Rozhkov et al. explore the mechanisms by which Piwi restricts transposon expression in two cell types: germ cells and somatic support cells. Using genome-wide analyses and cell type-specific knockdown approaches, they find that in somatic cells, Piwi silencing occurs by regulating the transcription of targeted elements. In germ cells, Piwi acts via transcriptional gene silencing and also affects piRNAs that occupy Aubergine and Ago3, thus affecting post-transcriptional gene silencing—revealing the complexity of Piwi interactions. A related paper by Le Thomas et al. appears in this issue.
Keywords: Drosophila, transposon, piRNA, steady state, nascent
Abstract
Silencing of transposons in the Drosophila ovary relies on three Piwi family proteins—Piwi, Aubergine (Aub), and Ago3—acting in concert with their small RNA guides, the Piwi-interacting RNAs (piRNAs). Aub and Ago3 are found in the germ cell cytoplasm, where they function in the ping-pong cycle to consume transposon mRNAs. The nuclear Piwi protein is required for transposon silencing in both germ and somatic follicle cells, yet the precise mechanisms by which Piwi acts remain largely unclear. We investigated the role of Piwi by combining cell type-specific knockdowns with measurements of steady-state transposon mRNA levels, nascent RNA synthesis, chromatin state, and small RNA abundance. In somatic cells, Piwi loss led to concerted effects on nascent transcripts and transposon mRNAs, indicating that Piwi acts through transcriptional gene silencing (TGS). In germ cells, Piwi loss showed disproportionate impacts on steady-state RNA levels, indicating that it also exerts an effect on post-transcriptional gene silencing (PTGS). Piwi knockdown affected levels of germ cell piRNAs presumably bound to Aub and Ago3, perhaps explaining its post-transcriptional impacts. Overall, our results indicate that Piwi plays multiple roles in the piRNA pathway, in part enforcing transposon repression through effects on local chromatin states and transcription but also participating in germ cell piRNA biogenesis.
Transposons represent one of the most fundamental embodiments of the host–parasite relationship (Slotkin and Martienssen 2007). Their imperative is to propagate by multiplying specifically in the genomes of host germ cells. In turn, hosts respond to restrict the genomic burden represented by transposons. Failure to control transposon activity can have catastrophic consequences. In addition to a long-term mutagenic burden and consequent loss of overall fitness, escape of even a single mobile element from its normal restrictions can cause sterility. Perhaps the most singular example of this phenomenon is hybrid dysgenesis in Drosophila, wherein the inability to silence, for example, P or I elements renders animals completely sterile (Kidwell et al. 1977; Bregliano et al. 1980).
Families of mobile elements are unrelated by primary sequence and use different strategies for their propagation, making it a formidable challenge to discriminate transposons from endogenous genes and selectively silence parasitic DNA (Levin and Moran 2011). Over the past several years, it has become apparent that a small RNA-based innate immune system, the Piwi-interacting RNA (piRNA) pathway, addresses the challenge of transposon recognition and suppression by exploiting the one property uniquely shared by all transposons: their mobility (Brennecke et al. 2007; Ghildiyal and Zamore 2009; Malone and Hannon 2009; Ishizu et al. 2012).
At the core of the piRNA pathway are small RNA generative loci, piRNA clusters, which comprise the molecular definition of what an organism recognizes as transposons (Aravin et al. 2007; Brennecke et al. 2007). In Drosophila, these clusters are mainly formed of transposon fragments, many of which are ancient and thus diverged from active consensus copies. According to current models, piRNA clusters also acquire new information about active transposon loads. As elements move throughout the genome, they ultimately land in piRNA clusters, consequently adding their sequences to small RNA repertoires. This supposition is supported by studies of P elements and Penelope, a Drosophila virilis transposon artificially introduced into Drosophila melanogaster. Acquisition of the ability to control these elements was associated with their integration into piRNA clusters (Ronsseray et al. 1998; Brennecke et al. 2008; Khurana et al. 2011; Rozhkov et al. 2013).
piRNA clusters give rise to long, single-stranded transcripts that are processed through at least two nucleolytic steps into primary piRNAs. Recent studies have implicated Zucchini, an HKD (Hys–Lys–Asp) family nuclease, in the production of the 5′ ends of primary piRNAs (Haase et al. 2010; Olivieri et al. 2010; Ipsaro et al. 2012; Nishimasu et al. 2012; Voigt et al. 2012). Biochemical studies in Bombyx cell extracts have provided support for a model in which the 3′ ends of piRNAs are formed by exonucleolytic resection of longer RNAs following their loading into Piwi family proteins, although the enzymes responsible for this reaction are unknown (Kawaoka et al. 2011).
In germ cells, primary piRNAs are loaded into Piwi and Aubergine (Brennecke et al. 2007). In Aubergine, primary piRNAs join maternally inherited Aub complexes in priming the ping-pong cycle, a loop of nucleolytic reactions, which simultaneously degrades transposon mRNAs and generates new, secondary piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007). The cycle initiates as a piRNA guides Aubergine to a target, which it cleaves using the nuclease motif present within its Piwi domain (Cox et al. 1998, 2000). This event creates the 5′ end of a new, secondary piRNA, derived directly from the transposon transcript, which becomes loaded into Ago3. The Ago3 complex cleaves its target, likely a piRNA cluster transcript, generating a new antisense piRNA resident in Aubergine. Thus, transposon mRNAs form an additional input to the adaptive loop, which drives piRNA populations toward those matching actively transcribed elements. The impacts of Ago3 mutations indicate that cleavage of transposon mRNAs within the ping-pong cycle is important to their silencing in germ cells by post-transcriptional gene silencing (PTGS) (Li et al. 2009).
In both germ and follicle cells, loading of primary piRNAs into Piwi promotes its translocation into the nucleus (Saito et al. 2009, 2010). Follicle cells rely exclusively on Piwi for silencing elements active in the soma, and germ cells require Piwi in addition to Aub and Ago3 for transposon repression (Li et al. 2009; Malone et al. 2009). However, the precise mechanisms through which Piwi acts are unknown. Although Piwi contains the catalytic triad necessary for Argonaute proteins to function as nucleases, several factors, including its nuclear localization, have pointed to the possibility that it might regulate targets by repressing their transcription.
Early studies of spindle-E mutants, which lose essentially all piRNAs, reported chromatin changes at several classes of repeats. These included loss of the repressive mark H3K9me3 and increases in a signature mark of active promoters, H3K4me2 (Klenov et al. 2007). A two-hybrid screen raised the possibility of an association between Piwi and HP1, an interaction that was supported by coimmunoprecipitation of these proteins from fly ovaries (Brower-Toland et al. 2007). Subsequent studies showed effects of HP1 dosage on the silencing of a lacZ transgene that also depended on elements of the piRNA pathway for its repression (Josse et al. 2007). Recent analysis of a germ cell-specific piwi knockdown continued to reinforce a connection between Piwi and HP1 (Wang and Elgin 2011). Patterns of HP1 association with the promoter regions of some transposons were altered in knockdown cells, with coincident changes in the presence of H3K9me3 marks. This joins another recent study implicating transcriptional gene silencing (TGS) in the regulation of telomeric transposons (Shpiz et al. 2011). spn-E mutation or germline-specific piwi knockdown resulted in transcription activation of HeT-A and TART. These also correlated with the presence of active chromatin marks.
Despite these indications, no genome-wide study had directly examined the mechanisms by which Piwi acts to restrict transposon expression in the two cell types in which it is essential for transposon silencing. We therefore probed the contributions of Piwi to transposon silencing in germ cells and in their somatic support cells using tissue-specific piwi knockdowns. Integrated analysis of steady-state RNA levels by RNA sequencing (RNA-seq), measurements of nascent RNA synthesis using global run-on sequencing (GRO-seq), chromatin marks by chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq), and small RNA levels indicated multiple roles for Piwi in the piRNA pathway. In the soma, Piwi enforces silencing by regulating the transcription of targeted elements, whereas in germ cells, our data revealed that Piwi both acts via TGS and exerts unexpected impacts on piRNAs that occupy Aubergine and Ago3, indirectly also reducing the capacity to silence transposons by PTGS.
Results
Cell type-specific knockdown of Piwi protein
Numerous studies have highlighted a role for Piwi in silencing transposons in Drosophila gonads (Kalmykova et al. 2005; Saito et al. 2006; Pelisson et al. 2007; Yin and Lin 2007; Malone et al. 2009; Gangaraju et al. 2011; Klenov et al. 2011; Sokolova et al. 2011). However, these have been based on genetic mutants, which lose Piwi function in both the germline and somatic compartments with consequent severe effects on ovary morphology. Only a few studies have addressed this problem with selective loss of Piwi in clonal cell populations within the ovary (Rangan et al. 2011) or using cell type-specific knockdown (Shpiz et al. 2011; Wang and Elgin 2011). We sought to use the latter approach as a basis for our studies of the mechanisms by which Piwi contributes to transposon silencing.
We obtained a line harboring a Gal4-responsive long inverted repeat derived from the piwi gene from the Vienna Drosophila RNAi Center (VDRC) and crossed this into two different backgrounds. In one, expression of the dsRNA is driven in somatic follicle cells by a Gal4 activator expressed from the traffic jam promoter (tj-Gal4). The second line harbors two transgenes. The first provides a germ cell-specific activator for the piwi dsRNA, specifically the Gal4 activator expressed from the nanos promoter (nos-Gal4). The second provides enforced expression of Dcr-2, which is essential for the ability of long dsRNA triggers to provoke an effective RNAi response in this cell type.
Because we were knocking down piwi in specific cell types within a complex tissue, it was impossible to quantify suppression using quantitative RT–PCR (qRT–PCR). We therefore took several approaches to evaluate the efficiency of our piwi knockdown. It is well established that the integrity of the piRNA pathway is essential for fertility and that mutations in piwi result in fully penetrant sterility. Similarly, piwi knockdown in either germ cells or somatic follicle cells led to complete sterility (data not shown). We also evaluated piwi knockdown and its tissue restriction by immunofluorescence. Staining with Piwi antisera revealed loss of detectable Piwi signal in the nuclei of follicle cells in animals where piwi dsRNA was driven by tj-Gal4 (Supplemental Fig. S1A). In animals where dsRNA expression was driven by nos-Gal4, nuclear Piwi signals were retained in follicle cells but were lost from the majority of germ cells (Supplemental Fig. S1B). We did see robust Piwi staining in germaria and early-stage egg chambers. This likely indicated that during germ cell development, accumulation of Piwi slightly preceded the mounting of an efficient RNAi response. Finally, we evaluated Piwi knockdown semiquantitatively by measuring levels of piRNAs derived from soma- and germ-specific piRNA clusters, flamenco and 42AB, respectively (Supplemental Fig. S1C).
Somatic cell piRNAs are provided exclusively by the flamenco cluster, whereas a variety of clusters are active in germ cells. Loss of piRNAs derived from germ cell or somatic loci in Piwi immunoprecipitates from tissues with cell type-specific piwi knockdown could provide an estimate of the degree of Piwi silencing. We calculated the relative ratio of somatic (flamenco-derived) to germline (42AB-derived) piRNAs in each sample. We saw a 75% reduction in somatic, Piwi-bound species in animals with tj-driven knockdown and a 70% reduction in germ cell, Piwi-bound species in animals with nos-driven knockdown (Supplemental Fig. S1C). Considered together, our data suggest that we achieved strong, cell type-specific depletion of Piwi.
Piwi enforces transposon silencing by transcriptional repression in the soma
To investigate the mechanisms by which Piwi contributes to transposon silencing in somatic, follicle cells, we generated three data sets from flies, which express piwi dsRNA under the control of the tj-Gal4 activator. We used GRO-seq to measure nascent RNA synthesis. We isolated nuclei from ovaries of piwi knockdown and control flies and allowed engaged RNA polymerase II to elongate in the presence of BrU. Following serial antibody purification of transcripts synthesized during the incubation, we measured nascent RNA levels by Illumina sequencing. For comparison, we also determined steady-state RNA levels by RNA-seq. In both cases, strand information was preserved. Finally, we generated small RNA libraries, which included piRNAs, microRNAs (miRNAs), and endo-siRNAs. Control libraries were prepared from a strain expressing a dsRNA targeting the white gene under the control of the tj-Gal4 activator.
For analysis, we mapped all reads from each library to consensus transposon sequences, allowing up to two mismatches. For GRO-seq and RNA-seq, each element was assigned an RPM (reads per million) value. For small RNA libraries, mapped reads were normalized among libraries using miRNAs as a reference. We then compared the behavior of transposons and small RNAs, targeting those elements in the piwi knockdown versus control animals.
Transposon families and individual elements differ in their activity and expression in different cell types. Our previous studies—integrating information on the content of small RNAs from tissue-specific clusters, responses of individual element transcripts to mutations that selectively affect the germ cell or somatic piRNA pathways, and the measurement of maternal deposition of piRNAs—permitted us to assign transposons to four classes (Malone et al. 2009). Transposons that are mainly active in somatic cells are enriched in gypsy family long terminal repeat (LTR) retrotransposons (Fig. 1, green). These are thought to propagate in germ cell genomes by being packaged into virus-like particles in follicle cells and infecting the underlying germline (Pelisson et al. 1994, 1997; Prud'homme et al. 1995; Desset et al. 2003; Mevel-Ninio et al. 2007). Elements that are active mainly in the germline (Fig. 1, red) are diverse, with a broad representation of LTR and non-LTR retroelements (Malone et al. 2009). Some elements are intermediate and appear to be active in both cell types (Fig. 1, yellow), and the remainder (Fig. 1, black) cannot yet be classified based on our existing data sets.
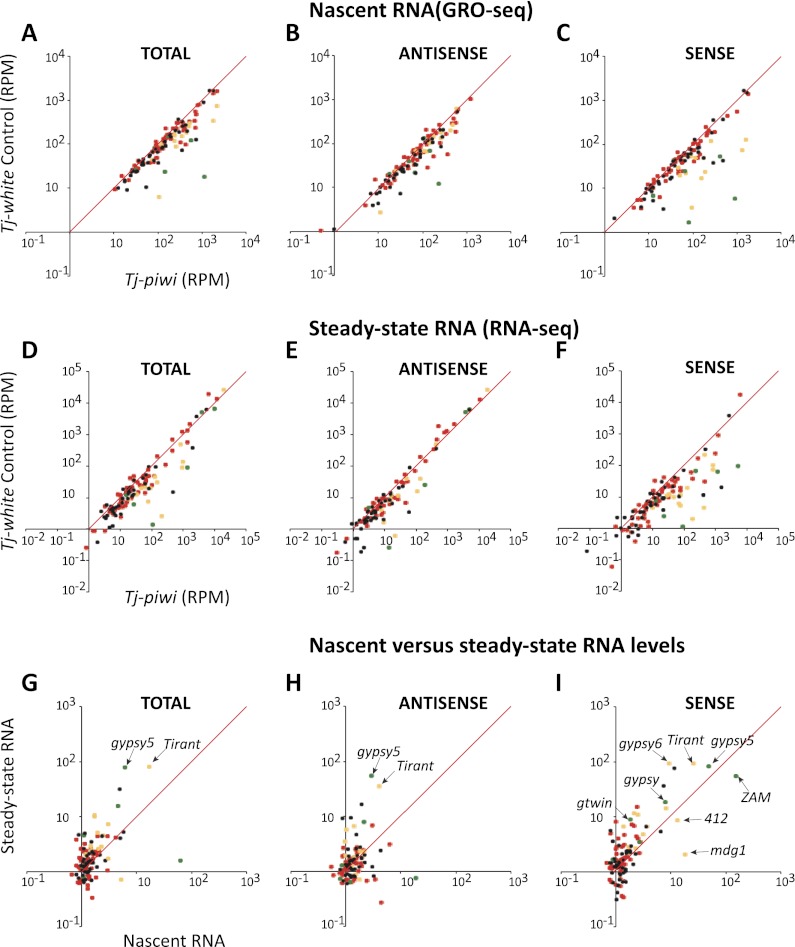
Figure 1.
Integrated analysis of nascent RNA synthesis and steady-state RNA levels upon piwi knockdown in the soma. (A–C) Comparisons are shown of RPM mapping to each transposon in the control knockdown (Y-axis) versus the Tj-driven piwi knockdown (X-axis). Red dots indicate germline-biased element, green dots indicate soma-biased elements, yellow dots indicate intermediate elements targeted in both compartments, and black dots indicate elements with “on” designation. Analysis of the total library is displayed in A, with specific analysis of reads mapping to sense and antisense strands shown in B and C, respectively. (D–F) Data are presented for steady-state RNA levels measured by RNA-seq and organized exactly as described for A–C. (G–I) Comparisons of the change in steady-state RNA levels (Y-axis) as compared with changes in nascent RNA levels (X-axis) for total reads (G), reads antisense to transposons (H), and reads sense to transposons (I). Elements falling along the diagonal show correlated changes; those left of the diagonal are dominated by effects on steady state, presumably representing changes in RNA stability, and those right of the diagonal are dominated by transcriptional effects. Selected transposons are indicated.
In the total GRO-seq and RNA-seq libraries, we saw a number of elements become derepressed upon Piwi loss. These were enriched for somatic and intermediate elements (Fig. 1, green and yellow), although a number of unclassified elements were also affected.
Transposon content in the genome represents both full-length, potentially active copies and fragmented and diverged transposons, some of which might be expressed as a result of integration near active promoters. To focus our attention on the silencing of potentially active elements, we decomposed our stranded libraries into reads that were sense or antisense with respect to each transposon (Fig. 1B,C,E,F). Considering only the sense-oriented reads magnified the effect seen in the total libraries. For the most part, the impacts of Piwi loss on antisense-oriented reads were minimal.
To investigate whether the impact of Piwi loss occurred mainly at the transcriptional or post-transcriptional level, we compared the relative change in each element in the GRO-seq and RNA-seq libraries (Fig. 1G–I). Considering antisense reads, we saw mainly low-level changes, with a few exceptions; notably, gypsy5, tirant, and springer (Fig. 1H). Effects were tipped to the left of the diagonal, indicating that any changes that do occur are mainly in steady-state levels. For sense-oriented reads that are more likely to represent transcription from intact transposons, a number of elements showed substantial derepression, and in almost every case, there was a rough correspondence between the fold changes seen in nascent and steady-state RNA levels (Fig. 1I). This strongly pointed to Piwi regulating these elements mainly at the transcriptional level. We did attempt to investigate changes in chromatin state upon somatic Piwi knockdown; however, the presence of abundant germ cell genomes in our mixed samples apparently masked our ability to detect statistically significant changes (Supplemental Fig. S2).
For a few elements, the effects on nascent transcripts were even greater than observed for steady-state RNAs. The most striking of these was ZAM, which showed a 55-fold increase in steady-state mRNA as compared with a 153-fold increase in nascent transcripts (Fig. 1I). At least one hypothesis to resolve this discordance came from analysis of small RNA levels.
siRNA responses to ZAM are enhanced in piwi knockdown
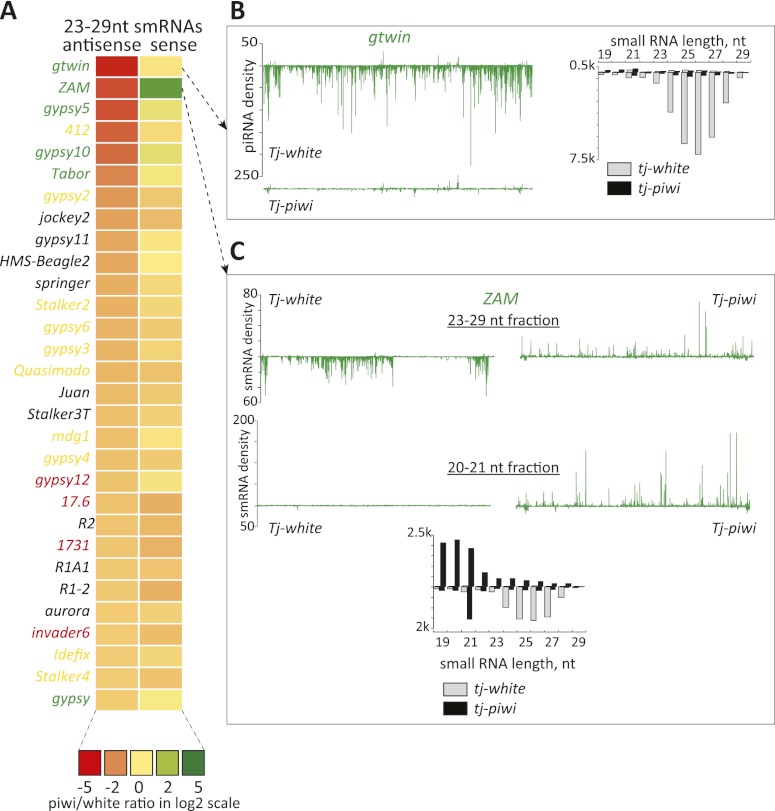
piRNAs produced in somatic cells are almost exclusively antisense and are stable only in the presence of their Piwi-binding partner. Thus, it was not surprising to note that antisense piRNAs were reduced in abundance for many of the elements that showed substantial changes in expression upon piwi knockdown (Fig. 2A). For example, gypsy twin (gtwin) showed twofold and ninefold changes in nascent and steady-state RNA levels, respectively (Fig. 1A), and piRNAs corresponding to this element were almost completely lost in the absence of Piwi (Fig. 2B).
Figure 2.
Effects of somatic piwi knockdown on piRNAs. (A) A heat map represents fold changes (key below) in presumed piRNAs (23–29 nt in length) that are sense or antisense to transposons. Elements are ranked according to the fold change (decrease) in antisense piRNAs. Color coding for names represents the cell type bias of the element, with the colors as defined in Figure 1. (B, top) As an example, a soma-biased transposon, gtwin, is shown. piRNA density in library from the control animals is displayed along a line representing the extent of the transposon consensus. Below is shown piRNA density observed in libraries from the knockdown animals. To the right, small RNAs are shown according to their size distributions, divided into sense (above the axis) and antisense (below the axis) species. Control and piwi knockdown libraries are as indicated. (C) An analysis of the ZAM transposon, similar to what is described in B, is shown. Here, both piRNA (23–29 nt, top) and siRNA (20–21 nt, bottom) fractions are plotted along the transposon consensus.
For ZAM, there was also a strong loss of antisense species in the piRNA size range. However, this was accompanied by a strong increase in sense-oriented small RNAs in knockdown animals (Fig. 2A,C). A closer inspection argued strongly against these being piRNAs. First, they were not enriched for U at position 1, as is characteristic of binding partners for Piwi. Second, their size distribution profile did not match Piwi-binding partners. Instead, we favor the hypothesis that these are degradation products that increase in their abundance because of the strong burst of sense ZAM expression that occurs in the absence of transcriptional repression by Piwi. A possible mechanism for the production of these RNA fragments was offered by the observation that there is a discrete peak of antisense small RNAs of 21 nucleotides (nt) that increases in abundance by approximately sixfold in piwi knockdown animals. These likely represented endogenous siRNAs that were formed from hybrids between sense and antisense ZAM transcripts and that may in turn target ZAM mRNAs for degradation by classical RNAi.
Piwi loss impacts both TGS and PTGS in the germline
Piwi is also required for transposon silencing in germ cells, wherein a wider variety of elements and element types are repressed by the piRNA pathway. To investigate the mechanisms of Piwi action in germ cells, we again compared piwi knockdown (driven by nos-Gal4) with control (white knockdown) animals.
Overall, nascent transcript levels increased modestly for a large number of elements, particularly if one focused analysis only on sense-oriented reads (Fig. 3A–C). Soma-specific elements remained largely unaffected (Fig. 3C, green dots), and there was an enrichment for increases in the transcription of elements strongly biased toward the germline (Fig. 3C, red dots). A number of intermediate and unclassified elements were also impacted.
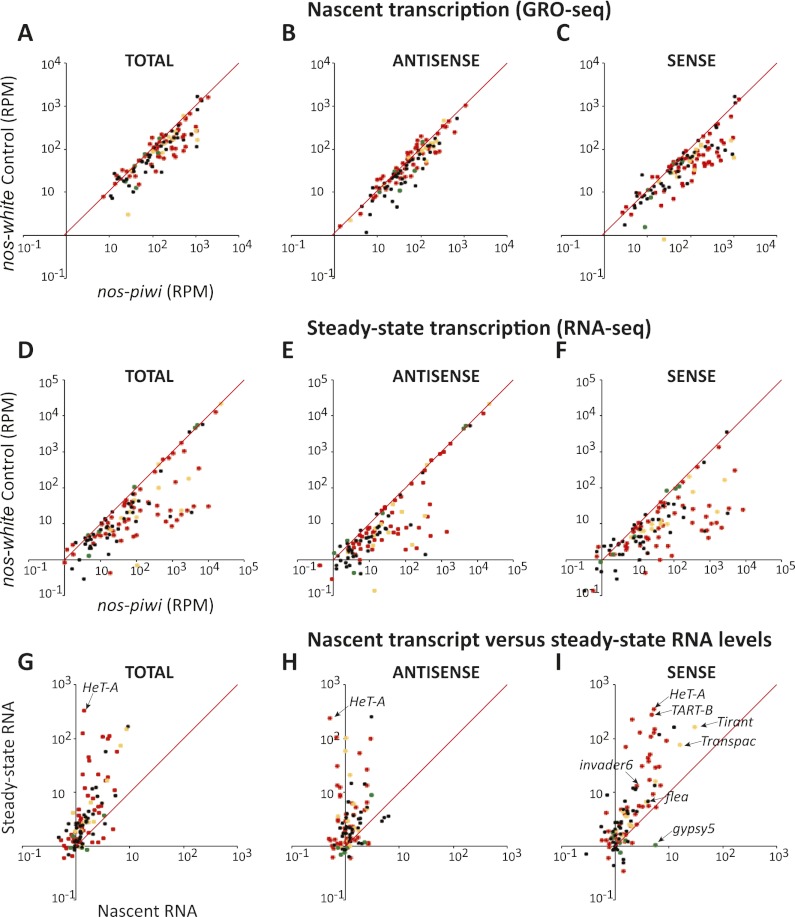
Figure 3.
Integrated analysis of nascent RNA synthesis and steady-state RNA levels upon piwi knockdown in the germline. (A–C) Comparisons are shown of RPM mapping to each transposon in the control knockdown (Y-axis) versus the nos-driven piwi knockdown (X-axis). Coloration of the dots and organization of the panels are as described in Figure 1. (D–F) Data are presented for steady-state RNA levels measured by RNA-seq and organized exactly as described for A–C. (G–I) Comparisons of the change in steady-state RNA levels (Y-axis) as compared with changes in nascent RNA levels (X-axis) as described in Figure 1. Selected transposons are indicated.
In general, much larger changes were observed in steady-state RNA levels (Fig. 3D–F) than in nascent transcripts upon germline piwi knockdown. Effects were apparent in analyses of total reads (cf. Fig. 3A,D), antisense reads (Fig. 3B,E), and sense reads that are expected to be enriched for transposon mRNAs (Fig. 3C,F). Comparing fold changes for individual elements in nascent versus steady-state RNA populations was consistent with Piwi loss exerting its impacts through effects on both transcriptional and post-transcriptional silencing mechanisms, as most elements appeared in the top left portion of the graphs in Figure 3, G–I. For a few elements, the discrepancies between increases in nascent RNA and levels of steady-state mRNA were >100-fold. For example, HeT-A nascent transcripts rose by fourfold, whereas sense RNAs rose in RNA-seq libraries by 350-fold (Fig. 3I). Considered together, these data indicate that Piwi functions differently in the germline and in the soma.
A role for Piwi in maintenance of piRNAs bound to Aub and Ago3
In germ cells, individual loss of any of the three Piwi proteins impairs transposon silencing and results in sterility. Primary piRNAs are derived from a variety of clusters and load both Piwi and Aubergine. Aub loaded in this manner combines with maternally deposited Aub complexes to initiate the ping-pong cycle by recognition of transposon mRNAs. Here, it acts in concert with Ago3 to repress transposons post-transcriptionally (Li et al. 2009). Given the strong post-transcriptional effects of Piwi loss on some elements, we investigated whether piwi knockdown might also impact Aub and Ago3 complexes.
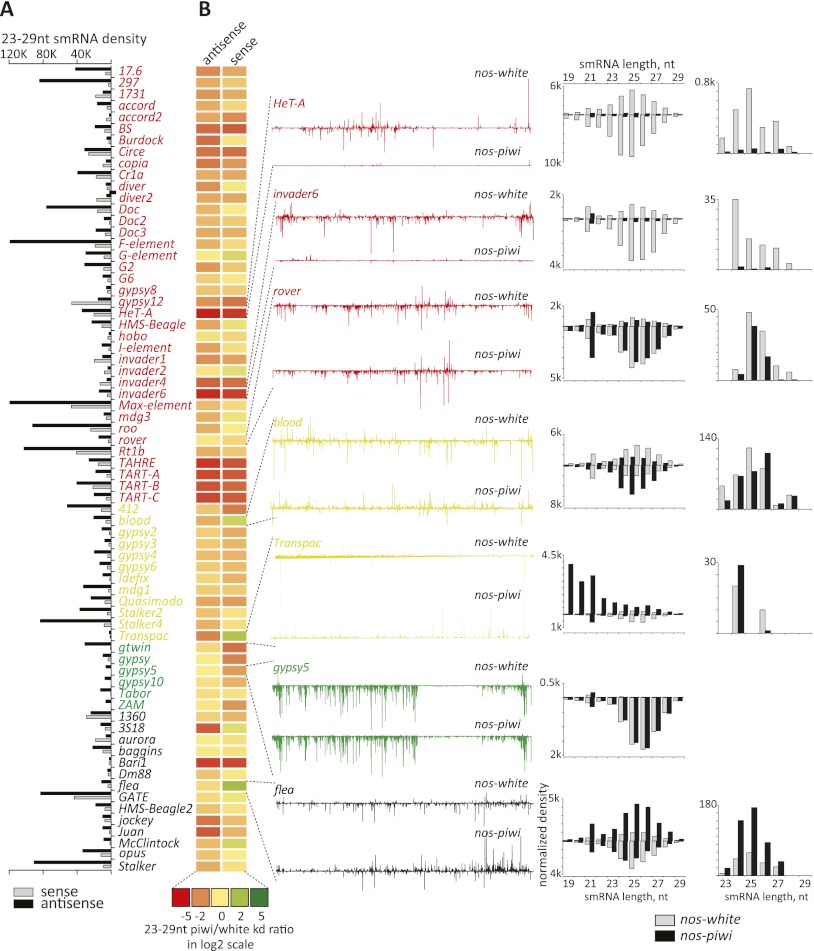
We first examined the abundance of presumed piRNAs, small RNAs of 23–29 nt, for the 70 most highly targeted elements (Fig. 4A,B). In the control animals, piRNA populations as a whole were generally enriched for antisense species, as previously reported (Fig. 4A). As expected, elements that are strongly soma-biased are largely unaffected by germline piwi knockdown (Fig. 4B, green with gypsy5 as an example). For elements that are strongly germ cell-biased (Fig. 4, red), there was a general trend toward an overall decrease in piRNAs. HeT-A was strongly affected, with an almost complete loss of small RNAs throughout the 23- to –29-nt size range. Other telomeric elements, including TART and TAHRE elements, also showed strong overall losses of both sense- and antisense-oriented piRNAs. These elements are unusual in that primary piRNAs appear to be derived from precisely the same loci that are targets for repression; specifically, the Drosophila telomeres. HeT-A, TART, and TAHRE also differ from others in that flies have domesticated these elements to provide a telomere maintenance strategy. Thus, at some point during development, limited transposition must be permitted in germ cells to protect chromosome ends. We therefore examined how other elements, which are more conventionally regulated by piRNA clusters, might behave.
Figure 4.
Impact of germline piwi knockdown on piRNAs mapping to transposons. (A) A bar graph represents the density of piRNA (RPM, strand as indicated) mapping to the 70 most highly targeted elements. These are sorted according to their cell type bias, with the coloration of the names as described in Figure 1. (Red) Germline-biased; (yellow) intermediate; (green) soma-biased; (black) undetermined. (B) A heat map represents changes in small RNAs (scale below) in comparisons of germline piwi knockdown versus control animals. To the left, densities of piRNAs along lines representing the extent of the transposon consensus are shown for the control (top) and piwi knockdown libraries (bottom) for selected elements (indicated). Adjacent to that are size distributions for small RNAs corresponding to those elements, with sense species displayed above the axis and antisense displayed below the axis (piwi knockdown and control as indicated). At the extreme right are similar plots of only that subset of piRNAs (23–29 nt) that is most probably bound to Ago3; namely; those sense to elements that lack a 5′ U and have an A at position 10.
For some elements, such as rover, we noted little change in piRNA populations upon germline piwi knockdown, although this element is not among the most highly targeted in control animals (Fig. 4A,B). For rover and many other elements, we do note an increase in endo-siRNA populations in piwi knockdown animals, which may indicate that, as for ZAM in the soma, increases in sense transcripts prime siRNA production, potentially leading to the generation of degradation products, which might confound our analysis of piRNA populations. This was particularly evident for Transpac, an element for which loss of piwi caused a burst of siRNA synthesis and the generation of what were apparently degradation products of the transposon mRNA (Fig. 4B).
For blood, which is an intermediate element, we saw modest reductions in antisense small RNAs and an increase in sense small RNAs, with a modest increase in siRNA production. In contrast, piRNA populations collapsed for invader6 nearly as completely as they did for HeT-A, again with an apparent increase in endo-siRNAs (Fig. 4B).
Small RNAs bound to each of the three Piwi proteins have a characteristic size distribution. If Piwi loss affected only those species that occupy that protein, we would expect the loss of the largest class of piRNAs, with a median size of 26 nt. However, for elements that show strong reductions in piRNAs, this occurs uniformly, affecting both sense and antisense species and piRNAs of sizes that predict their occupancy in Aub and Ago3 complexes. This suggested that Piwi loss does not simply impact Piwi-bound piRNAs but that at least for some elements, there are also secondary effects both on primary piRNAs loaded into Aub and, perhaps as a result, on the ping-pong cycle. To evaluate this possibility, we extracted those RNAs most likely to represent Ago3 partners. These were RNAs in the 23- to 29-nt size range that are sense to transposons, lack a U at position 1, and have an A at position 10. This class of piRNAs (Fig. 4B, right) was strongly reduced for elements that showed a substantial impact on small RNA levels in the face of Piwi loss.
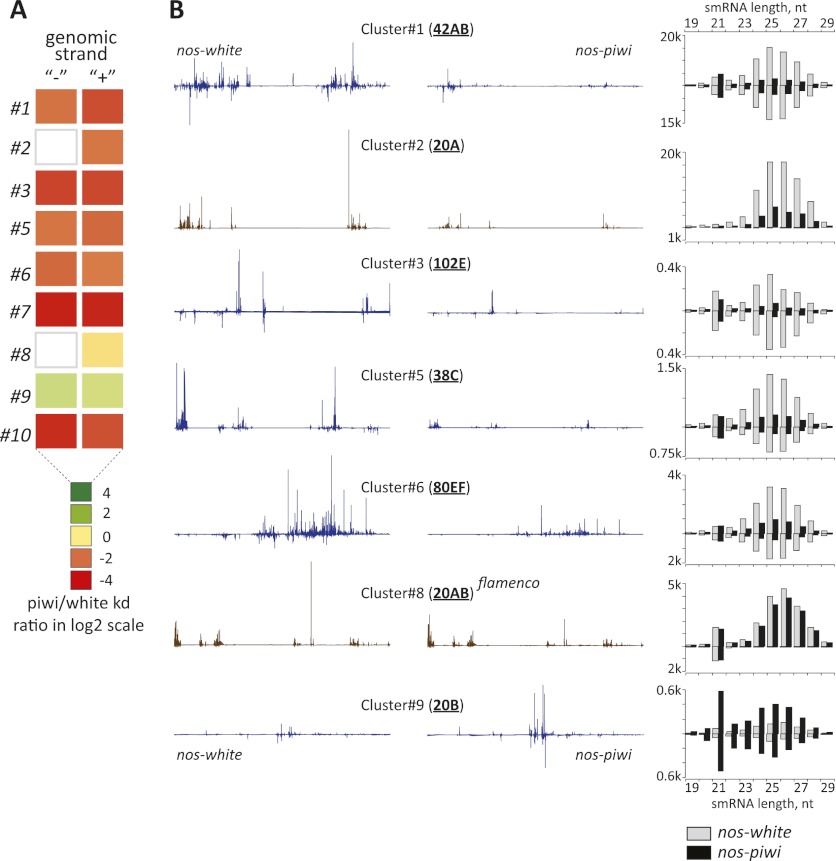
Support for a broad impact of Piwi loss on primary piRNAs came also from an analysis of RNAs that can be uniquely attributed to the most prominent piRNA generative loci. Among the 10 most active clusters, nine showed strong reductions in piRNA production across all size classes (Fig. 5A). The exception was, as expected, the flamenco cluster, which continued to load piRNAs into Piwi, whose expression persisted in somatic cells. Both dual-strand clusters, such as 42AB, and single-strand clusters, such as 20A, were affected.
Figure 5.
Effects of germline piwi knockdown on primary piRNA levels. (A) The relative change in piRNAs that can be uniquely mapped to any of the nine most productive piRNAs is shown, comparing levels in libraries derived from nos-driven piwi knockdown with the control animals (scale below). (B) For selected clusters, piRNA densities are displayed along a line representing the extent of the cluster for the control (left) or piwi knockdown (right) libraries. To the right are shown small RNA distributions corresponding to those clusters with species derived from the top strand shown above the axis and species from the bottom strand shown below the axis. Bars corresponding to control or piwi knockdown are as indicated.
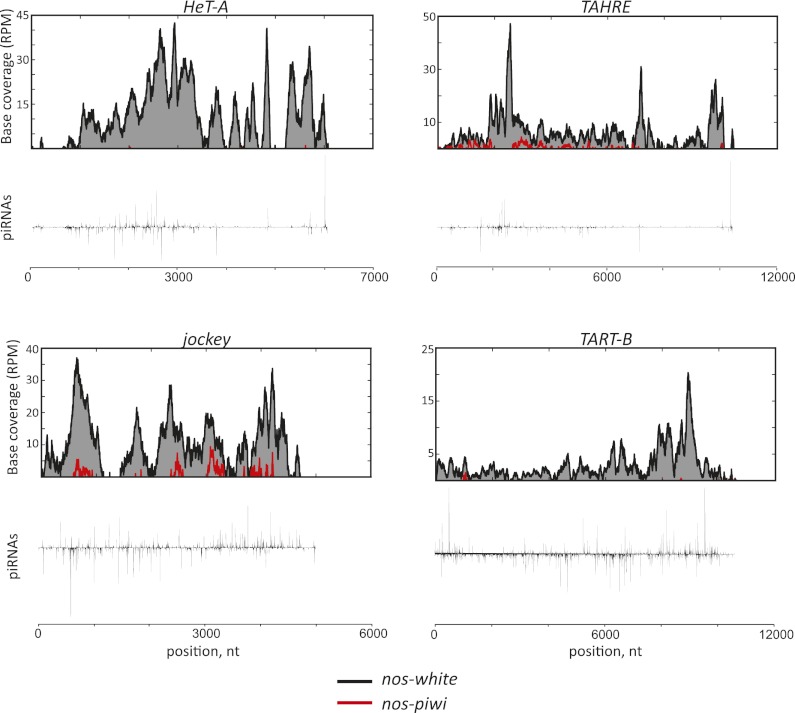
To investigate the mechanisms by which Piwi might contribute to transposon silencing, we used ChIP-seq to profile H3K9me3 marks in ovaries from animals with Piwi depleted from germ cells. Despite the presence of somatic nuclei, which still express Piwi, we were able to detect statistically significant changes on a number of elements. Among analyzed transposons, HeT-A, TAHRE, TART, and jockey showed the greatest changes in comparisons of piwi knockdown with controls (Fig. 6). While not much is known about jockey, the telomeric retrotransposons are among the best studied in the Drosophila genome due to their role in telomere elongation (Savitsky et al. 2006). The enrichment of repressive marks in control animals at these sites was anticipated, since the Drosophila telomeres have a distinctive chromatin structure and depend on HP1 loading for capping and elongation (Savitsky et al. 2002; Perrini et al. 2004; Frydrychova et al. 2008).
Figure 6.
Impact of piwi knockdown in the germline on H3K9me3 marks at transposons. Normalized densities of the repressive mark H3K9me3 (top plots) are compared with piRNA distributions (bottom plots) in control animals for HeT-A, TART, TAHRE, and jockey. Black and red are H3K9me3 enrichments over input in control (nos-white) and nos-piwi samples, respectively.
Not only are repressive H3K9me3 marks lost over these elements upon germline piwi knockdown, but the patterns along element consensus sequences of piRNAs and H3K9me3 ChIP signals also show remarkable similarity (Fig. 6; Supplemental Fig. S2). The latter is true even across somatic elements for which we were not able to detect H3K9me3 loss upon piwi knockdown, likely due to domination of our signals by germ cell genomes. Considered as a whole, these data implicate Piwi and its associated piRNAs in sequence-directed shaping of chromatin structure.
Discussion
Here, we used integrated, genome-wide profiles to examine the mechanisms by which Piwi impacts transposon silencing in the somatic and germ cell compartments within the Drosophila ovary. In the soma, a simplified version of the piRNA pathway primes Piwi alone with primary piRNAs derived from flamenco (Brennecke et al. 2007). Our data indicate that Piwi functions in this cell type by transcriptionally silencing its target elements. Prior studies that have indicated genetic and physical interactions between Piwi and HP1 provide a strong hint that perhaps TGS is mediated by the deposition of repressive histone modifications, particularly H3K9me3 (Brower-Toland et al. 2007). Given that we carried out our studies in vivo by knocking down piwi in a minority cell type in a mixed tissue, validation of this hypothesis by direct analysis of chromatin did not succeed. While this study was under review, Brennecke and colleagues (Sienski et al. 2012) provided strong support for a role of Piwi in directing the deposition of repressive H3K9me3 on somatic elements by analyses carried out in cultured ovarian somatic sheath cell lines. Additionally, they implicated Maelstrom, another well-established piRNA pathway component, in modulating the deposition of H3K9me3 marks in a Piwi-dependent fashion.
In germ cells, our data also indicate a role for Piwi in transcriptional silencing of transposons via directing deposition of H3K9me3 marks. However, we also observed a much broader impact, with effects on both post-transcriptional silencing mechanisms and populations of primary and secondary piRNAs that likely occupy Ago3 and Aub. This impact on piRNA biogenesis or stability is unexpected, particularly given a previous report that piwi knockdown in germ cells did not affect piRNA levels (Moshkovich and Lei 2010; Wang and Elgin 2011). In this case, analyses relied on Northern blotting for three individual piRNAs. Surprisingly, one of these was derived from HeT-A, which showed the strongest overall depletion of piRNAs upon germ cell piwi knockdown among all of the elements that we examined. piRNAs derived from roo are less sensitive to Piwi loss, perhaps explaining why a depletion was not observed for the roo piRNA that was analyzed.
How Piwi might impact piRNA populations generally is unclear. We observed an overall loss of primary piRNAs that can be uniquely mapped to clusters, perhaps implying an effect of Piwi on primary piRNA biogenesis. However, we did not observe an accumulation of long transcripts from clusters, presumed piRNA precursors, upon piwi knockdown as we did upon loss of other biogenesis factors, such as zucchini (data not shown).
Perhaps a clue can be drawn from a recent study in which maternally deposited piRNAs were able to convert a genomic locus that did not previously produce piRNAs into a piRNA cluster through paramutation (de Vanssay et al. 2012). Such observations strongly imply an interplay between piRNAs and cluster identity. This would represent a second important epigenetic role for maternally transmitted small RNAs in addition to their priming the ping-pong cycle. If Piwi is linked to the deposition of particular chromatin marks, these could provide part of the signal that marks a locus as a source of piRNAs. In this regard, an HP1 family protein, Rhino, is required for piRNA production from dual-strand germline clusters, at least in part by promoting cluster expression (Klattenhoff et al. 2009). Although one could speculate that Piwi-induced deposition of H3K9me3 marks might help to recruit Rhino, creating a loop that maintains cluster identity, the impacts of Piwi and Rhino loss are not identical. In particular, production of germ cell piRNAs from the single-strand cluster at 20A depends on Piwi but not on Rhino. Moreover, we did not observe differences in H3K9me3 densities on germline-active clusters upon piwi knockdown. It is also not yet clear why Piwi loss disproportionately affects some elements more than others. A particular example is found in those telomeric elements where piRNA clusters and their targets are essentially one and the same. However, other elements that show more conventional relationships to well-annotated piRNA clusters are also strongly impacted.
Considered together with prior studies, our data provide strong support for the notion that nuclear Piwi protein enforces transposon silencing through effects on transcription. This provides parallels to the effector mechanisms by which piRNAs silence transposons in mammals. In mice, a presumed Piwi ortholog, Miwi2, localizes to developing germ cell nuclei, where it is essential for the ultimate deposition of repressive DNA methylation marks on transposons (Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). Although flies lack DNA methylation (Schaefer and Lyko 2010; Zemach et al. 2010), it seems likely that the proximate effects of Piwi and Miwi2 on chromatin might be shared and deeply conserved among animals, although the ultimate output of their recruitment of shared repressive factors to their sites of action might differ.
Materials and methods
Fly husbandry
Fly stocks used in the study were as follows: tj-GAL4 (Drosophila Genetic Resource Center, stock no. 104055), nos-GAL4 (Bloomington Stock Collection, stock no. 25751), dsRNA-white (transformant ID 30033), and dsRNA-piwi (transformant ID 101658) (VDRC). Crosses were set up en masse on standard medium at 25°C. Ovaries were dissected from 3- to 5-d-old flies and used as described.
Immunohistochemistry
Ovaries were fixed in 4% PFA/PBS supplemented with 0.2% Triton X-100 (PBSTx) for 20 min at room temperature and washed three times (10 min each) with PBSTx. Fixed ovaries were blocked in PBSTx with 1% BSA for 1 h at room temperature and incubated with primary anti-Piwi (rabbit polyclonal) (Brennecke et al. 2007) at 1:1000 overnight at 4°C. The next day, ovaries were washed and incubated with secondary antibodies (Life Technologies, AlexaFluor-568) at 1:500 for ∼3–4 h at room temperature, washed again, and incubated in 1uM DAPI for 20 min. Images were acquired on a Perkin Elmer UltraVIEW spinning-disk confocal microscope.
Small RNA cloning and library normalization
Small RNA libraries used in the analysis were previously published (Preall et al. 2012). However, in the present study, a different normalization strategy was used. Here, small RNA reads were normalized such that each library had the equivalent of 1 million miRNA reads. Transposon-derived small RNAs were revealed by mapping to a set of canonical transposon sequences from D. melanogaster (version 9.4.1).
GRO-seq library preparation
The overall procedure was as described (Core et al. 2008) with modifications as described (Shpiz et al. 2011; Core et al. 2012). Briefly, ∼100 freshly dissected ovaries were homogenized on ice in 350 μL of HB-0.35 buffer (15 mM HEPES at pH 7.5, 10 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 0.05% NP40, 0.35 M sucrose). This and downstream steps were performed in a cold room on ice using ice-cold buffers supplemented with 1 mM DTT and protease inhibitors (Roche). Nuclei were purified by centrifugation (10,000g for 10 min) through a sucrose cushion containing two phases—the bottom was 800 μL of HB-0.8, and the top was 350 μL of HB-0.35—containing 0.8 M and 0.35 M sucrose, respectively. This step was done twice. The nuclear pellet was resuspended in 500 μL of GRO freezing buffer (50 mM Tris-HCl at pH 8.3, 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA), centrifuged at 10,000g for 3 min, resuspended again in 100 μL of the same buffer, and either kept at −80°C or immediately used for run-on reactions. The run-on reaction was done by adding 100 μL of freshly prepared reaction buffer (10 mM Tris-HCl at pH 8.0; 5 mM MgCl2; 300 mM KCl; 1% sarkosyl, 500 μM ATP, GTP, and Br-UTP; 2.3 μM CTP; 1 mM DTT; 20 U of SUPERase In) and was carried out for 5 min at 30°C. Base-hydrolyzed run-on transcripts were cloned using the TruSeq approach and were sequentially purified with anti-deoxy-BrU (Santa Cruz Biotechnology). SuperScript III (Life Technologies) was used for cDNA synthesis according to its protocol. PCR was performed using 10 μL of cDNA and Phusion high-fidelity PCR master mix (New England Biolabs) with 15 cycles of amplification. The amplified libraries were purified twice by resolution on 6% PAGE/TBE (∼150- to 350-base-pair [bp] region) and quantified on Bioanalyzer. The resulting libraries were sequenced on the Illumina platform with SE50 reads.
ChIP-seq library preparation
ChIP was done as described (Ram et al. 2011) with the following modifications. About 50 freshly dissected ovaries were cross-linked in 1.8% paraformaldehyde/PBS for 10 min at room temperature, quenched with 125 mM glycine for 5 min, and washed three times with ice-cold PBS (1 min each). Lysis was performed in 500 μL of ChIP lysis buffer (50 mM Tris-HCl at pH 8, 10 mM EDTA, 1% SDS) for 10 min on ice, sheared on a Bioruptor (Diagenode) for 14 min (two cycles at 7 min each) at “High” with 30-sec ON/OFF pulses, and cleared by centrifugation at 13,000g for 10 min. The resulting “chromatin extract” was diluted with ChIP dilution buffer (16.7 mM Tris-HCl at pH 8, 1.2 mM EDTA, 167 mM NaCl, 1.1% Triton X-100, 0.01% SDS) to bring the SDS concentration to 0.1% (500 μL of chromatin extract + 4.5 mL of ChIP dilution buffer) and used for ChIP experiments. A mix of Protein A and G Dynabeads (25 μL of each) was blocked in ChIP-blocking buffer (0.5% Tween 20, 5 mg/mL BSA in PBS) for 30 min at 4°C and conjugated with 5 μg of anti-H3K9me3 (Abcam, ab8898) in ChIP-blocking buffer for 2 h in a cold room. Finally, chromatin extract and anti-H3K9me3-conjugated beads were combined and incubated overnight on a rotating wheel at 4°C. The chromatin-immunoprecipitated samples were sequentially washed six times with ChIP-RIPA buffer (10 mM Tris-HCl at pH 8, 1 mM EDTA, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% DOC), twice with ChIP-RIPA/500 buffer (RIPA supplemented with 500 mM NaCl), twice with ChiP-LiCl buffer (10 mM Tris-HCl at pH 8, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% DOC), and twice with TE buffer (10 mM Tris-HCl at pH 8, 1 mM EDTA) and eluted in 50 μL of elution buffer (10 mM Tris-HCl at pH 8, 5 mM EDTA, 300 mM NaCl, 0.1% SDS) for 6 h at 65°C. The ChIP buffers used after the cross-linking-quenching steps were ice-cold and supplemented with protease inhibitors (Roche). The eluate was sequentially treated with 20 μg of RNase A (Epicentre) for 30 min at 37°C and 100 μg of Proteinase K (New England Biolabs) for 2 h at 37°C. ChIP-seq libraries were prepared using “with-bead SPRI method” as described (Fisher et al. 2011). PCR was performed using Phusion high-fidelity PCR master mix (New England Biolabs) and 15 cycles of amplification. The libraries were sequenced on the Illumina platform with PE70 reads.
RNA-seq library preparation
Total RNA isolation was done using Trizol reagent. The Encore Complete RNA-seq DR Multiplex system (Nugen) was used to prepare stranded RNA-seq libraries. The libraries were sequenced on the Illumina platform with SE36 reads.
Databases used for mapping and annotation
A set of canonical transposon sequences for D. melanogaster (version 9.4.1) was obtained from FlyBase (http://flybase.org). The dm3 release of the D. melanogaster genome was obtained from the University of California at Santa Cruz genome database. To this genome database, we also added the X-TAS heterochromatic sequences, which represent GenBank entry L03284 and comprise X-telomeric TAS repeats from the OregonR strain. For genomic annotations, we used the D. melanogaster release 5.46 gene annotation file in gff format from FlyBase (http://flybase.org). For piRNA cluster boundaries, we used the coordinates given in Brennecke et al. (2007).
Sequencing data preprocessing
Any identifiable sequencing barcodes (at the 5′ end of reads) and adaptors (Illumina TruSeq adaptors, present at the 3′ end of the reads) were clipped using the fastx toolkit version 0.0.13. For GRO-seq and RNA-seq data, sequences <20 nt in length after clipping were discarded. For small RNA samples, all reads at least 16 nt in length were retained.
GRO-seq data analysis
The GRO-seq data were mapped first to the transposon database using the Bowtie 2 software package (version 2.0.0-beta17) (Langmead and Salzberg 2012). Due to the repetitive nature of transposon sequences, reads were allowed to map to up to 10 different canonical transposons, with read counts distributed evenly among all potential matches. For all samples, the percentage of reads that mapped exactly one time to the canonical transposon database was always more than the percentage of reads that aligned to multiple transposon sequences. To obtain an accurate estimate of the “library size” for each sample, we used all reads that failed to align to the transposon database and mapped just those sequences to the genome using the same alignment criteria (two mismatches per read, 10 total alignments). We then added together the total number of sequenced reads that were mappable to either the transposon or genome database and used this number as a normalization factor for all subsequent analyses, reporting all feature abundances as RPM mapped. To obtain accurate estimates of genomic features that might overlap partial transposon sequences (such as the piRNA clusters), we remapped all reads to the genome using more stringent criteria (one mismatch per read, 10 total alignments) but still retaining the normalization factor obtained above as the global normalization factor. For piRNA clusters, we required that the reads align exactly once. For other features, such as genes, we allowed multiple mapped reads, evenly distributing the read counts between all equivalently aligned matches (up to 10 alignments for this study, which is more stringent than the default for Tophat, which allows up to 20 alignments per read). Feature abundances were obtained by annotating each alignment with a genomic feature using intersectBed from BedTools (version-2.10.1), dividing each alignment by the total number of matches as described above, summing the alignments with the BEDTools “groupBy,” and maintaining the strandedness of all reads. For differential expression analysis, we used the DESeq package with parameters advisable for unreplicated data sets (fit, ”local”; method, ”blind”; and sharingMode, “fit-only”), accepting only changes greater than twofold, with adjusted P-values <0.05 and counts per feature >100.
mRNA-seq data analysis
The RNA-seq data were mapped as described above except that Tophat (version 1.3.2) was used any time reads were aligned to the genome to allow for reads that cross splice junctions. Feature abundances were obtained also as described above. For differential expression analysis of the mRNA-seq data sets, which were fully replicated, we used the DESeq package with default parameters, accepting only changes greater than twofold, with adjusted P-values <0.05 and counts per feature >100. For differential expression analysis of the small RNA-seq data sets, we used the DESeq parameters as described above for GRO-seq data sets.
ChIP-seq data analysis
The ChIP-seq data were mapped first to the transposon database using the Bowtie 2 software package (version 2.0.0-beta7) (Langmead and Salzberg 2012). Due to the repetitive nature of transposon sequences, reads were allowed to map to up to 10 different canonical transposons, with read counts distributed evenly among all potential matches. For all samples, the percentage of reads that mapped exactly one time to the canonical transposon database was always more than the percentage of reads that aligned to multiple transposon sequences. To obtain an accurate estimate of the “library size” for each sample, we used all reads that failed to align to the transposon database and mapped just those sequences to the genome using the same alignment criteria (two mismatches per read, 10 total alignments). We then added together the total number of sequenced reads that were mappable to either the transposon or genome database and used this number as a normalization factor for all subsequent analyses, reporting all feature abundances as RPM mapped. Enrichments for all of the H3K9me3 libraries were calculated relative to the corresponding input control sample. For differential enrichment analysis, we used the DESeq package, with parameters advisable for unreplicated data sets (fit, ”local”; method, ”blind”; and sharingMode, “fit-only”), accepting only differential enrichment changes greater than twofold, with adjusted P-values <0.05 and counts per feature >100. For genomic H3K9me3 enrichment calculations, we mapped the reads separately to the genome, accepting only uniquely mapped reads. We removed presumptive PCR duplicates with the same mapped coordinates. We used the SICER software package (version 1.1) (Zang et al. 2009) to call significantly enriched peaks at a false discovery rate (FDR) of 0.05 relative to the input control samples. For the SICER peak-calling program, we additionally set the parameters such that reads were binned in windows of 500 bp, aggregating all peaks within a distance of 500 bp.
High-throughput data deposition
The GRO-seq, RNA-seq and ChIP-seq data are available at the Gene Expression Omnibus database under GSE43360.
Acknowledgments
We thank Emily Lee for help with sequencing, Elena Rozhkova for fly husbandry, Assaf Gordon for bioinformatics support, and Alon Goren and Brad Bernstein (Massachusetts General Hospital and the Broad Institute) for help with ChIP-seq experiments. We are grateful to the entire Hannon laboratory for sharing reagents, productive discussions, and support. This work was supported by grants from the NIH (to G.J.H.) and by a kind gift from Kathryn W. Davis. G.J.H. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.209767.112.
References
- Aravin AA, Hannon GJ, Brennecke J 2007. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregliano JC, Picard G, Bucheton A, Pelisson A, Lavige JM, L'Heritier P 1980. Hybrid dysgenesis in Drosophila melanogaster. Science 207: 606–611 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H 2007. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev 21: 2300–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT 2012. Defining the status of RNA polymerase at promoters. Cell Rep 2: 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H 2000. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127: 503–514 [DOI] [PubMed] [Google Scholar]
- Desset S, Meignin C, Dastugue B, Vaury C 2003. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics 164: 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vanssay A, Bouge AL, Boivin A, Hermant C, Teysset L, Delmarre V, Antoniewski C, Ronsseray S 2012. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 490: 112–115 [DOI] [PubMed] [Google Scholar]
- Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, et al. 2011. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol 12: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydrychova RC, Mason JM, Archer TK 2008. HP1 is distributed within distinct chromatin domains at Drosophila telomeres. Genetics 180: 121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju VK, Yin H, Weiner MM, Wang J, Huang XA, Lin H 2011. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat Genet 43: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev 24: 2499–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ 2012. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H, Siomi H, Siomi MC 2012. Biology of PIWI-interacting RNAs: New insights into biogenesis and function inside and outside of germlines. Genes Dev 26: 2361–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse T, Teysset L, Todeschini AL, Sidor CM, Anxolabehere D, Ronsseray S 2007. Telomeric trans-silencing: An epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genet 3: 1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA 2005. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res 33: 2052–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y 2011. 3′ End formation of PIWI-interacting RNAs in vitro. Mol Cell 43: 1015–1022 [DOI] [PubMed] [Google Scholar]
- Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE 2011. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell 147: 1551–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Kidwell JF, Sved JA 1977. Hybrid dysgenesis in Drosophila melanogaster: A syndrome of aberrant traits including mutation, sterility and male recombination. Genetics 86: 813–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA 2007. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res 35: 5430–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, Gvozdev VA 2011. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci 108: 18760–18765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin HL, Moran JV 2011. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet 12: 615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevel-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A 2007. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 175: 1615–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovich N, Lei EP 2010. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet 6: e1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. 2012. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491: 284–287 [DOI] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J 2010. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 29: 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Song SU, Prud'homme N, Smith PA, Bucheton A, Corces VG 1994. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J 13: 4401–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Teysset L, Chalvet F, Kim A, Prud'homme N, Terzian C, Bucheton A 1997. About the origin of retroviruses and the co-evolution of the gypsy retrovirus with the Drosophila flamenco host gene. Genetica 100: 29–37 [PubMed] [Google Scholar]
- Pelisson A, Sarot E, Payen-Groschêne G, Bucheton A 2007. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol 81: 1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, Berloco M, Turano C, Ferraro A, Pimpinelli S 2004. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol Cell 15: 467–476 [DOI] [PubMed] [Google Scholar]
- Preall JB, Czech B, Guzzardo PM, Muerdter F, Hannon GJ 2012. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA 18: 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme N, Gans M, Masson M, Terzian C, Bucheton A 1995. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139: 697–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. 2011. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell 147: 1628–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R 2011. piRNA production requires heterochromatin formation in Drosophila. Curr Biol 21: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Marin L, Lehmann M, Anxolabehere D 1998. Repression of hybrid dysgenesis in Drosophila melanogaster by combinations of telomeric P-element reporters and naturally occurring P elements. Genetics 149: 1857–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Schostak NG, Zelentsova ES, Yushenova IA, Zatsepina OG, Evgen'ev MB 2013. Evolution and dynamics of small RNA response to a retroelement invasion in Drosophila. Mol Biol Evol 30: 397–408 [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky M, Kravchuk O, Melnikova L, Georgiev P 2002. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol Cell Biol 22: 3204–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V 2006. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev 20: 345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Lyko F 2010. Lack of evidence for DNA methylation of Invader4 retroelements in Drosophila and implications for Dnmt2-mediated epigenetic regulation. Nat Genet 42: 920–921 [DOI] [PubMed] [Google Scholar]
- Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, Kalmykova A 2011. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res 39: 8703–8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J 2012. Transcriptional silencing of transposons by piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Sokolova OA, Iakushev E, Stoliarenko AD, Mikhaleva EA, Gvozdev VA, Klenov MS 2011. [The interplay of transposon silencing genes in the Drosophila melanogaster germline]. Mol Biol (Mosk) 45: 633–641 [PubMed] [Google Scholar]
- Voigt F, Reuter M, Kasaruho A, Schulz EC, Pillai RS, Barabas O 2012. Crystal structure of the primary piRNA biogenesis factor Zucchini reveals similarity to the bacterial PLD endonuclease Nuc. RNA 18: 2128–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Elgin SC 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci 108: 21164–21169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Lin H 2007. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450: 304–308 [DOI] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W 2009. A clustering approach for identification of enriched domains from histone modification ChIP-seq data. Bioinformatics 25: 1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]