (AUB), and Argonaute3 (AGO3). In the cytoplasm, AUB and AGO3 work together to repress transposons through cleavage of transposon transcripts, which are recognized through sequence complementarity by the associated piRNAs (Vagin et al. 2006; Agger et al. 2007; Brennecke et al. 2007; Gunawardane et al. 2007).
In both Drosophila and mammals, one member of the Piwi clade proteins localizes to the nucleus. Analogously to small RNA pathways in plants, the mouse piRNA pathway is required for de novo DNA methylation and silencing of TEs (Carmell et al. 2007; Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008); however, the exact mechanism of this process is unknown. In Drosophila, DNA methylation is absent; however, several studies indicate that elimination of Piwi from the nucleus causes changes in histone marks on TEs (Klenov et al. 2011; Pöyhönen et al. 2012), yet a genome-wide analysis of Piwi's effect on chromatin marks and transcription is lacking.
Here we show that Piwi interacts with chromatin on polytene chromosomes in nurse cell nuclei. We found that Piwi exclusively represses loci that are targeted by piRNAs. We show that Piwi-mediated silencing occurs through repression of transcription and correlates with installment of repressive chromatin marks at targeted loci.
Results
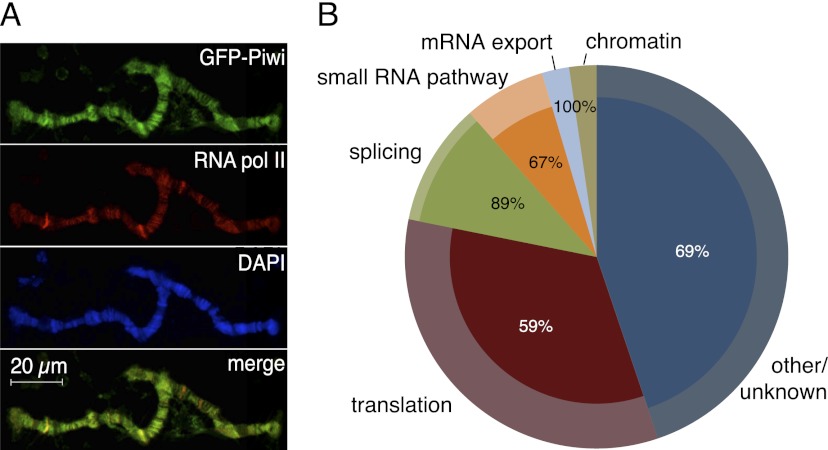
To analyze the role of Piwi in the nucleus, we generated transgenic flies expressing a GFP-tagged Piwi protein (GFP-Piwi) under the control of its native regulatory region. GFP-Piwi was expressed in the ovary and testis in a pattern indistinguishable from the localization of native Piwi and was able to rescue the piwi-null phenotype as indicated by ovarian morphology, fertility, transposon expression, and piRNA levels. GFP-Piwi was deposited into the mature egg and localized to the pole plasm; however, contrary to a previous observation (Brower-Toland et al. 2007), we did not detect Piwi expression outside of the ovary and testis in third instar larvae or adult flies. We also did not observe the association of Piwi with polytene chromosomes in salivary gland cells of third instar larvae. In both follicular and germline cells of the Drosophila ovary, GFP-Piwi localized exclusively in the nucleus, with slightly higher concentrations apparent in regions enriched for DAPI, indicating a possible interaction with chromatin. To gain further insight into Piwi localization in the nucleus, we took advantage of the fact that nurse cell chromosomes are polytenized and can be visualized on the otu mutant background (Mal'ceva et al. 1997). Analysis of polytene chromosomes from nurse cells demonstrated that GFP-Piwi associates with chromatin in a specific banding pattern. Interestingly, coimmunostaining showed that a GFP-Piwi signal on polytene chromosomes generally overlaps with the RNA polymerase II (Pol II) signal, which marks sites of active transcription (Fig. 1A).
Figure 1.
Piwi associates with chromatin and nuclear transcripts. (A) Polytene chromosomes from Drosophila nurse cells expressing GFP-Piwi on the otu[7]/otu[11] background. Piwi pattern on chromosomes correlates with Pol II staining. (B) Mass spectrometry analysis of Piwi interaction partners. Piwi complexes were precipitated in the presence and absence of RNase A. The outer circle represents classification of Piwi-associated proteins based on GO term analysis. The inner pies represent the fraction of each group whose association with Piwi depends on RNA (percentage indicated). Note that chromatin, splice, and mRNA export factors are virtually absent after RNase A treatment.
In order to identify factors that might be responsible for targeting Piwi to chromatin, we immunoprecipitated Piwi complexes from the Drosophila ovary and analyzed Piwi interaction partners by mass spectrometry. We purified Piwi complexes from ovaries of three different transgenic lines expressing GFP-Piwi, myc-Piwi, or Flag-Piwi using antibodies against each respective tag. As a control, we used flies expressing free GFP in the ovary. We identified >50 factors that showed significant enrichment in all three Piwi purifications but were absent in the control. We were unable to identify chromatin-associated factors that directly associate with Piwi but identified several RNA-binding proteins that associate with nascent transcripts, such as splicing (Rm62, Pep, Ref1, Yps, CG9684, CG31368, CG5728, and Mago) and nuclear export (Tho2 and Hpr1) factors (Fig. 1B). Upon RNase A treatment prior to immunoprecipitation, the presence of most of these RNA-binding proteins in purified Piwi complexes was eliminated.
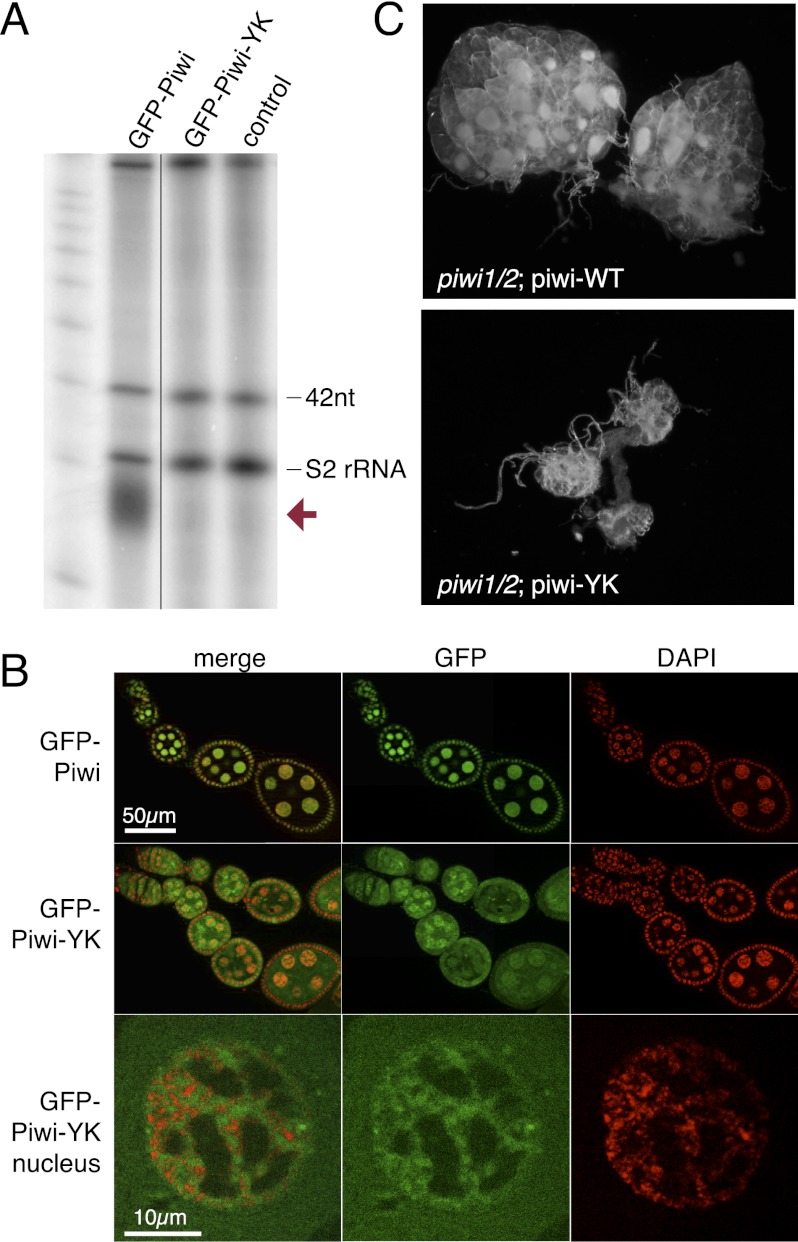
Piwi proteins are believed to find their targets through sequence complementarity of the associated piRNA. In fact, it has been proposed that lack of the associated piRNA leads to destabilization of piwi proteins and to Piwi's inability to localize to the nucleus (Saito et al. 2009; Haase et al. 2010; Olivieri et al. 2010; Handler et al. 2011; Ishizu et al. 2011). On the other hand, Piwi has been proposed to have functions that are independent of its role in transposon control by regulating stem cell niche development (Cox et al. 1998; Klenov et al. 2011). To address the role of piRNA in translocation of Piwi into the nucleus and its function, we generated transgenic flies expressing a point mutant Piwi—referenced as Piwi-YK—that is deficient in piRNA binding due to a substitution of two conserved amino acid residues (Y551L and K555E) in the 5′ phosphate-binding pocket (Kiriakidou et al. 2007; Djuranovic et al. 2010). The Piwi-YK mutant was expressed in Drosophila follicular and germ cells at levels similar to that of wild-type Piwi but was completely devoid of associated piRNA (Fig. 2A). In contrast to wild-type Piwi, Piwi-YK could be found in the cytoplasm, supporting the existence of a quality control mechanism that prevents entrance of unloaded Piwi into the nucleus (Ishizu et al. 2011). Nevertheless, a significant amount of piRNA-deficient Piwi localized to the nucleus (Fig. 2B). Similar to wild-type Piwi, Piwi-YK seemed to associate with chromatin, as indicated by its localization in DAPI-stained regions of the nuclei, and this is consistent with fluorescence loss in photobleaching (FLIP) experiments that demonstrated reduced nuclear mobility compared with free diffusion (Supplemental Fig. S1). Based on sterility and ovarian morphology, the piwi-YK transgene was unable to rescue the piwi-null phenotype despite its nuclear localization (Fig. 2C), indicating that while piRNA binding is not absolutely essential for stability and nuclear localization of Piwi, it is required for Piwi function.
Figure 2.
Piwi function, but not its nuclear localization, requires piRNA association. (A) The Piwi-YK mutant does not associate with piRNA. Immunoprecipitation of Piwi–piRNA complexes was performed with GFP antibody on ovaries from GFP-Piwi and GFP-Piwi-YK transgenic flies and a control strain. Small RNAs were isolated, 5′-labeled, and resolved on a denaturing gel. The same amount of 42-nucleotide RNA oligonucleotides was spiked into all samples prior to RNA isolation to control for loss of RNA during isolation and labeling. piRNAs (red arrow) are absent in the Piwi-YK complex. (B) GFP-Piwi-YK is present in the nuclei of nurse cells and colocalizes with chromatin (DAPI-stained areas). (C) The Piwi-YK mutant does not rescue the morphological changes caused by the piwi-null mutation. Dark-field images of ovaries where either the wild-type piwi or the piwi-YK transgene has been backcrossed onto the piwi-null background.
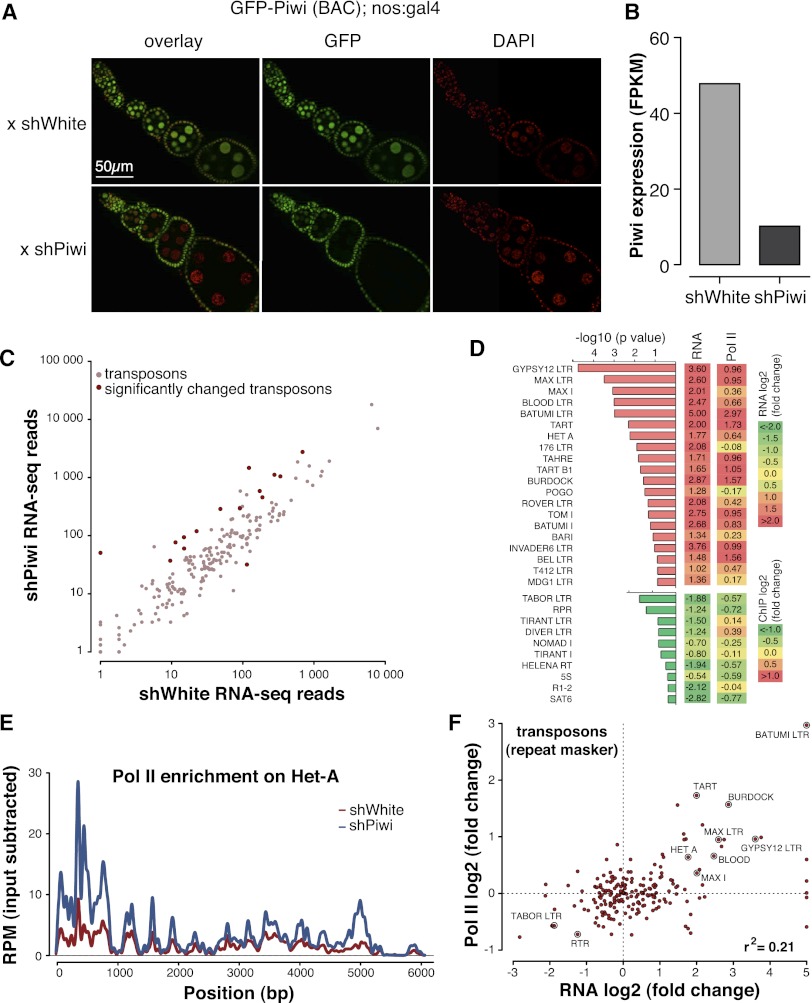
To directly test the function of Piwi in the nucleus, we analyzed the effect of Piwi deficiency on gene expression and chromatin state on a genome-wide scale. Piwi mutant females have atrophic ovaries caused by Piwi deficiency in somatic follicular cells (Lin and Spradling 1997; Cox et al. 1998), which precludes analysis of Piwi function in null mutants. Instead, we used RNAi knockdown to deplete Piwi in germ cells while leaving it functionally intact in somatic follicular cells. The Piwi knockdown flies did not exhibit gross morphological defects in the ovary; however, they showed drastic reduction in GFP-Piwi expression in germ cells and were sterile (Fig. 3A,B). To analyze the effect of Piwi deficiency on the steady-state transcriptome as well as the transcription machinery, we performed RNA sequencing (RNA-seq) and Pol II chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) experiments from Piwi knockdown and control flies.
Figure 3.
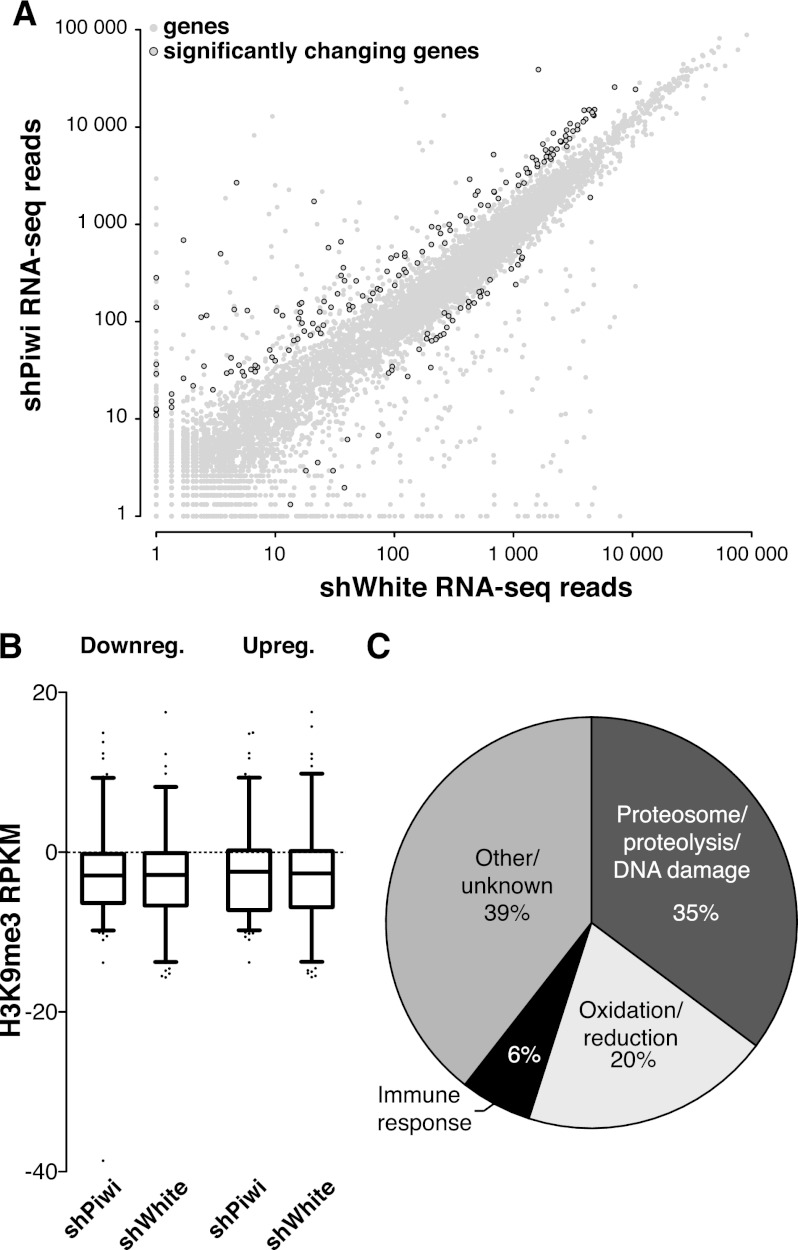
Piwi transcriptionally represses TEs. (A) Piwi knockdown is efficient and specific to ovarian germ cells as indicated by GFP-Piwi localization. GFP-Piwi; Nanos-Gal4-VP16 flies were crossed to control shRNA (shWhite) or shPiwi lines. Piwi is specifically depleted in germ cells and not in follicular cells, consistent with expression of the Nanos-Gal4-VP16 driver. (B) Piwi expression as measured by RNA-seq in the Piwi knockdown and control lines. Note that Piwi expression is unaffected in follicular cells, leading to relatively weak apparent knockdown in RNA-seq libraries from whole ovaries. (C) Effect of Piwi knockdown on the expression of TEs. Two biological replicate RNA-seq experiments were carried out, and differential expression was assessed using DESeq. Transposons that show significant change (P < 0.05) are indicated by dark-red circles. Out of 217 individual RepeatMasker-annotated TEs, 15 show a significant increase in expression upon Piwi knockdown. (D) The change in the levels of TE transcripts and Pol II occupancy on their promoters upon Piwi knockdown. Twenty up-regulated and 10 down-regulated transposons with the most significant changes in expression level are shown. Note the low statistical significance for down-regulated transposons. For a complete list of transposons, see Supplemental Figure S2. (E) Pol II signal over the Het-A retrotransposon in control flies (shWhite; red) and upon Piwi knockdown (shPiwi; blue). (F) Increased abundance of transposon transcripts upon Piwi depletion correlates with increased Pol II occupancy over their promoters (r2 = 0.21). Note that the majority of elements do not show significant change in either RNA abundance or Pol II occupancy.
In agreement with previous observations that implicated Piwi in transposon repression (Saito et al. 2006; Aravin et al. 2007; Brennecke et al. 2007), we found that steady-state transcript levels of several TEs were increased upon Piwi knockdown in germ cells (Fig. 3C,D; Supplemental Fig. S2). We found little to no change of RNA levels for transposons whose activity is restricted to follicular cells of the ovary, indicating that the observed changes are indeed due to loss of Piwi in the germline (Supplemental Fig. S2). The analysis of Pol II ChIP-seq showed that Pol II occupancy increased over promoters of multiple TEs (Fig. 3D–F; Supplemental Fig. S3). Indeed, the change in steady-state levels of transposon transcripts upon Piwi depletion correlated with changes of Pol II occupancy (Fig. 3F). This result demonstrates that Piwi ensures low levels of transposon transcripts through a repressive effect on the transcription machinery.
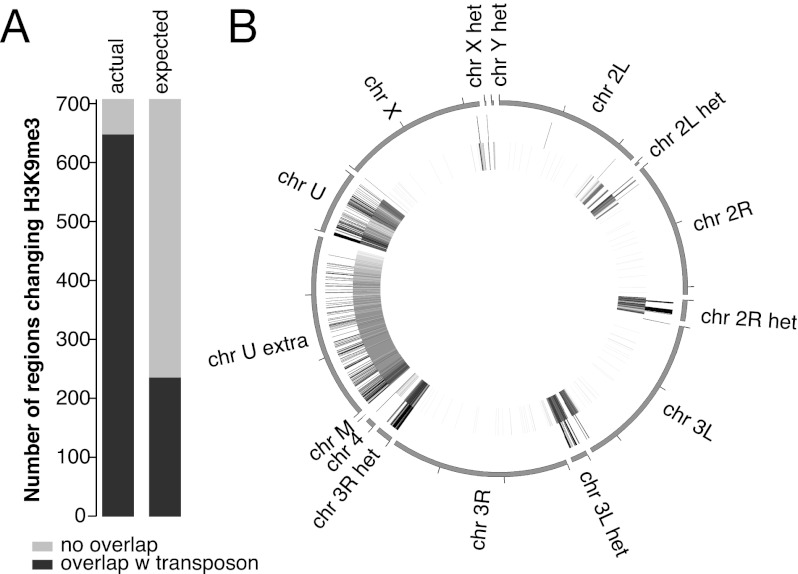
To test whether Piwi-mediated transcriptional repression is accompanied by a corresponding change in chromatin state, we used ChIP-seq to analyze the genome-wide distribution of the repressive H3K9me3 mark in the ovary upon Piwi knockdown. We identified 705 genomic loci at which the level of H3K9me3 significantly decreased. More than 90% of the regions that show a decrease in the H3K9me3 mark upon Piwi depletion overlapped TE sequences, compared with the 33% that is expected from random genome sampling (Fig. 4A). Furthermore, these regions tend to be located in the heterochromatic portions of the genome that are not assembled on the main chromosomes (Fig. 4B). Only 20 of the identified regions localized to the euchromatic parts of the genome. Of these, 15 (75%) contained potentially active annotated copies of transposons. Taken together, our results indicate that Piwi is required for installment of repressive H3K9me3 chromatin marks on TE sequences of the genome.
Figure 4.
Piwi-induced transcriptional repression correlates with establishment of a repressive chromatin state. (A) Overlap between genomic regions of H3K9me3 depletion upon Piwi knockdown and TEs. Two replicates of H3K9me3 ChIP-seq experiments were carried out on control and Piwi-depleted ovaries, and enriched regions were identified using DESeq (see the Materials and Methods for details). A total of 705 regions show significant (P < 0.05) decrease in H3K9me3 occupancy upon Piwi knockdown, while only 30 regions showed a similarly significant increase. Out of the 705 regions that show a decrease in H3K9me3 marks upon Piwi knockdown, 91% (646) overlap with TE sequences compared with the 33% expected from random genome sampling. (B) Genomic positions of H3K9me3-depleted regions upon Piwi depletion (outer circle) and RepeatMasker-annotated transposons (inner circle). Note that almost all regions are localized in heterochromatic and repeat-rich portions of the genome (Het, chrU, and chrUExtra chromosomes).
While the vast majority of protein-coding host genes did not show significant changes in transcript level or Pol II occupancy upon Piwi knockdown, the expression of a small set of protein-coding genes (150 genes with a P-value <0.05) was significantly increased (Fig. 5A; Supplemental Table 1). There are several possible explanations for Piwi's effect on host gene expression. First, failure in the piRNA pathway might cause up-regulation of several genes that generate piRNAs in wild-type ovaries (Robine et al. 2009; Saito et al. 2009). However, the genes up-regulated in Piwi-deficient ovaries were not enriched in piRNAs compared with other genes. Second, H3K9me3 marks installed on TE sequences in a Piwi-dependent manner might spread into neighboring host genes and repress their transcription, as was recently demonstrated in a follicular cell culture model (Sienski et al. 2012). To address this possibility, we analyzed genomic positions of the genes whose expression was increased upon Piwi knockdown relative to genomic regions that showed a decrease in H3K9me3 marks. We found that up-regulated genes did not show a significant change in the H3K9me3 mark (Fig. 5B; Supplemental Fig. S4). Furthermore, the few genes located close to the regions that show a decrease in H3K9me3 signal had unaltered expression levels upon Piwi knockdown. Next, we analyzed the functions of up-regulated genes using gene ontology (GO) term classifications and found significant enrichment for proteins involved in protein turnover and stress and DNA damage response pathways (Fig. 5C). Particularly, we found that 31 subunits of the proteasome complex were overexpressed. Therefore, our analysis indicates that up-regulation of specific host genes is likely a secondary response to elevated transposon levels and genomic damage.
Figure 5.
Piwi does not directly repress protein-coding genes. (A) Effect of Piwi knockdown on the expression of genes. Two replicate RNA-seq experiments were carried out, and differential expression was assessed using DESeq. Genes that show significant change (P < 0.05) are indicated by black circles. The vast majority of genes does not change significantly upon germline Piwi knockdown (shPiwi) compared with control (shWhite). (B) H3K9me3 mark density does not change over genes that show a significant change in expression upon Piwi knockdown (see Fig. 3C). Up-regulated and down-regulated genes are plotted separately. Signal indicated is after background subtraction. (C) Functional analysis of up-regulated genes by the Database for Annotation, Visualization, and Integrated Discovery (DAVID) reveals activation of the protein degradation and DNA damage response pathways. Percentages of all up-regulated genes are indicated.
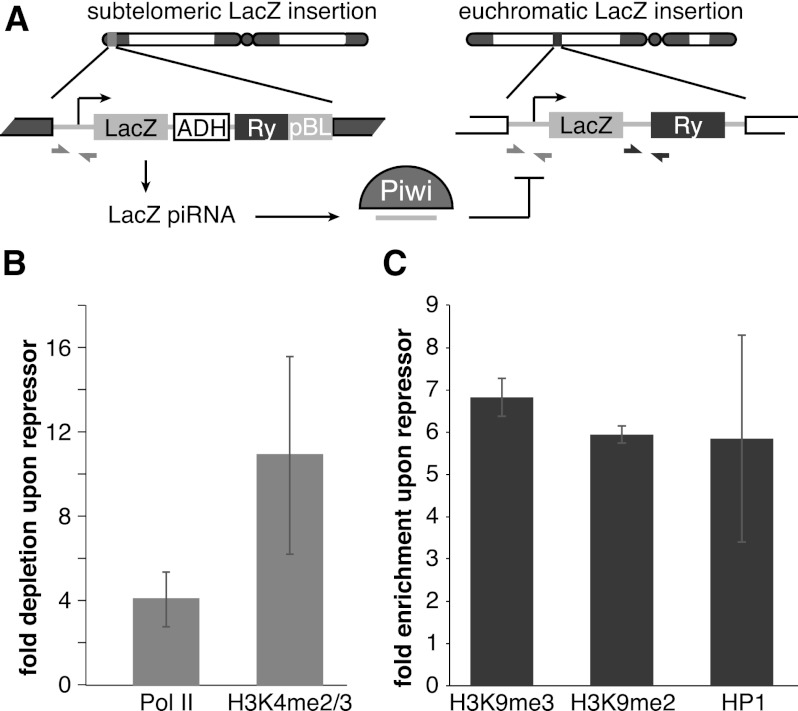
In contrast to host genes, transcripts of TEs are targeted by piRNA. To directly address the role of piRNA in Piwi-mediated transcriptional silencing, we took advantage of a fly strain that expresses artificial piRNAs against the lacZ gene, which are loaded into Piwi complexes and are able to repress lacZ reporter expression in germ cells (Fig. 6A; Josse et al. 2007; Muerdter et al. 2012). Expression of piRNAs that are antisense to the reporter gene caused transcriptional silencing of the lacZ gene as measured by Pol II occupancy (Fig. 6B). Furthermore, we found that piRNA-induced silencing of the reporter gene was associated with an increase in the repressive H3K9me3 mark and HP1 occupancy and a decrease in the abundance of the active H3K4me2/3 marks at the reporter locus (Fig. 6C). This result is in good agreement with the genome-wide effect of Piwi depletion on distribution of the H3K9me3 mark and suggests that transcriptional silencing correlates with the establishment of a repressive chromatin structure and is mediated by piRNAs that match the target locus.
Figure 6.
piRNA-dependent targeting of Piwi to a reporter locus leads to establishment of a repressive chromatin state and transcriptional silencing. (A) The mechanism of trans-silencing mediated by artificial piRNA and a schematic representation of the repressor and reporter lacZ constructs. The repressor construct is inserted in a subtelomeric piRNA cluster, leading to generation of piRNA from its sequence. Primers mapping to both constructs used for the Pol II and H3K4me2/3 ChIP-quantitative PCR (qPCR) are shown by light-gray arrows; primers specific to the reporter locus used for the H3K9me3, H3K9me2, and HP1 ChIP-qPCR are indicated by dark-gray arrows. (B) piRNAs induce transcriptional repression of the lacZ reporter. Pol II and H3K4me2/3 signals decreased on the lacZ promoter in the presence of artificial piRNAs as measured by ChIP-qPCR. Shown is the fold depletion of signal in flies that carry both repressor and reporter constructs compared with control flies that have only the reporter construct. The signal was normalized to RP49. (C) piRNAs induce an increase in H3K9me3 and H3K9me2 marks and HP1 binding as measured by ChIP-qPCR. Shown is the fold increase of corresponding ChIP signals downstream from the lacZ reporter in flies that carry both repressor and reporter constructs compared with control flies that have only reporter construct. The signal was normalized to RP49.
Discussion
Little is known about the function of nuclear piwi proteins. The nuclear piwi in mice (Miwi2) affects DNA methylation of TEs (Carmell et al. 2007; Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). Several recent reports implicate Drosophila Piwi in regulation of chromatin marks on transposon sequences (Lin and Yin 2008; Klenov et al. 2011; Wang and Elgin 2011; Sienski et al. 2012). The mechanism of these processes is unknown in both organisms. Previously, Piwi was shown to associate with polytene chromosomes in salivary gland cells and colocalize with HP1, a chromodomain protein that binds to heterochromatin and a few loci in euchromatin, suggesting that HP1 mediates Piwi's interaction with chromatin (Brower-Toland et al. 2007). However, recent results showed that the putative HP1-binding site on Piwi is dispensable for Piwi-mediated transposon silencing (Wang and Elgin 2011).
We did not detect Piwi expression outside of the ovary and testis, including in salivary gland cells, using a GFP-Piwi transgene expressed under native regulatory elements. We detected GFP-Piwi on polytene chromosomes in ovarian nurse cells that have a germline origin; however, it localizes in a pattern that largely does not overlap with HP1. FLIP experiments with GFP-Piwi indicated a relatively fast rate of fluorescence redistribution as compared with histone H2A (Supplemental Fig. S1), implying a transient interaction of Piwi with chromatin. Our proteomic analysis of Piwi complexes isolated from Drosophila ovaries did not identify chromatin-associated factors but revealed several RNA-binding proteins, such as splicing and nuclear export factors that bind nascent RNA transcripts (Fig. 1B). Importantly, the interaction of most of these RNA-binding proteins with Piwi was dependent on RNA, indicating that Piwi associates with nascent transcripts. As Piwi itself lacks DNA- and RNA-binding domains (beyond the piRNA-binding domain), it is likely that the recruitment of Piwi to chromatin is through interactions with other RNA-binding proteins or sequence-specific interactions between Piwi-bound piRNA and nascent transcripts.
Using specific Piwi knockdown in germ cells of the Drosophila ovary, we analyzed the effect of Piwi depletion on gene expression, the transcription machinery, and H3K9me3 chromatin marks genome-wide. In agreement with previous results (Klenov et al. 2011), we found up-regulation of several TEs upon Piwi knockdown (Fig. 3C). The TEs that did not change their expression upon germline knockdown of Piwi might be expressed exclusively in somatic follicular cells of the ovary, such as the gypsy retrotransposon. Alternatively, some elements present in the genome might not have transcriptionally active copies, or the cytoplasmic AUB/AGO3 proteins may efficiently silence them at the post-transcriptional level.
The increase in steady-state levels of RNA upon Piwi depletion strongly correlates with an increase in Pol II occupancy on the promoters of transposons (Fig. 3D,F; Supplemental Fig S2). This result suggests that Piwi represses transposon expression at the transcriptional level, although we cannot completely exclude the possibility of an additional post-transcriptional effect. It was shown previously that depletion or mutation of Piwi leads to depletion of the repressive H3K9me3 mark and an increase in the active H3K4me2/3 marks on several transposon sequences (Klenov et al. 2011; Wang and Elgin 2011). Our ChIP-seq data extend these results to a genome-wide scale, proving that transposons are indeed the sole targets of Piwi, and demonstrate that changes in histone marks directly correlate with transcriptional repression.
Piwi depletion in the germline does not affect expression of the majority of host genes, although a small fraction of genes changes expression (Fig. 5A). One possible mechanism of the effect Piwi has on host genes is the spreading of repressive chromatin structure from transposon sequences to adjacent host genes. Indeed, such a spreading and the resulting repression of host gene transcription were observed in an ovarian somatic cell (OSC) culture model (Sienski et al. 2012). However, we did not find significant changes in the H3K9me3 mark for genes that are up-regulated upon germline depletion of Piwi, arguing against this mechanism playing a major role in host gene regulation. Instead, we found that the majority of host genes whose expression is increased as a result of Piwi depletion participate in protein turnover (e.g., proteasome subunits) and stress and DNA damage response pathways, indicating that they might be activated as a secondary response to cellular damage induced by transposon activation. The different effect of Piwi depletion on host gene expression in ovary and cultured cells might be explained by the fact that silencing of host genes due to transposon insertion would likely have a strong negative effect on the fitness of the organism but could be tolerated in cultured cells. Accordingly, new transposon insertions that cause repression of adjacent host genes should be eliminated from the fly population but can be detected in cultured cells. In agreement with this explanation, the majority of cases of repressive chromatin spreading in OSCs were observed for new transposon insertions that are absent in the sequenced Drosophila genome. Indeed, it was shown that the vast majority of new transposon insertions is present at a low frequency in the Drosophila population, likely due to strong negative selection (Petrov et al. 2003). Such selection was primarily attributed to the ability of TE sequences to cause recombination and genomic rearrangements. We propose that in addition to the effects on recombination, the selection against transposons can be driven by their negative impact on host gene expression in the germline linked to Piwi-mediated chromatin silencing.
How does Piwi discriminate its proper targets—transposons—from host genes? In the case of cytoplasmic Piwi proteins AUB and AGO3, recognition and post-transcriptional destruction of TE transcripts is guided by associated piRNAs. Our results indicate that piRNAs provide guidance for transcriptional silencing by the nuclear Piwi protein as well. First, in contrast to host genes that are not targeted by piRNAs, TE transcripts, which are regulated by Piwi, are recognized by antisense Piwi-bound piRNA (Brennecke et al. 2007). Second, a Piwi mutant that is unable to bind piRNA failed to rescue the piwi-null mutation despite its ability to enter the nucleus. Finally, expression of artificial piRNAs that target a reporter locus induced transcriptional silencing associated with an increase in repressive H3K9me3 and HP1 chromatin marks and a decrease in the active H3K4me2/3 marks (Fig. 6B,C). In contrast, the tethering of Piwi to chromatin in a piRNA-independent fashion by fusing Piwi with the lacI DNA-binding domain that recognizes lacO sequences inserted upstream of a reporter gene did not lead to silencing of the reporter (data not shown). Together, our results demonstrate that piRNAs are the essential guides of Piwi to recognize its targets for transcriptional repression.
It is tempting to propose that, similar to Argonautes in fission yeast, Drosophila Piwi directly recruits the enzymatic machinery that establishes the repressive H3K9me3 mark on its targets. Establishment of repressive marks can lead to stable chromatin-based transcriptional silencing that does not require further association of Piwi with target loci. This model explains why we found that Piwi is relatively mobile in the nucleus, indicative of only a transient interaction with chromatin. The Piwi-mediated transcriptional silencing has an interesting parallel in Caenorhabditis elegans, where the Piwi protein PRG-1 and associated 21U RNAs are able to induce stable transgenerational repression that correlates with formation of silencing chromatin marks on target loci. Interestingly, PRG-1 and 21U RNAs are necessary only for initial establishment of silencing, while continuing repression depends on siRNA and the WAGO group of Argonautes (Ashe et al. 2012; Bagijn et al. 2012; Buckley et al. 2012; Shirayama et al. 2012). Future studies should reveal the pathway that leads to transcriptional repression downstream from Piwi in Drosophila and the differences from and similarities to other species.
Materials and methods
Drosophila stocks
Nanos-Gal4-VP16 (BL4937), UASp-shWhite (BL33623), UASp-shPiwi (BL 33724), and Chr. I and II Balancer (BL7197) were purchased from the Bloomington Stock Center. GFP-Piwi-expressing flies (see below) were backcrossed onto the piwi1/piwi2 (available from Bloomington Stock Center) background or the otu7/otu11 (available from Bloomington Stock Center) background, respectively. LacZ reporter lines were a generous gift from S. Ronsseray.
Generation of transgenic fly lines
The GFP-Piwi, 3xFlag-HA-Piwi, and myc-Piwi constructs were generated using bacterial recombineering (Gene Bridges Counter Selection kit) to insert the respective tag after the start codon of the Piwi genomic region cloned in BAC clone BACN04M10. The KpnI–XbaI genomic fragment that contains the Piwi gene and flanking sequences was transferred to corresponding sites of the pCasper4 vector to create pCasper4/tagged Piwi.
The pCasper4/GFP-Piwi construct was used to generate pCasper4/GFP-Piwi-YK with two point mutations, Y551I and K555E. Mutations were introduced by PCR, amplifying products corresponding to a 3.1-kb upstream fragment and a 2.58-kb downstream fragment. The upstream fragment included a unique XbaI site at the 5′ end of the amplicon and overlapped 39 base pairs (bp) with the downstream fragment, which included a unique BamHI site at its 3′ end. The single XbaI–BamHI fragment was generated by overlap PCR with outside primers and cloned into corresponding sites of pCasper4/GFP-Piwi to replace the wild-type fragment. Transgenic flies were generated by P-element-mediated transformation (BestGene).
Immunoprecipitation of Piwi proteins and RNA gel of piRNA
Dissected ovaries were lysed in lysis buffer (20 mM HEPES at pH 7.0, 150 mM KCl, 2.5 mM MgCl, 0.5% Triton X-100, 0.5% Igepal, 100 U/mL RNasin [Promega], EDTA-free Complete Protease Inhibitor Cocktail [Roche]) and supernatant clarified by centrifugation. Supernatant was incubated with anti-eGFP polyclonal antibody (Covance) conjugated to Protein-G Dynabeads at 4°C. Beads were spiked with 5 pmol of synthesized 42-nucleotide RNA oligomer to assess purification efficiency, proteinase K-digested, and phenol-extracted. Isolated RNA was CIP-treated, radiolabeled using PNK and γ-P32-labeled ATP, and run on a 15% urea-PAGE gel. Western blots of ovary lysate and anti-eGFP immunoprecipitates were obtained from 8% SDS-PAGE gels and probed with polyclonal rabbit anti-eGFP antibody to confirm expression of the full-length transgene.
Mass spectrometric analysis of Piwi interaction partners
Lysis and clarification of ovary samples were performed as described above using lysis buffer with reduced detergent (0.1% Triton X-100, 0.1% Igepal). Piwi proteins with Flag, Myc, or GFP tag were purified from Drosophila ovaries using corresponding antibodies covalently coupled to M-270 epoxy Dynabeads (Invitrogen) (Cristea et al. 2005). Immunoprecipitation of free GFP from GFP-expressing ovaries was used as a negative control. Immunoprecipitations were performed in the presence or absence of RNase A (100 μg/mL; 30 min at 25C). Piwi and copurified interacting proteins were resolved on NuPAGE Novex 4%–12% Bis-Tris gels and stained with colloidal Coomassie blue. Gel fragments that contained protein bands were excised and in-gel-trypsinized, and the peptides were extracted following the standard protocol of the Proteome Exploration Laboratory at California Institute of Technology. Peptide analyses were performed on an LTQ-FT Ultra (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Thermo Fisher Scientific) connected to an EASY-nLC. Fractionation of peptides was performed on a 15-cm reversed-phase analytical column (75-μm internal diameter) in-house-packed with 3-μm C18 beads (ReproSil-Pur C18-AQ medium; Dr. Maisch GmbH). Acquired spectra were searched against the Drosophila melanogaster proteome using the search engine Mascot (Matrix Science, version 2.2.06), and protein inferences were performed using Scaffold (Proteome Software, version 3). For an Excel file of Piwi interaction partners, see the Supplemental Material.
ChIP, ChIP-seq, and RNA-seq
ChIP was carried out using standard protocols (Moshkovich and Lei 2010). ChIP-seq and RNA-seq library construction and sequencing were carried out using standard protocols following the general principles described by Johnson et al. (2007) and Mortazavi et al. (2008), respectively. Data analysis was carried out using a combination of publicly available software tools and custom-written python scripts. Additional details regarding high-throughput data analysis are described in the Supplemental Material. For quantitative PCR (qPCR) primers, see Supplemental Table 2. GO term analysis of genes up-regulated upon Piwi knockdown was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Huang et al. 2009a,b) and FlyBase for additional assignment of GO terms. Sequencing data is available through Gene Expression Omnibus (accession no. GSE43829).
Antibodies
eGFP antibody (rabbit polyclonal serum; Covance) was affinity-purified in our laboratory. Anti-myc (Millipore), anti-Flag (Sigma), Pol II (ab5408), and Pol II pSer5 (ab5131) are commercially available.
Imaging of ovaries
Ovaries were fixed in 4% PFA in PBS for 20 min, permeabilized in 1% Triton X-100 in PBS, DAPI-stained (Sigma-Aldrich), washed, and mounted in 50% glycerol/PBS. Images were captured using an AxioImager microscope; an Apotome structured illumination system was used for optical sections (Carl Zeiss).
FLIP
FLIP time series were captured on an LSM510 confocal microscope equipped with a 40×/0.9 NA Imm Corr multi-immersion objective. Ovaries were dissected into halocarbon 700 oil (Sigma) and mounted under a 0.17-mm coverslip (Carl Zeiss) immediately before imaging. Two initial baseline images were captured, followed by 80–100 iterations consisting of two bleach iterations at 100% laser power (488 nm or 543 nm for GFP- and RFP-tagged proteins, respectively), followed by two images with reduced illumination intensity. FLIP series were cropped and median-filtered with a 2-pixel radius to reduce noise using FIJI (Schindelin et al. 2012) and the “Rigid Body” function of the StackReg plugin (Thévenaz et al. 1998) to correct drift when needed. Using Matlab software (The Mathworks), images were background-subtracted and corrected for acquisition bleaching. A value representing the true loss of intensity relative to the initial prebleach images, where 0 indicates no change in intensity and 1 represents complete photobleaching, was calculated for each pixel and each bleach/capture cycle and plotted with a color lookup table and calibration bar. Scale bars and annotations were made in Inkscape (http://inkscape.org).
Preparation of polytene squashes for immunofluorescence
Flies carrying the GFP-Piwi BAC construct were backcrossed onto the otu[7] and otu[11] background. Progeny from the cross of the two lines were grown at 18°C. Stage 7–12 egg chambers were separated and transferred to a polylysine-coated microscopic slide into PBST. From here, the “smush” protocol was followed (Johansen et al. 2009), but PFA cross-linking was reduced to 10 min. Slides were imaged using an AxioImager microscope and a 63× oil immersion objective (Carl Zeiss).
Acknowledgments
We are grateful to Evelyn Stuwe from the Aravin laboratory for purifying the GFP antibody; I. Antoshechkin of the Millard and Muriel Jacobs Genetics and Genomics Laboratory for sequencing; D. Trout, H. Amrhein, and S. Upchurch for computational assistance; the Bloomington Stock Center for fly stocks; and S. Hess, B. Graham, and M Sweredoski from the Proteome Exploration Laboratory at the Beckmann Institute, California Institute of Technology, for assistance with the mass spectrometry experiments. We thank members of the Aravin laboratory for critical comments on the manuscript. We thank Barbara Wold and members of the Wold laboratory for helpful discussions on ChIP protocols and analysis. A.K.R. and E.M.P. are supported by the Institutional Training Grant NIH/NRSA 5T32 GM07616, and E.M.P. is additionally supported by the Gordon Ross Medical Foundation. G.K.M. is supported by The Beckman Foundation, the Donald Bren Endowment, and NIH grant U54 HG004576. This work was supported by grants from the National Institutes of Health (R01 GM097363, R00 HD057233, and DP2 OD007371A to A.A.A.), the Searle Scholar Award (to A.A.A.), and the Ellison Medical Foundation New Scholar in Aging Award (to K.F.T.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.209841.112.
References
- Agger K, Cloos P, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini A, Helin K 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau J-C et al. 2012. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 19: 998–1004 [DOI] [PubMed] [Google Scholar]
- Aravin A, Hannon G, Brennecke J 2007. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick E-M, Mitchell J, Bagijn M, Cording A, Doebley A-L, Goldstein L, Lehrbach N, Le Pen J et al. 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150: 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn M, Goldstein L, Sapetschnig A, Weick E-M, Bouasker S, Lehrbach N, Simard M, Miska E 2012. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337: 574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin A, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon G 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Findley S, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin S, Lin H 2007. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev 21: 2300–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley B, Burkhart K, Gu S, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S 2012. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489: 447–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M, Girard A, van de Kant H, Bourc’his D, Bestor T, de Rooij D, Hannon G 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Chung W-J, Okamura K, Martin R, Lai E 2008. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol 18: 795–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D, Chao A, Baker J, Chang L, Qiao D, Lin H 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea IM, Williams R, Chait BT, Rout MP 2005. Fluorescent proteins as proteomic probes. Mol Cell Proteomics 4: 1933–1941 [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Zinchenko M, Hur J, Nahvi A, Brunelle J, Rogers E, Green R 2010. Allosteric regulation of Argonaute proteins by miRNAs. Nat Struct Mol Biol 17: 144–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R, Farh K, Burge C, Bartel D 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann J, Imler J-L 2006. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat Immunol 7: 590–597 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich M, Li C, Du T, Lee S, Xu J, Kittler E, Zapp M, Weng Z et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S, Jia S 2007. Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Gunawardane L, Saito K, Nishida K, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi M 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Haase A, Fenoglio S, Muerdter F, Guzzardo P, Czech B, Pappin D, Chen C, Gordon A, Hannon G 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev 24: 2499–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber F, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J 2011. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J 30: 3977–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman B, Lempicki R 2009a. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman B, Lempicki R 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard M 2008. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Ishizu H, Nagao A, Siomi H 2011. Gatekeepers for Piwi–piRNA complexes to enter the nucleus. Curr Opin Genetic Dev 21: 484–490 [DOI] [PubMed] [Google Scholar]
- Johansen K, Cai W, Deng H, Bao X, Zhang W, Girton J, Johansen J 2009. Polytene chromosome squash methods for studying transcription and epigenetic chromatin modification in Drosophila using antibodies. Methods 48: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, Mortazavi A, Myers R, Wold B 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Josse T, Teysset L, Todeschini A-L, Sidor C, Anxolabéhère D, Ronsseray S 2007. Telomeric trans-silencing: An epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genetic 3: 1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Adaixo R, Stunnenberg R, Woolcock KJ, Hiller S, Buhler M 2012. HP1(Swi6) mediates the recognition and destruction of heterochromatic RNA transcripts. Mol Cell 47: 215–227 [DOI] [PubMed] [Google Scholar]
- Kiriakidou M, Tan G, Lamprinaki S, De Planell-Saguer M, Nelson P, Mourelatos Z 2007. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 129: 1141–1151 [DOI] [PubMed] [Google Scholar]
- Klenov M, Sokolova O, Yakushev E, Stolyarenko A, Mikhaleva E, Lavrov S, Gvozdev V 2011. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci 108: 18760–18765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri T et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling A 1997. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124: 2463–2476 [DOI] [PubMed] [Google Scholar]
- Lin H, Yin H 2008. A novel epigenetic mechanism in Drosophila somatic cells mediated by Piwi and piRNAs. Cold Spring Harb Symp Quant Biol 73: 273–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Almouzni G 2004. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5: 296–304 [DOI] [PubMed] [Google Scholar]
- Mal’ceva N, Belyaeva E, King R, Zhimulev I 1997. Nurse cell polytene chromosomes of Drosophila melanogaster otu mutants: Morphological changes accompanying interallelic complementation and position effect variegation. Dev Genetic 20: 163–174 [DOI] [PubMed] [Google Scholar]
- Matzke M, Aufsatz W, Kanno T, Daxinger L, Papp I, Mette M, Matzke A 2004. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim Biophys Acta 1677: 129–141 [DOI] [PubMed] [Google Scholar]
- Mette M, Aufsatz W, van der Winden J, Matzke M, Matzke A 2000. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J 19: 5194–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams B, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Moshkovich N, Lei E 2010. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genetics 6: e1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Olovnikov I, Molaro A, Rozhkov N, Czech B, Gordon A, Hannon G, Aravin A 2012. Production of artificial piRNAs in flies and mice. RNA 18: 42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice J, Strahl B, Allis C, Grewal S 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Olivieri D, Sykora M, Sachidanandam R, Mechtler K, Brennecke J 2010. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 29: 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag J, Ream T, Costa Nunes P, Pontes O, Pikaard C 2005. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Petrov DA, Aminetzach YT, Davis JC, Bensasson D, Hirsh AE 2003. Size matters: Non-LTR retrotransposable elements and ectopic recombination in Drosophila. Mol Biol Evol 20: 880–892 [DOI] [PubMed] [Google Scholar]
- Pöyhönen M, de Vanssay A, Delmarre V, Hermant C, Todeschini A, Teysset L, Ronsseray S 2012. Homology-dependent silencing by an exogenous sequence in the Drosophila germline. G3 (Bethesda) 2: 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N, Lau N, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower M, Lai E 2009. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol 19: 2066–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida K, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi M 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi M 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al. 2012. Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, Conte D, Mello C 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J 2012. Transcriptional silencing of transposons by piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M, Sato K, Pezic D, Aravin A 2011. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang M, Jacobsen S, Schubert I, Fransz P 2002. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J 21: 6549–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Verdel A, Moazed D, Grewal S 2005. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci 102: 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenaz P, Ruttimann U, Unser M 1998. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7: 27–41 [DOI] [PubMed] [Google Scholar]
- Vagin V, Sigova A, Li C, Seitz H, Gvozdev V, Zamore P 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Wang S, Elgin S 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci 108: 21164–21169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-H, Aliyari R, Li W-X, Li H-W, Kim K, Carthew R, Atkinson P, Ding S-W 2006. RNA interference directs innate immunity against viruses in adult Drosophila. Science 312: 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon RA, Vakharia VN, Wu LP 2006. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol 8: 880–889 [DOI] [PubMed] [Google Scholar]