To understand the ribonucleoprotein remodeling events that occur during spliceosome assembly, Fourmann et al. create a purified splicing system that reproduces the entire ordered disassembly phase of the spliceosome with purified components. Using biochemical analysis and mass spectrometry, they characterize the proteins of the intron-lariat spliceosome (ILS) and identify the role of Prp43 in promoting the dissociation of the ILS.
Keywords: intron-lariat spliceosome, disassembly, Prp22, Prp43, Brr2
Abstract
The spliceosome is a single-turnover enzyme that needs to be dismantled after catalysis to both release the mRNA and recycle small nuclear ribonucleoproteins (snRNPs) for subsequent rounds of pre-mRNA splicing. The RNP remodeling events occurring during spliceosome disassembly are poorly understood, and the composition of the released snRNPs are only roughly known. Using purified components in vitro, we generated post-catalytic spliceosomes that can be dissociated into mRNA and the intron-lariat spliceosome (ILS) by addition of the RNA helicase Prp22 plus ATP and without requiring the step 2 proteins Slu7 and Prp18. Incubation of the isolated ILS with the RNA helicase Prp43 plus Ntr1/Ntr2 and ATP generates defined spliceosomal dissociation products: the intron-lariat, U6 snRNA, a 20–25S U2 snRNP containing SF3a/b, an 18S U5 snRNP, and the “nineteen complex” associated with both the released U2 snRNP and intron-lariat RNA. Our system reproduces the entire ordered disassembly phase of the spliceosome with purified components, which defines the minimum set of agents required for this process. It enabled us to characterize the proteins of the ILS by mass spectrometry and identify the ATPase action of Prp43 as necessary and sufficient for dissociation of the ILS without the involvement of Brr2 ATPase.
Pre-mRNA splicing proceeds by way of two phosphoester transfer reactions and is catalyzed by the spliceosome, which consists of the U1, U2, U4/U6, and U5 small nuclear ribonucleoproteins (snRNPs) and numerous non-snRNP proteins (Will and Lührmann 2011). The snRNPs are involved in recognizing short conserved sequences of the pre-mRNA, including the 5′ and 3′ splice sites (SS) and the branch site (BS), and in positioning the reactive nucleotides for catalysis. The spliceosome is a dynamic molecular machine, undergoing several major structural rearrangements during its functional cycle. These events are mediated by at least eight conserved DExD/H-box NTPases (hereafter termed RNA helicases) that act at discrete stages of the splicing pathway (Staley and Guthrie 1998; Cordin et al. 2012).
Spliceosome assembly is initiated by the binding of U1 and U2 snRNPs to the 5′ and the BS, respectively. This is followed by the recruitment of the U4/U6.U5 tri-snRNP, forming the precatalytic B complex. Catalytic activation of the spliceosome (leading to complex Bact) involves the displacement of U1 and U4 snRNAs from the spliceosome and the formation of new base pair interactions between the U6 and U2 snRNAs and the 5′ SS (Staley and Guthrie 1998). The resulting RNA structure plays a central role in catalyzing both steps of splicing (Nilsen 1998).
These RNA–RNA rearrangements are remodeled by the combined actions of the RNA helicases Prp28 and Brr2, which disrupt the base-pairing between U1 snRNA and the 5′ SS and between the U4 and U6 snRNAs, respectively (Laggerbauer et al. 1998; Raghunathan and Guthrie 1998; Staley and Guthrie 1999; Small et al. 2006). In addition, the GTPase Snu114, which together with Brr2 is associated with the U5 snRNP, is also required for the activation step and is thought to regulate Brr2 activity (Small et al. 2006). The transition from the B to the Bact complex involves a significant change in the spliceosome's protein composition, with numerous polypeptides such as the U1 and U4/U6 proteins being displaced, while ∼20 others, including those of the NTC (nineteen complex) are stably recruited to the Bact complex (Chan et al. 2003; Fabrizio et al. 2009; Warkocki et al. 2009). For catalytic activation of the spliceosome, the DEAH-box helicase Prp2 binds together with a cofactor, Spp2, and restructures the Bact complex in an ATP-dependent manner, yielding the B* complex (Roy et al. 1995; Silverman et al. 2004); this leads to, among other things, destabilization and/or disruption of the binding of several spliceosomal proteins (Warkocki et al. 2009; Lardelli et al. 2010; Ohrt et al. 2012). Upon recruitment of the step 1 factor Cwc25 to the B* complex, step 1 catalysis occurs, whereby the adenosine at the BS attacks the 5′ SS, generating the cleaved exon 1 and intron-lariat (IL)–3′ exon intermediates (Chiu et al. 2009; Warkocki et al. 2009). The catalytic center of the resulting C complex is remodeled by the DEAH-box helicase Prp16 as a prerequisite for the second step of splicing; i.e., exon ligation, a process that also requires the action of the step 2 factors Slu7 and Prp18 (Horowitz 2012). However, the latter factors do not appear to be required if the distance between the pre-mRNA's BS and 3′ SS consensus sequences is short (i.e., ∼7 nucleotides [nt]) (Brys and Schwer 1996).
The post-catalytic spliceosome must be dismantled to both release the mature mRNA, snRNPs, and splicing factors and subsequently recycle the latter two for subsequent rounds of pre-mRNA splicing. In the first step of this disassembly, the DEAH-box RNA helicase Prp22 translocates in a 3′-to-5′ direction along the mature mRNA and promotes its release from the intron-lariat spliceosome (ILS) by disrupting the interaction between loop I of the U5 snRNA and the mRNA in an ATP-dependent manner (Company et al. 1991; Schwer 2008). During this step, Slu7 and Prp18 are expelled, but whether this is a prerequisite for disruption of the U5–mRNA interaction is not known. Indeed, whether the ATP-dependent action of Prp22 leads to the release of other spliceosomal proteins is not clear. Prp22 has an additional ATP-independent role in promoting efficient step 2 catalysis in whole yeast cell extracts in vitro (Schwer and Gross 1998) and in contributing, by an ATP-dependent mechanism, to the fidelity of exon ligation (Mayas et al. 2006).
The dissociation of the IL RNA from the ILS requires the DEAH-box RNA helicase Prp43 (Arenas and Abelson 1997; Martin et al. 2002; Tsai et al. 2005). At the same time, the remaining spliceosomal core, containing the U2/U6 and U5 snRNPs, is also disassembled (Tsai et al. 2005). Yeast Prp43 interacts with its cofactors, Ntr1 (also called Spp382) and Ntr2, forming the NTR complex (Tsai et al. 2005; Boon et al. 2006). Ntr2 forms a tight complex with Ntr1, while Ntr1 interacts with Prp43 through its G-patch domain (Tsai et al. 2005; Tanaka et al. 2007). The latter interaction is crucial for the stimulation of Prp43's otherwise weak RNA helicase activity, which is essential for IL release (Tanaka et al. 2007). Ntr2 can also bind to Brr2, and it has been suggested that this interaction may help recruit Prp43 to the ILS (Tsai et al. 2007). In yeast, Brr2 helicase activity also appears to be required for ILS disassembly, as has been shown in vitro for the dissociation of purified endogenous spliceosomes (Small et al. 2006), with the consequent proposal that Brr2 and Prp43 act together but on different targets in the ILS.
While the requirement of the disassembly of the yeast spliceosome for Prp22 and Prp43 activity is well established, less is understood about the effect of the remodeling activities of these helicases on the structure and biochemical composition of the ILS and the products that arise during its disassembly. For example, it is not known which proteins of the post-catalytic spliceosome remain bound to the ILS and which dissociate simultaneously from the ILS together with Slu7/Prp18 upon Prp22-mediated release of the mRNA from the post-catalytic spliceosome. Moreover, the protein composition of the U2, U5, and U6 snRNPs released from the ILS are only poorly understood. In humans, a 35S U5 snRNP containing the core U5 proteins hPrp8, hBrr2, and hSnu114; all proteins of the human NTC complex; and additional NTC-related proteins can be isolated, and it was suggested that this 35S U5 snRNP might be a disassembly intermediate of the post-catalytic human spliceosome (Makarov et al. 2002). However, it remains unclear whether, in yeast, NTC proteins remain associated with particular snRNPs during the disassembly process. Other particularly interesting questions are whether the Prp43 and Brr2 helicases act sequentially or cooperatively and whether the inactivation of one of these helicases may lead to the appearance of high-molecular-weight RNP disassembly intermediates.
To address these and further questions, we recapitulated the disassembly steps of yeast splicing using purified Bact spliceosomes and recombinantly expressed proteins needed for splicing catalysis and subsequent disassembly of the spliceosome. We investigated the factor requirements for disassembly of the post-catalytic spliceosome by Prp22 and show that Slu7 and Prp18 are not required for the stable binding of the mRNA to the post-catalytic spliceosome, the efficient recruitment of Prp22 to the post-catalytic spliceosome, or its subsequent dissociation. We also identify those proteins released from the spliceosome during Prp22-mediated disassembly by characterizing the purified ILS by mass spectrometry (MS). We show that upon addition of the NTR complex plus ATP, the isolated ILS is dissociated into the IL RNA, the U6 snRNA, a 20–25S U2 snRNP, and an 18S U5 snRNP (all of which sediment separately during glycerol gradient centrifugation) and characterize in detail the biochemical composition of these released components. Finally, we demonstrate that in our purified splicing system, the helicase activity of Prp43 is both necessary and sufficient to promote the dissociation of the ILS and that the ATPase activity of Brr 2 is not required for this.
Results
Purification of ILSs
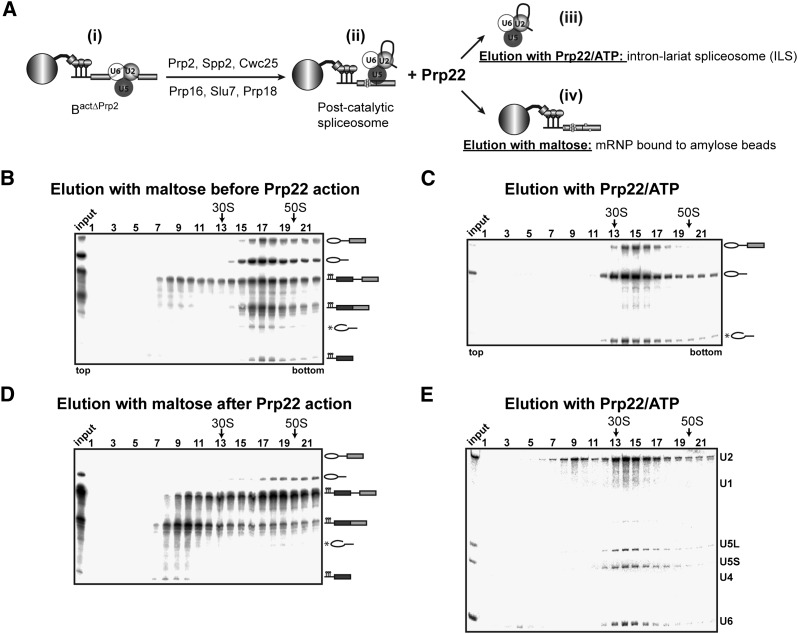
The strategy that we used to reproduce in vitro the disassembly steps of the yeast spliceosome using purified components is illustrated schematically in Figure 1A. The key to this strategy was our recent observation that in our extract-free, in vitro splicing system (Warkocki et al. 2009), the second catalytic step proceeds very efficiently in the absence of the RNA helicase Prp22 (Supplemental Fig. S1, note that there is no difference in the yield of step 2 products in the presence or absence of Prp22; T Ohrt, P Odenwälder, J Dannenberg, M Prior, Z Warkocki, J Schmitzová, R Karaduman, I Gregor, J Enderlein, P Fabrizio, et al., in prep.). In this way, we were able to uncouple the generation of the post-catalytic spliceosome from its Prp22-mediated dissociation into the ILS and mRNA.
Figure 1.
The ILS is released by the action of Prp22. (A, panel i) Schematic of the reconstitution strategy, starting from affinity-purified BactΔprp2 spliceosomes bound to the amylose matrix (large ball). (Panel ii) Post-catalytic spliceosomes bound to the matrix were obtained by adding recombinant Prp2, Spp2, Cwc25, Prp16, Slu7, and Prp18. (Panel iii) Subsequent addition of Prp22 triggers the release of the ILS from the amylose matrix. (Panel iv) The matrix was washed, and the bound material (i.e., mRNA products) was eluted with maltose. (B) BactΔprp2 spliceosomes were incubated with ATP plus recombinant Prp2, Spp2, Cwc25, Prp16, Slu7, and Prp18 to obtain post-catalytic spliceosomes, which were then eluted with maltose from the matrix and separated on a 10%–30% glycerol gradient. (C) Post-catalytic spliceosomes bound to the amylose matrix were incubated with Prp22 plus ATP, and the released ILS was separated on a 10%–30% glycerol gradient. (D) mRNA and other complexes still bound to the matrix after incubation with Prp22 and ATP were eluted with maltose and separated on a 10%–30% glycerol gradient. (B–D) 32P-labeled pre-mRNA and splicing products/intermediates (indicated on the right) from the gradient fractions shown were recovered, separated by denaturing PAGE, and visualized by autoradiography. (E) Northern blot analysis of the snRNAs associated with the ILS. RNA identities are indicated on the right. Sedimentation coefficients were determined by analyzing the UV absorbance of a reference gradient containing prokaryotic ribosomal subunits. Input lanes indicate the samples before sedimentation.
To isolate the ILS, we first assembled spliceosomes that were stalled before step 1 (i.e., yielding BactΔPrp2 complexes) by performing splicing in vitro with Actin7 (Act7) pre-mRNA containing three binding sites for the MS2 phage coat protein upstream of the first exon (M3Act7). In Act7 pre-mRNA, the distance between the BS and the 3′ SS consensus sequences is shorter than in wild-type actin pre-mRNA (7 nt vs. 38 nt), and thus one or more of the step 2 factors Slu7, Prp18, and Prp22 are not required (Brys and Schwer 1996). Splicing was performed with heat-inactivated extracts derived from a yeast strain expressing the temperature-sensitive Prp2 mutant (prp2-1) as described previously (Warkocki et al. 2009). BactΔPrp2 complexes were then purified in a two-step procedure via the MS2-MBP fusion protein (MS2 fused to maltose-binding protein) that allows the BactΔPrp2 complexes to be retained on an amylose affinity matrix (see Fig. 1A). The two-step splicing reaction was then initiated by incubating the matrix-bound BactΔPrp2 spliceosomes with recombinant step 1 factors Prp2, Spp2, and Cwc25 plus step 2 factors Prp16, Slu7, and Prp18 in the presence of ATP for 45 min at 23°C (Fig. 1A).
Under these conditions, post-catalytic spliceosomes composed of the ILS in complex with the M3Act7 mRNA were efficiently formed and remained bound to the amylose matrix. In the experiment shown in Figure 1B, spliceosomal complexes eluted with maltose from the affinity column (on which the splicing reaction with reconstituted components had been performed, as shown in Fig. 1A) were subjected to glycerol gradient centrifugation, and the distribution of their RNAs was analyzed. The spliced mRNA comigrated together with the ILS, forming the post-catalytic spliceosome, at an S value of ∼40 (Fig. 1B, fractions 16–19). While the latter is the most abundant spliceosomal complex among the reaction products, low amounts of earlier-stage Bact and C complexes, which contained unspliced M3Act7 pre-mRNA (the Bact complex) or M3Act7 splicing intermediates (the C complex), were also eluted from the column; these exhibit S values similar to those of the post-catalytic spliceosomes (Fig. 1B). Analysis of the minor amounts of spliceosomal particles migrating in fractions 7–14 revealed that these were disassembled spliceosomes containing only a subset of snRNPs (data not shown). In fractions 16–19 of the gradient in Figure 1B, an RNA band that migrates slightly faster than the mRNA is visible. This is linearized IL RNA that migrates as an ∼280-nt-long linear RNA (see also Supplemental Fig. S1, where a size marker has been added to an RNA gel). This RNA species always migrates in gradients together with the native IL RNA (see below) and is labeled with an asterisk in Figure 1, B–D.
Next, we tested whether the ILS could be released from the affinity matrix-bound post-catalytic spliceosome by the action of Prp22, a process in which the M3Act7 mRNA should be retained on the matrix. For this purpose, the affinity matrix containing the post-catalytic spliceosomes was washed thoroughly and then incubated with recombinant Prp22 and ATP for 10 min at 23°C. The liquid phase was collected and then subjected to glycerol gradient centrifugation. The RNA content from each fraction of the gradient was subsequently analyzed by denaturing PAGE (Fig. 1C). The predominant RNA species in the gradient was the IL RNA, which peaked in fractions 13–17, corresponding to the 35S region of the gradient. As no mRNA product was detected in the gradient, the 35S complex containing the IL must have been the ILS liberated from the post-catalytic spliceosome by Prp22/ATP. The 35S ILS peak was contaminated with a small amount of the IL–3′ exon intermediate (generally <5%) that dissociated spontaneously during incubation with Prp22. Note that usually ∼5%–10% of the 35S ILS contains the linearized IL RNA (Fig. 1C). Dissociation of the 35S ILS from the post-catalytic spliceosome required Prp22-mediated ATP hydrolysis, as in the absence of ATP, no ILS is released from the affinity matrix (data not shown).
Northern blotting showed that the 35S ILS also contains (in addition to the IL RNA) the U2, U5, and U6 snRNAs (Fig. 1E, fractions 13–17). Small quantities of free U2, U5, and U6 snRNPs, which probably dissociated spontaneously from the spliceosomal complexes during incubation with Prp22, migrated at low S values (Fig. 1E, fractions 2–11). Glycerol gradient fractionation of the material eluted from the affinity matrix with maltose (after the action of Prp22 and subsequent washing) (Fig. 1D) revealed that most of the eluted mRNA migrated in fractions 8–11 with an S value of ∼20. Only a small portion remained associated with the ILS and thus was found in the 40S region of the gradient, confirming that the ILS was efficiently released by Prp22 from the post-catalytic spliceosome. In summary, these results show that our purified splicing system allows efficient Prp22-mediated dissociation of post-catalytic spliceosomes into ILS and mRNA/RNP.
Protein requirements for dissociation of the ILS from the mRNA
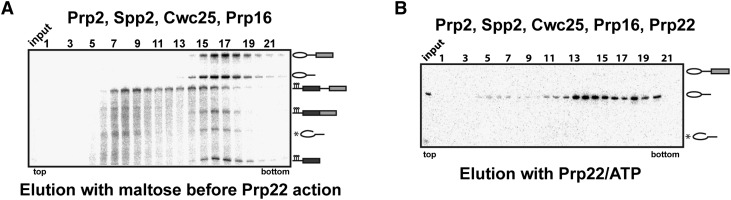
Immunodepletion of Slu7 from splicing extracts still allows step 2 catalysis if pre-mRNAs have a short distance between their BS and 3′ SS, such as the Act7 substrate used here (Brys and Schwer 1996). In our purified splicing system, none of the step 2 factors Slu7, Prp18, and Prp22, which are completely absent from our purified BactΔPrp2 spliceosomes, are required for step 2 catalysis when spliceosomes are assembled on Act7 pre-mRNA, although the yield of second-step products is higher in the presence of Slu7 and Prp18 (T Ohrt, P Odenwälder, J Dannenberg, M Prior, Z Warkocki, J Schmitzová, R Karaduman, I Gregor, J Enderlein, P Fabrizio, et al., in prep.). To test whether, in the absence of these three step 2 factors, the newly spliced Act7 mRNA remains stably bound to the post-catalytic spliceosome, we reconstituted the post-catalytic spliceosome as described above—this time, however, in the absence of Slu7/Prp18 (and Prp22). When the reaction mixture was eluted from the column with maltose and subjected to glycerol gradient centrifugation, the spliced Act7 mRNA quantitatively cosedimented together with the ILS (Fig. 2A), demonstrating that the newly formed mRNA remains tightly bound to the spliceosome even in the absence of Slu7 and Prp18. Addition of Prp22 and ATP to the reconstituted post-catalytic spliceosome formed in the absence of Slu7 and Prp18 led to efficient dissociation of the ILS from the Act7 mRNA (Fig. 2B). Thus, Slu7 and Prp18 are not required for the recruitment of Prp22 to its target site in the post-catalytic spliceosome or for Prp22′s catalytic activity at this stage.
Figure 2.
Prp22 dissociates the post-catalytic spliceosome in the absence of Slu7 and Prp18. (A) BactΔprp2 spliceosomes in solution were incubated with ATP plus recombinant Prp2, Spp2, Cwc25, and Prp16 and then separated on a 10%–30% glycerol gradient. (B) BactΔprp2 spliceosomes bound to the matrix were incubated with ATP plus recombinant Prp2, Spp2, Cwc25, and Prp16. The matrix was washed and then incubated with Prp22 plus ATP, and the released ILS was separated on a 10%–30% glycerol gradient. Input lanes indicate the samples before sedimentation.
Protein composition of the isolated 35S ILS
Little is known about the protein composition of the ILS produced in vitro. We therefore isolated 35S ILSs as described above and purified them by glycerol gradient centrifugation. ILS-containing gradient fractions (13–17) were pooled (see Fig. 1C), and proteins were separated by SDS-PAGE and identified by MS. In Table 1, the number of peptides for each protein detected in the ILS is shown in column 5. We also analyzed the protein composition of the top five fractions (pooled) from the same gradient (Table 1, column 4). These top fractions should contain those proteins that lose their affinity for the spliceosome during the Prp22-mediated dissociation of the ILS from the mRNA and that are therefore released from the affinity matrix along with the ILS. For comparison, we also eluted 40S post-catalytic spliceosomes from the amylose beads with maltose before incubation with Prp22, subjected them to glycerol gradient centrifugation (see fractions 15–19 of Fig. 1B), and analyzed the spliceosome-containing fractions and the top five fractions (pooled) of the gradient by MS (Table 1, columns 3 and 2, respectively). Finally, proteins associated with isolated BactΔPrp2 spliceosomes were also analyzed (Table 1, column 1).
Table 1.
Proteins identified by MS in BactΔPrp2 complexes, supplemented stepwise with the recombinant proteins indicated
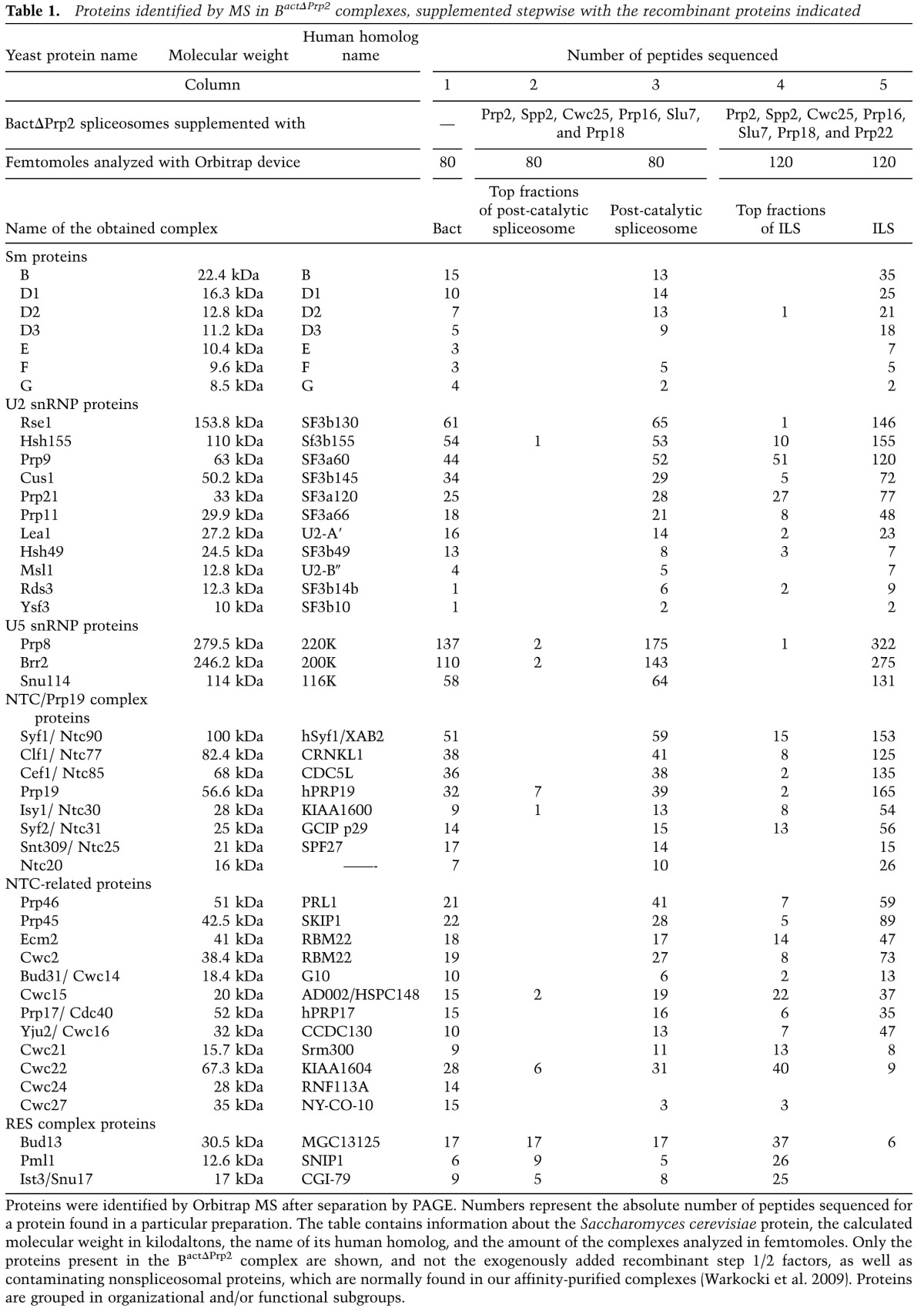
The 35S ILS contained—in addition to the Sm proteins—all U2 and three U5 proteins, plus all components of the NTC complex in abundances comparable with those found in the post-catalytic and BactΔPrp2 spliceosomes (Table 1). No Sm or U5 proteins were found in the top fractions of the ILS gradient, demonstrating that they are stably integrated into the ILS. In contrast, proteins of the RES complex were found almost quantitatively in the top fractions of the ILS gradient, with only trace amounts associated with the 35S ILS (Table 1). As the RES proteins were present in significant amounts in the post-catalytic spliceosome (Table 1, column 3), these results suggest that the RES proteins dissociate from the spliceosome upon Prp22-mediated release of the ILS (see also the Discussion). A similar loss of spliceosomal binding affinity upon Prp22-mediated dissociation of the post-catalytic complex was also found for the NTC-related proteins Cwc21 and Cwc22; the majority of both proteins was found in the top fractions of the 35S ILS gradient (Table 1, columns 4, 5), while both remained stably bound to the post-catalytic spliceosome (Table1, columns 2, 3). In contrast, the majority of Yju2 remained stably associated with the ILS (Table 1). Cwc24 and Cwc27 were not detected in the 35S ILS, consistent with the fact that they dissociate from the spliceosome already during Prp2-mediated catalytic activation of the Bact complex (Warkocki et al. 2009; Ohrt et al. 2012). We note that Cwc23 was not detected by MS either here or in previous studies (Warkocki et al. 2009). Cwc23 was previously implicated in the disassembly of spliceosomes but is not essential for this step (Pandit et al. 2006, 2009). We found that in our reconstitution system, Cwc23 addition has no effect on the efficiency of ILS displacement from the mRNA by Prp22 or on that of the NTR-mediated dissociation of the purified ILS (data not shown). Possibly, Cwc23 may assist in the disassembly of only a subset of post-catalytic spliceosomes in vivo. In summary, the isolated 35S ILS contains most of the proteins present in the post-catalytic spliceosome, including all U2 and U5 proteins, the NTC complex, and most NTC-related proteins, while Cwc21, Cwc22, and the RES proteins dissociate from the spliceosome or are less stably associated at this stage.
A 20–25S U2 snRNP, 18S U5 snRNP, and naked U6 snRNA are released upon NTR-mediated disassembly of the ILS
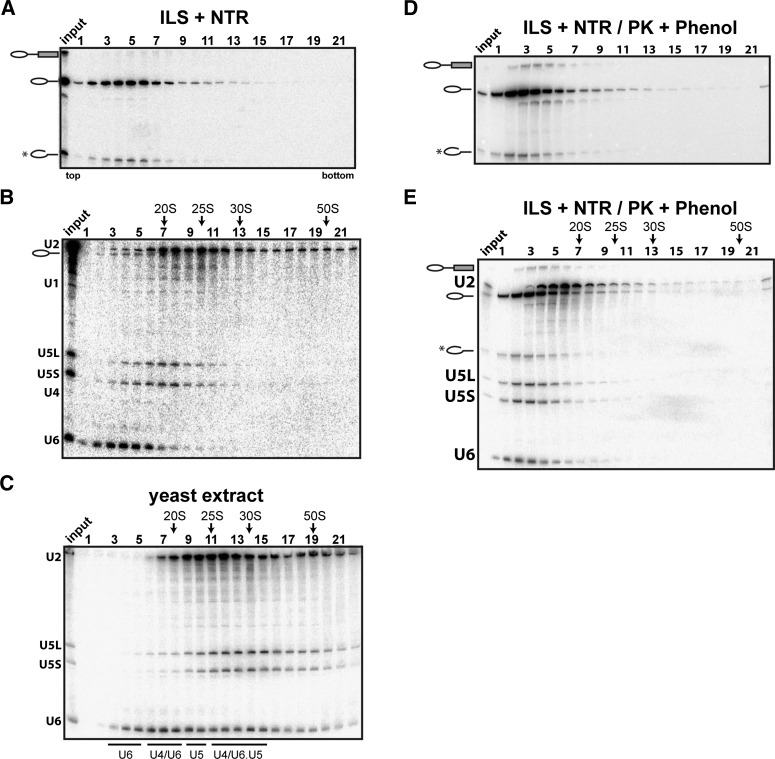
We next investigated whether our isolated 35S ILS complex can be dissociated by the action of the RNA helicase Prp43. We incubated the isolated 35S ILS (in each case, the entire amount of ILSs eluted from the matrix with Prp22/ATP) with recombinant NTR together with ATP and then fractionated the mixture by glycerol gradient centrifugation. Figure 3A shows that the IL RNA is released quantitatively from the ILS and sediments near the top of the gradient in fractions 2–6. The same was observed for the linearized IL RNA (Fig. 3A). We note that a certain excess of NTR proteins over ILS was required in our purified system. In pilot experiments (data not shown), disassembly occurred only to a minor extent (<5%) in the presence of ATP and low concentrations of NTR. Efficient disassembly was only observed at NTR concentrations >30 nM. Therefore, we generally used an NTR concentration of 50 nM in order to achieve nearly complete disassembly of the ILS input. We ascribe the requirement for a certain excess of recombinant proteins in our purified disassembly system to the need to attain sufficient saturation of binding—of the components of the NTR complex to one another and/or of this complex to the ILS—that is not arbitrarily strong. An alternative/additional possibility that we cannot exclude at present is that not the entire fraction of all of the recombinant proteins is correctly folded. The dissociation of the ILS required not only Prp43, but also Ntr1, Ntr2, and ATP (Supplemental Fig. S2). We conclude that the highly purified ILS, like endogenous ILSs, can be dissociated in vitro in an NTR-specific manner.
Figure 3.
The purified ILS was disassembled in solution by the action of Prp43 plus Ntr1 and Ntr2. (A) The ILS was incubated in solution with ATP and recombinant Prp43, Ntr1, and Ntr2 and separated on a 10%–30% glycerol gradient. RNA from the gradient fractions shown was recovered, separated by denaturing PAGE, and visualized by autoradiography (A) and Northern blot analysis (B). (C) Total yeast extract was subjected to sedimentation on a 10%–30% glycerol gradient, and RNA was extracted from each fraction and analyzed by Northern blotting. (D) After incubation with Prp43, Ntr1, and Ntr2, proteins associated with the ILS were digested by Proteinase K (PK) and subjected to phenol extraction, and the ILS-associated RNAs were separated on a 10%–30% glycerol gradient. RNAs from the gradient fractions shown were analyzed as in A or E by Northern blot. RNA identities are indicated on the left. Sedimentation coefficients were determined by analyzing the UV absorbance of a reference gradient containing prokaryotic ribosomal subunits and human snRNAs. Input lanes indicate the samples before sedimentation.
To identify the Prp43-mediated dissociation products of the ILS, RNAs were extracted from the gradient fractions and analyzed by Northern blotting with U2-, U5-, and U6-specific probes. As shown in Figure 3B, U2 snRNA migrated in several fractions, with the majority in fractions 7–12. U5 snRNA was found in fractions 4–10, with a clear peak in fractions 6–8, whereas U6 snRNA was found at the top of the gradient, peaking in fractions 3–5. The high S values (∼16–18S for U5 and ∼20–25S for U2) indicate that these particles are associated with proteins, as protein-free (proteinase K-digested and phenolized) U2 and U5 snRNAs have much lower S values (Fig. 3D,E). When total yeast splicing extract was subjected to glycerol gradient centrifugation, endogenous U2 snRNPs also migrated with S values of ∼20–25S (Fig. 3C), whereas most of the U5 snRNP appeared to be mainly present as part of the 25S tri-snRNP particle. Thus, the U2 snRNP released from the ILS by the NTR exhibits an S value similar to that of U2 present in splicing extract. U6 exists as part of the U6 snRNP, U4/U6 di-snRNP, and tri-snRNP and thus migrates very broadly with S values of 10–25S.
While the majority of the U2, U5, and U6 snRNPs sediment independently in the glycerol gradient (Fig. 3B), some comigration was seen between the U5 and U2 snRNPs. Likewise, U6 snRNA and some U2 snRNA comigrated with the IL RNA in the top fractions of the gradient. This raised the possibility that there may be some residual higher-order complexes formed between the RNPs/RNAs after the NTR-mediated dissociation of the ILS. To investigate this, we conducted a series of pull-down experiments with the gradient-fractionated RNPs/RNAs (Fig. 4). First, fractions 2–6 of the gradient were pooled, and aliquots were incubated with biotinylated antisense oligonucleotides complementary to either U6 snRNA or IL RNA, followed by precipitation with streptavidin-agarose beads. Coprecipitated RNAs were separated by denaturing gel electrophoresis and then analyzed by either direct autoradiography (for the 32P-labeled IL RNA) (Fig. 4A, top panel) or Northern blotting (for snRNAs) (Fig. 4A, bottom panel). Only small amounts of U2, U6, and IL snRNPs/RNAs were precipitated by the beads alone, while much higher background precipitation was observed for U5 snRNP, despite extensive washing (Fig. 4A, lane 4). The antisense oligonucleotide against the IL RNA precipitated significant quantities of IL RNP/RNA (Fig. 4A, lane 2) and minor amounts of U2 snRNA, while the amount of U6 snRNA coprecipitated was only slightly above the background level (Fig. 4A, lanes 2,4). Furthermore, whereas the anti-U6 oligonucleotide precipitated U6, no IL RNA, U2, or U5 snRNA was coprecipited above background levels (Fig. 4A, lanes 2–4). Thus, upon dissociation of the ILS, U6 snRNA/RNP and IL RNA/RNP are separated from both one another and also the bulk of the U2 and the U5 snRNPs.
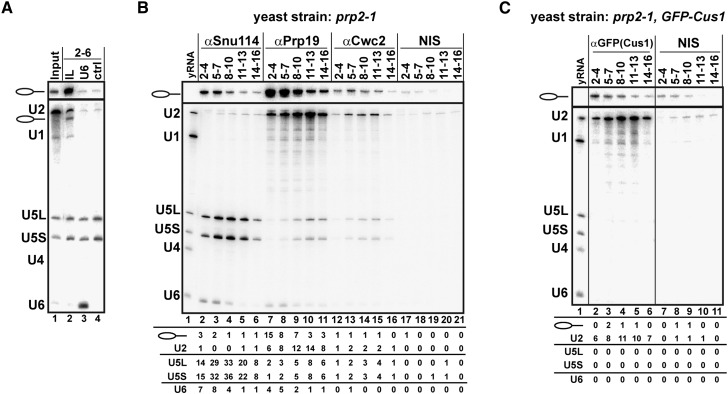
Figure 4.
Identification of proteins associated with the U2 and U5 snRNAs and the IL after ILS dissociation by the NTR complex. (A) The ILS was incubated with Prp43 plus Ntr1 and Ntr2 and then separated on a 10%–30% glycerol gradient. Fractions 2–6 were pooled and subjected to pull-down with bio-TEG RNA oligonucleotides complementary to U6 and the IL or without oligos (ctrl). (Lane 1) Input lane indicates the sample before sedimentation. (Bottom panel) snRNAs were visualized by autoradiography after Northern blotting. (B) The ILS was incubated with Prp43 plus Ntr1 and Ntr2 and separated on a 10%–30% glycerol gradient. Every three fractions from 2 to 16 (2–4, 5–7, etc.) were combined and immunoprecipitated with anti-Snu114, anti-Prp19, and anti-Cwc2 antibodies or with nonimmunoserum (NIS), respectively. (Lane 1) Total yeast RNA (yRNA) was used as a marker. (C) The ILS was reconstituted and affinity-purified from the yeast strain prp2-1 carrying the U2 protein Cus1 tagged with GFP. Subsequently, the ILS was incubated with Prp43 plus Ntr1 and Ntr2 and separated on a 10%–30% glycerol gradient. Every three fractions from 2 to 16 were combined and immunoprecipitated with anti-GFP antibodies or NIS. (Lane 1) Total yeast RNA (yRNA) was used as a marker. Quantifications were performed with ImageQuant software (Molecular Dynamics). Numbers represent the percentage of RNA precipitated by the antibody relative to the total amount of each RNA in the corresponding fractions. Aliquots of each fraction before immunoprecipitation were analyzed on denaturing PAGE, and these gels were used to quantify the total amount of each RNA present in each fraction (data not shown).
To determine whether the U2 and U5 snRNPs are also completely separated from one another upon ILS disassembly, we pooled three gradient fractions starting at fraction 2 (2–4, 5–7, etc.) and performed immunoprecipitations with antibodies against the U5 protein Snu114 and Protein A-Sepharose beads. An analysis of the RNA content of the immunoprecipitates showed that whereas U5 snRNPs were precipitated very efficiently, no U2 snRNPs were coprecipitated above background levels (Fig. 4B, cf. lanes 2–6 and nonimmunoserum [NIS] control lanes 17–21), indicating that these snRNPs do not interact with each other. The anti-Snu114 immunoprecipitates from the pooled fractions 2–4 and 5–7 contained only low amounts of IL and U6 snRNA, (i.e., 1%–3% and ∼7% of the respective input RNA). As no antibodies against a U2 protein were available, we performed a similar experiment but with a yeast strain expressing the U2-specific Cus1 protein tagged with GFP (green fluoresent protein) (Ohrt et al. 2012). Antibodies against GFP precipitated the U2 snRNP, and not U5, from the pooled gradient fractions (Fig. 4C, cf. lanes 2–6 and the NIS control in lanes 7–11). In summary, these experiments show that the U2, U5, and U6 snRNPs and the IL RNA/RNP are released as separate components during the NTR-mediated dissociation of the ILS.
The S values of the U2 and U5 particles released by the NTR-mediated dissociation of the ILS and their precipitability with anti-Cus1 or anti-Snu114 antibodies indicated that at least some of the snRNP proteins remain specifically associated with their respective snRNAs. To learn more about the protein composition of the released U2 and U5 snRNPs, we dissociated the ILS with the NTR, separated the released RNPs by glycerol gradient centrifugation, and then analyzed the gradient fractions pooled pairwise (1+2, 3+4, etc.) by MS. The majority of the U5 proteins Prp8, Brr2, and Snu114 were found in fractions 7 and 8 of the gradient (Table 2), where the U5 snRNA also peaks (see Fig. 3B), indicating that all of the U5 proteins present in the ILS remain stably and specifically associated with the U5 snRNA after ILS disassembly by the NTR. Likewise, most of the U2 proteins appear to remain associated with the U2 snRNA, as the largest peptide numbers for U2 proteins were found in fractions 9–12, where the U2 snRNA also peaked (Table 2; Fig. 3B).
Table 2.
Protein composition of reconstituted ILS subsequently disassembled by the action of Prp43, Ntr1, and Ntr2
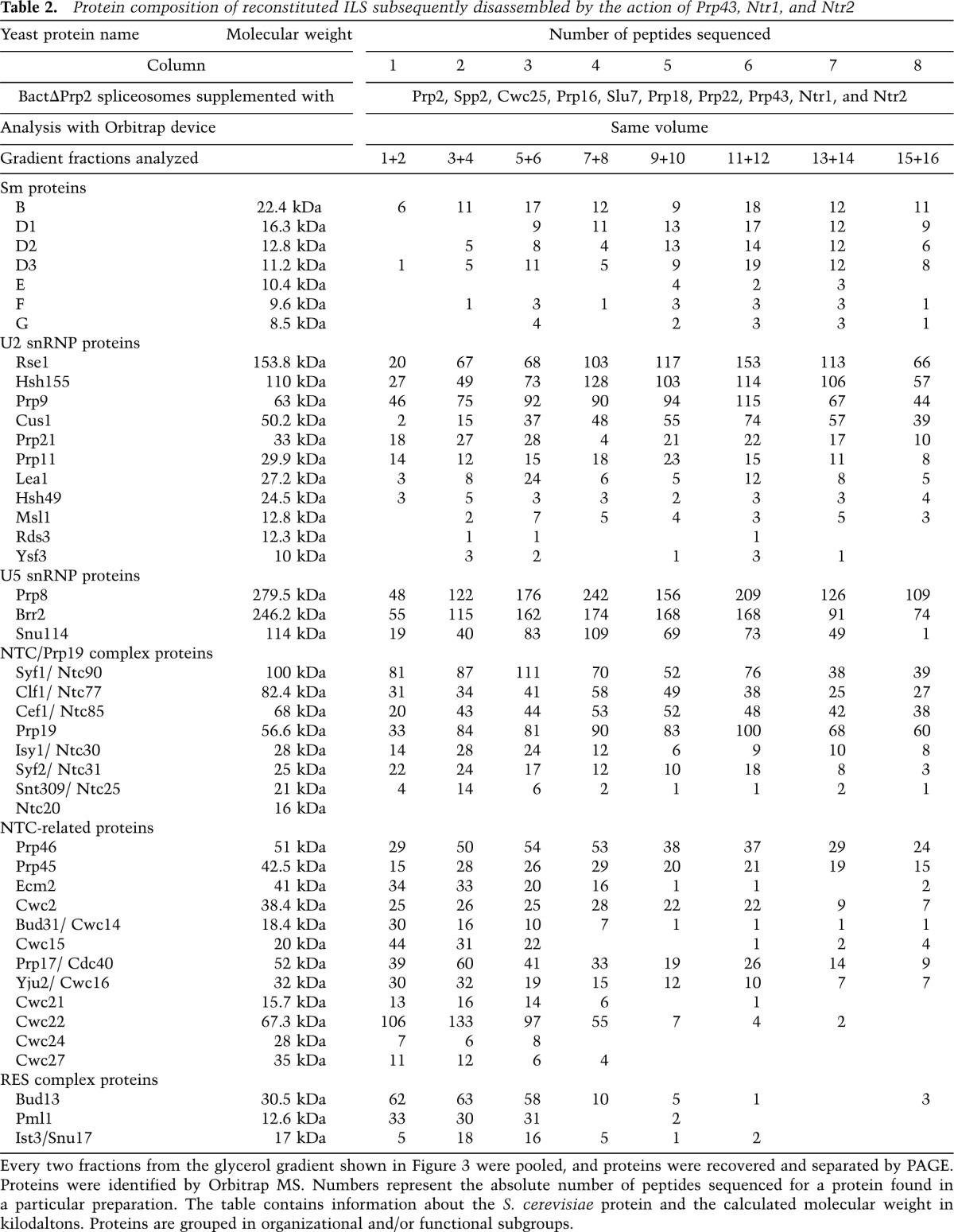
Upon disassembly of the ILS, NTC proteins remain predominantly associated with the U2 snRNP and the IL RNA
Of particular interest was the association behavior of the NTC and NTC-related proteins after ILS dissociation. Prp19, the scaffold protein of the NTC, was found in high abundance (based on peptide numbers) over a wide range of the gradient (Table 2). All other core NTC proteins—except for Ntc20, for which no peptides were found—were also widely distributed (Table 2). Interestingly, the NTC-related proteins Prp45, Prp46, and Cwc2 also appeared to comigrate with the core NTC proteins in the gradient, while the majority of all of the other NTC-related proteins migrated predominantly at the top of the gradient (Table 2). The presence of significant amounts of NTC proteins in those fractions containing the U5 and U2 snRNPs suggested a possible association with these RNP particles. To test this, we performed immunoprecipitation experiments with antibodies against Prp19 and pooled fractions from a gradient on which the NTR-mediated dissociation products of the ILS had been fractionated (Fig. 4B). The anti-Prp19 antibodies coprecipitated significant amounts of the U2 snRNA with a peak observed in fractions 9–12 (Fig. 4B, lanes 9–11), where the majority of U2 snRNA is also found (Fig. 3B). U5 snRNPs were also precipitated, albeit in significantly lower amounts than U2 snRNPs but clearly above the background level observed in a NIS control. Interestingly, anti-Prp19 antibodies also precipitated significant quantities of IL RNA from the top fractions of the gradient (Fig. 4B, lanes 7–9). Similar results were also obtained with antibodies specific for another core NTC protein, Syf1 (Supplemental Fig. S3). In contrast, anti-Cwc2 antibodies coprecipitated only small amounts of U2, U5, and IL RNA but essentially no U6 snRNA (Fig. 4B). Cwc2 also comigrated with Prp19 and other core NTC proteins in the glycerol gradient (Table 2). However, it is only loosely associated with the NTC complex (Ohi and Gould 2002), which may explain the low precipitation efficiency of the U2 snRNP and IL RNA. Taken together, our immunoprecipitation results indicate that a part of the NTC complex remains associated with a fraction of the U2 and U5 snRNPs as well as the IL RNA after ILS dissociation by the NTR complex.
Prp43 activity is necessary and sufficient to dissociate the purified ILS
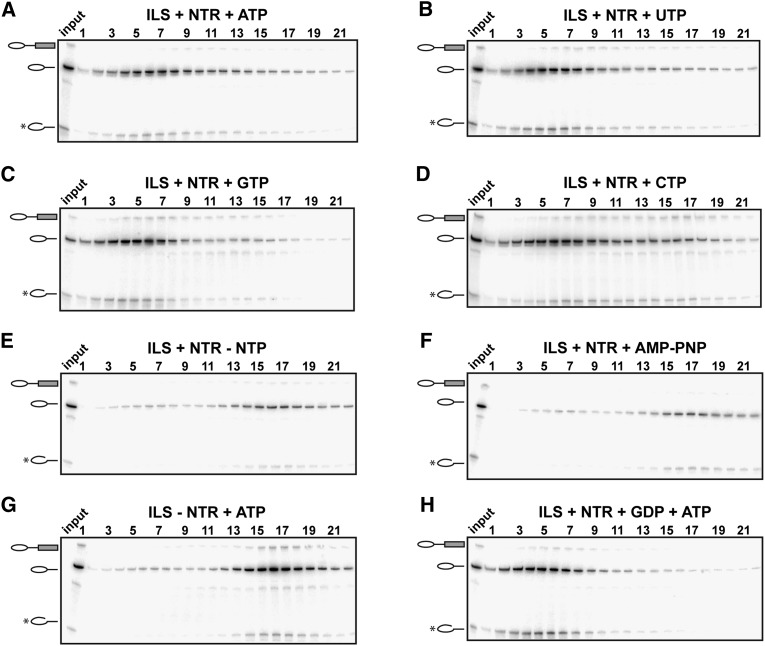
Recent data suggest that the RNA helicase Brr2 may also be involved in the dissociation of the yeast ILS (Small et al. 2006). We therefore set out to investigate whether Brr2 and Prp43 have different, perhaps sequential, targets in the ILS and, if so, whether potential “disassembly intermediates” of the ILS are detectable by inactivating one of the helicases. As Brr2 helicase activity is strictly ATP-dependent, while Prp43 is active in the presence of any rNTP, we incubated ILS (released from the matrix-bound post-catalytic spliceosome by Prp22) with Prp43 and Ntr1/Ntr2 plus UTP as the sole energy source (i.e., under conditions where Prp43 is active but not Brr2) (Fig. 5). Before addition of UTP, the ILS preparation was totally depleted of ATP by preincubation with hexokinase and glucose (see the Materials and methods). The reaction mixture was subsequently subjected to glycerol gradient centrifugation.
Figure 5.
The ILS was dissociated by NTR in the presence of any rNTP. Post-catalytic spliceosomes bound to the matrix were reconstituted in the presence of ATP and recombinant Prp2, Spp2, Cwc25, Prp16, Slu7, and Prp18. ILSs were eluted with Prp22 and ATP. Then, ATP was depleted by adding hexokinase and glucose to the ILS in solution before the addition of Prp43, Ntr1, and Ntr2 along with the indicated rNTP (A–D), without NTP (E), or with AMP-PNP (F); in further controls, ATP was present but without NTR proteins (G), or the NTR proteins were present along with a mixture of GDP and ATP (H). Each reaction was separated on a 10%–30% glycerol gradient. RNA from the gradient fractions shown was recovered, separated by denaturing PAGE, and visualized by autoradiography. The positions of the splicing intermediates and products are indicated on the left. Input lanes indicate the samples before sedimentation.
Surprisingly, in the presence of UTP, the ILS was disassembled as efficiently as it was with ATP as energy source (Fig. 5A,B; see also Figs. 1C, 5G for migration behavior of ILS before dissociation). Moreover, complete dissociation of the ILS was also observed in the presence of GTP or CTP (Fig. 5C,D). Northern blotting showed that the U2, U5, and U6 snRNPs were likewise released as distinct entities with equal efficiency regardless of the NTP used to dissociate the ILS (Supplemental Fig. S4). Incubation of ATP-depleted ILS in solution with the NTR complex in the absence of any rNTP or the presence of the nonhydrolysable ATP analog AMP-PNP failed to disrupt the ILS, as seen in Figure 5, E and F, and Supplemental Fig. S4. The weaker ILS signal in Figure 5, E and F, compared with Figure 5, A–D, was associated with a greater amount of aggregation of the ILS during the centrifugation (and thus more of the ILSs were pelleted) (Fig. 5, cf. also the input lanes where the signals are equal) when NTR was added in the absence of any NTP or in the presence of AMP-PNP (Fig. 5E,F). Moreover, the addition of ATP in the absence of NTR to ATP-depleted ILS did not promote its disassembly either (Fig. 5G).
Finally, we investigated whether Snu114 might inhibit the ATP-driven disassembly of the ILS in the presence of GDP, as reported previously (Small et al. 2006). However, disassembly of the purified ILS was not affected by the presence of GDP (Fig. 5H). To rule out the possibility that our NTP preparations were contaminated with ATP, we tested the unwinding of base-paired U4/U6 duplexes by recombinant yeast Brr2 in the presence of the various rNTPs. As shown in Supplemental Figure S5, U4/U6 snRNA is only unwound by purified Brr2 in the presence of ATP and not with UTP, CTP, or GTP. This documents the purity of the NTP preparations and confirms the high ATP specificity of Brr2 helicase activity.
As a further control experiment, we performed the entire splicing reaction with reconstituted components, including the catalytic activation of the isolated BactΔPrp2 complex by Prp2 and the two catalytic steps, in the presence of UTP as the sole energy source. In addition, in this experiment, we assembled the spliceosome on wild-type actin pre-mRNA. Even under these conditions, the ILS was completely dissociated (Supplemental Fig. S6). Moreover, MS analysis of disassembled ILS in the presence of ATP or UTP revealed the same distribution of proteins after glycerol gradient centrifugation and the same protein composition (data not shown). Taken together, these results show that in our purified splicing system, Brr2 helicase activity is not required. Instead, Prp43 activity is both necessary and sufficient for the complete dissociation of the ILS into its subunits.
Discussion
In this study, we investigated the active dissociation of the yeast spliceosome in a purified and well-defined in vitro system. As a starting point, post-catalytic spliceosomes were generated by complementation of isolated, activated BactΔPrp2 spliceosomes with recombinant step 1 and step 2 factors. The subsequent addition of Prp22 and ATP led to almost quantitative dissociation of the ILS from the mRNA (Supplemental Fig. S7). Furthermore, incubation of the isolated ILS with the NTR complex and ATP led to its quantitative disassembly into the IL RNA and individual U2, U5, and U6 snRNPs. Thus, our highly purified system faithfully recapitulates the dissociation stages of pre-mRNA splicing that are required to release spliceosome components for subsequent rounds of splicing. Using this system, we were able to (1) investigate in detail the factor requirements for the disassembly of the post-catalytic spliceosome by Prp22, (2) obtain insight into the protein dynamics of the remodeling steps that are induced by Prp22 or Prp43, and (3) characterize for the first time the protein composition of the isolated ILS and its dissociation products. We also demonstrate that Brr2 helicase activity is not strictly required for disassembly of the ILS but that this can be achieved by Prp43 activity alone.
Prp22 efficiently releases mRNA from the post-catalytic spliceosome in the absence of Slu7 and Prp18
The step 2 factors Slu7 and Prp18 have various functions in the second catalytic step of splicing—first and foremost, to help to bring the 3′ SS into the second-step catalytic center (Horowitz 2012). After the second catalytic step, Prp22 interacts with the newly ligated mRNA downstream from the splice junction (Schwer 2008). In a process driven by ATP hydrolysis, Prp22 then moves along the mRNA in a 3′-to-5′ direction, displaces Slu7 and Prp18 from the spliceosome, and disrupts the binding between the U5 snRNP and the mRNA, which leads to separation of the mRNA from the ILS (Schwer 2008; Horowitz 2012). To our knowledge, it has not been determined whether (1) Slu7 and Prp18 are required for the stable interaction of the newly ligated mRNA with the U5 snRNP in the post-catalytic complex or (2) the ejection of Slu7 and Prp18 from the spliceosome by Prp22 actually leads to the disruption of the U5 snRNP–mRNA interaction or is a result of it. In our studies, we assembled spliceosomes on the Act7 pre-mRNA, which has a short distance between the BS and the 3′ SS consensus sequences. Because, with this pre-mRNA substrate, the second catalytic step can be accomplished in our purified system with reasonable efficiency in the absence of added Slu7, Prp18, and Prp22 and the purified Bact spliceosomes used in these studies completely lacked these factors (data not shown), we were able to address the questions raised above. Indeed, we show that Slu7 and Prp18 are not required for stable retention of the newly spliced mRNA in the post-catalytic spliceosome. Moreover, Prp22 also efficiently disrupts the U5 snRNP–mRNA interaction in the absence of Slu7 and Prp18. Thus, the Prp22-mediated displacement of the mRNA from the ILS must be due primarily to disruption of the U5 snRNP–mRNA interaction and not to loss of Slu7 and Prp18 from spliceosomes. Finally, these results also show that Slu7 and Prp18 are not required for either the efficient recruitment of Prp22 to its target site in the post-catalytic spliceosome or Prp22′s catalytic activity at this stage.
Only a handful of proteins are displaced from the spliceosome upon Prp22 action
RNA helicase-mediated spliceosome remodeling steps occurring during catalytic activation and splicing catalysis involve the release of proteins from the spliceosome. For example, we showed recently that catalytic activation mediated by the RNA helicase Prp2 is accompanied by the almost quantitative dissociation of Cwc24 and Cwc27 from the spliceosome (Ohrt et al. 2012). Comparison of the protein composition of post-catalytic spliceosomes and the isolated ILS revealed that only a handful of proteins are displaced upon Prp22 action (Supplemental Fig. S7). Although the RES proteins are abundant in post-catalytic spliceosomes, their affinity for the ILS is severely reduced (Table 1). In addition to stimulating the splicing process, the RES proteins play an important part in retaining spliceosomes in the cell nucleus (Dziembowski et al. 2004). Their complete dissociation from the spliceosome after Prp22-mediated dissociation of the ILS from the spliced mRNA is consistent with such a role, as the released mRNA has to be exported from the nucleus to the cytoplasm, and the ILS in the nucleus is subsequently dissociated into its subunits by the action of the NTR complex.
A similarly drastic loss of affinity for the ILS was observed for Cwc22, which is abundant in the post-catalytic spliceosome, but only a minor fraction of which remains associated with the ILS (Table 1; Supplemental Fig. S7). Cwc22 is an integral protein of the Bact complex and is required for the catalytic activation of the spliceosome (Fabrizio et al. 2009; Yeh et al. 2011). As Cwc22 remains stably associated with the spliceosome during its catalytic phases and only leaves upon displacement of the mRNA, it could conceivably play a part in stabilizing the binding of the mRNA to the spliceosome. Recently, Cwc22 in the human spliceosome was shown to function as a binding platform for the exon junction complex (EJC) proteins (Barbosa et al. 2012; Steckelberg et al. 2012). As the EJC proteins are deposited in the post-catalytic spliceosome—or during its Prp22-mediated dissociation—upstream of the exon–exon junction, it seems likely that hCwc22 is located close to the exon junction in the spliceosome. It will thus be interesting to determine in the future whether Cwc22 contacts the newly spliced mRNA at the exon junction. Cwc21 is also displaced during disassembly of the post-catalytic spliceosome. It is not an essential protein but nonetheless interacts directly with Prp8 (Grainger et al. 2009), influencing the fidelity of the splicing reaction. It will thus be interesting to see whether it lies near the interaction interface between the mRNA and U5 snRNP in the post-catalytic spliceosome.
Apart from the proteins mentioned above, all other proteins of the post-catalytic spliceosome are retained in the ILS (Supplemental Fig. S7). These include the U5, the NTC, most of the NTC-related, and all U2 proteins. The latter is surprising, as it has been reported that the U2 SF3a/b proteins dissociate from the spliceosome already at the catalytic stage of splicing (Lardelli et al. 2010). It has been suggested that dissociation of U2 SF3a/b proteins might be necessary for catalysis (Lardelli et al. 2010). Our data clearly show that this is not the case. The apparent different association behavior of the U2 proteins observed here and in previous studies is likely due to differences in the stringency of the conditions used, as we showed recently in a detailed study (Ohrt et al. 2012).
Characterization of the ILS dissociation products generated by the NTR
Incubation of the isolated ILS with the NTR complex led to quantitative dissociation of not only the IL RNA from the U2, U5, and U6 snRNPs, but also the snRNPs from one another, as we show here by combining glycerol gradient centrifugation of the dissociation products with immunoprecipitation experiments and MS. This allowed us to characterize the biochemical composition of the released particles for the first time. Interestingly, a U2 snRNP with an S value of 20–25 containing all U2 proteins is released that exhibits an S value similar to that of endogenous U2 (Fig. 3C). Thus, under the conditions of our purified splicing system, the U2 proteins remain associated specifically with the U2 snRNA not only during the entire catalytic and post-catalytic phases of splicing, but also after the dissociation of the ILS (Supplemental Fig. S7). This also indicates that the presence of U2 proteins in the spliceosome does not impede the action of Prp22 and Prp43 in any way. Our results are thus consistent with the idea that an intact U2 snRNP is released from the ILS in vivo as well. It was reported previously that the U2 snRNP dissociated from the ILS by the NTR complex is likely missing some proteins or has an altered conformation (Tsai et al. 2005). The different association behavior of the U2 proteins observed here is probably due to differences in the stringencies of the experimental conditions used in these two studies.
In our purified system, U6 snRNA appears to leave the spliceosome as naked RNA. Cwc2—which in catalytically active spliceosomes can be cross-linked to the U6 snRNA (McGrail et al. 2009; Rasche et al. 2012)—also appears to no longer bind to the U6 snRNA after the dissociation of the ILS (Supplemental Fig. S7). However, it is likely that in the cell, the released U6 snRNA is bound immediately by Prp24 and the LSM2–8 proteins as a prerequisite for its integration into the U4/U6 di-snRNP particle (Bell et al. 2002; Rader and Guthrie 2002).
The majority of the U5 snRNP dissociates from the ILS as an ∼18S particle that contains the U5 proteins Prp8, Brr2, and Snu114 (Supplemental Fig. S7). In earlier work, we isolated a 35S U5 snRNP particle from a HeLa nuclear extract that contained— alongside hPrp8, hBrr2, and hSnu114—proteins of the human NTC complex and a number of NTC-related proteins (Makarov et al. 2002). It was suggested that this 35S U5 particle might be a disassembly intermediate of the human ILS. We have no evidence that an equivalent high-molecular-weight U5 particle is generated as a disassembly intermediate in our purified yeast splicing system. Even after short periods of incubation of the isolated ILS with the NTR complex, the main U5-containing product that we observed is invariably an 18S particle (data not shown). Nevertheless, in our immunoprecipitation experiments, we detected a small portion of U5 snRNPs that were associated with NTC proteins after dissociation of the ILS (Fig. 4). The association of NTC proteins with the human 35S U5 snRNP complex might be stabilized by 35S U5 proteins such as IBP160, MGC23918, and several peptidylprolyl isomerases that do not possess yeast homologs (Makarov et al. 2002).
Interestingly, a large proportion of the NTC proteins remain associated with both the U2 snRNP and the IL RNA, and a smaller proportion also remain associated with the U5 snRNP, as evidenced by the results of immunoprecipitation with antibodies against the NTC proteins Prp19 and Syf1 (Supplemental Fig. S7). These results further support the idea that the core proteins of the NTC complex remain associated with one another after disassembly of the ILS. At present, it is unclear to what extent this also applies to the other NTC proteins and some of the NTC-related proteins such as Prp45, Prp46, or Cwc2, all of which comigrate with Prp19 and Syf1 in glycerol gradients. How can we explain the fact that the same proteins are associated with both U2 and U5 snRNPs and also the IL RNA (albeit to different extents) even though the snRNPs and IL RNA no longer associate with one another? One possibility is that the NTC complex in the intact ILS engages in various protein–protein and protein–RNA contacts with the snRNPs and the IL RNA and that these hold the spliceosome together. Such a scaffolding function of the NTC was indicated by previous experiments showing that stable integration of the snRNPs in the activated spliceosome requires the NTC complex (Chan et al. 2003; Chan and Cheng 2005). After Prp43-mediated disassembly of the ILS, some of these contacts are retained, yielding a distribution of the NTC between the U2 and U5 snRNPs and IL RNA that may reflect its different binding affinities for these components.
The released snRNPs must be remodeled prior to their participation in subsequent rounds of splicing
Since each assembled spliceosome is a single-turnover enzyme, the snRNPs must be recycled after each round of splicing. Thus, the efficient disassembly of the ILS and recycling of its snRNP and protein components are essential for the assembly of new spliceosomes. Here we show for the first time the protein composition of the snRNPs dissociated from the ILS. As the U2 snRNP appears to remain intact during the splicing cycle, the main tasks remaining for the recycling of the U2 snRNP may be the reorganization of its snRNA and dissociation of associated NTC proteins. It is not clear whether the NTC interacts directly with one or more U2 proteins and/or a specific region of the U2 snRNA and whether it recognizes a specific conformation of the U2 snRNP/snRNA. It is likely that the conformation of the U2 snRNA must be remodeled by the helicases Sub2 and Prp5, which are involved in early steps of spliceosome assembly, and the action of these helicases could also lead to the removal of the NTC proteins from the U2 snRNP. The released U5 snRNP was shown here to contain only Prp8, Brr2, and Snu114 after dissociation and thus needs to reassociate minimally with the remaining three U5-specific proteins (i.e., Prp6, Lin1, and Dib1) to form a complete U5 snRNP (Stevens et al. 2001), which can then associate with the U4/U6 di-snRNP and additional proteins to reform the U4/U6.U5 tri-snRNP (Gottschalk et al. 1999). However, little is currently known about tri-snRNP-specific recycling factors. While Prp24 and the U6-associated Lsm2–8 protein complex indeed assist reassociation of U4 and U6 snRNAs after each round of splicing (Rader and Guthrie 2002), no recycling factors for di-snRNP-specific proteins have so far been characterized as well. Thus, several questions remain regarding the downstream steps required to reform fully functional snRNPs that participate in a new round of spliceosome assembly.
Prp43 helicase activity is sufficient to induce the dissociation of the ILS in vitro without evidence for the participation of Brr2's ATPase activity
The essential role of Prp43 in the disassembly of the ILS has long been established (Martin et al. 2002; Tsai et al. 2005). However, recent experiments in vitro indicated that the helicase activity of Brr2 also plays a role in this process (Small et al. 2006). Furthermore, the GTPase Snu114 is thought to down-regulate or up-regulate the Brr2 helicase activity, depending on whether its G-binding center carries a GDP or a GTP. An important rationale of the present work was to identify, by the selective use of ATP or UTP as the energy source, the respective roles of Brr2 and Prp43 in the ILS disassembly process. Our observation that the isolated ILS dissociates completely in the presence of any rNTP was therefore very surprising in that it indicated that the ATP-specific Brr2 helicase/ATPase is not required for the ILS disassembly. The recently determined X-ray crystal structure of Brr2 reveals a configuration of amino acid side chains at the nucleotide-binding pocket that exclusively allows binding of ATP (Santos et al. 2012). It can thus be ruled out that Brr2 loses its ATP specificity in the context of the spliceosome. One could imagine that trace quantities of ATP remain in the reaction mixture despite hexokinase and glucose treatment to remove any ATP. However, we can exclude the possibility that such minor amounts are sufficient to drive Brr2 helicase activity, as the Km value of Brr2 for ATP is relatively high (i.e., 0.3 ± 0.1 mM), and even an ATP concentration of 50 μM would not be sufficient to drive Brr2-mediated dissociation of the spliceosome. Finally, we were able to show that the entire catalytic phase (starting from purified and ATP-depleted Bact spliceosomes that were assembled on wild-type actin pre-mRNA) and the disassembly phase can take place efficiently with UTP as the sole exogenic energy source, confirming that ILS disassembly is catalyzed exclusively by Prp43 activity and that the ATPase activity of Brr2 is not needed (Supplemental Fig. S6). A further piece of evidence that Brr2 is not involved in this process in our system is that in the presence of ATP, where Brr2 could, in principle, be active, the loading of Snu114 with GDP (which should inhibit Brr2) does not lead to any inhibition of the disassembly of the ILS (Fig. 5H), in contrast to previous reports (Small et al. 2006).
At present, we have no explanation for the differences in factor requirements found in our study and that observed in the studies by Small et al. (2006). One possibility is that the partially purified spliceosome preparations used in those studies contained one or more additional factors that wholly or partly suppress the activity of Prp43 and thus made Brr2 helicase activity indispensable. However, biochemical and genetic data have so far not revealed the existence of any such factors. A methodological difference was that Small et al. (2006) carried out all of their disassembly assays in vitro at 4°C. We therefore repeated our own assays at low temperature but again found no requirement for Brr2 helicase activity (data not shown). A second difference from the result of Small et al. (2006) is the more rapid disassembly found by those investigators (cf. 10–20 sec and 10 min in our purified system). Importantly, the disassembly rate the we observed was independent of whether we used ATP (allowing Brr2 to act) or UTP (allowing only Prp43 to act), indicating that the difference in kinetics between the study by Small et al. (2006) and our study cannot be due to the lack of Brr2 action. The difference can plausibly be explained by the fact that Small et al. (2006) purified yeast spliceosomes with TAP-tagged Prp43; i.e., they used endogenous ILSs that were stalled immediately before disassembly. In contrast, in our study, we added recombinant NTR proteins to the purified ILS; i.e., under conditions where the recruitment of the NTR proteins to the ILS to form a productive disassembly complex might need more time.
We should emphasize that our experiments rule out only the direct participation of the Brr2 helicase in the disassembly of the isolated ILS and do not exclude the possibility that this protein may have some other helper function in spliceosome disassembly. For example, Tsai et al. (2007) have shown in a yeast two-hybrid assay that Ntr2 binds to Brr2, and, as Brr2 also binds to U5 snRNP and Prp43 binds to Ntr1, this would suggest a functional link among these components and the possibility that the formation of this complex would stimulate the helicase activity of Prp43 (Tanaka et al. 2007).
In our purified splicing system, the helicase activity of Prp43 is sufficient for not only releasing the IL RNA from the other components of the spliceosome, but also separating the snRNPs from one another. This suggests the existence of a cooperative disassembly process in which the initial actions of Prp43 initiate a cascade of events that leads to complete separation of several components. The target structures of Prp43 in the ILS are unknown at present. It is likely that in the ILS, the U2/U6 helices I and II are still present and that these are unwound by Prp43. Recently, a UV cross-linking study in yeast cells revealed that Prp43 can be cross-linked, among other species, to the U6 snRNA (Bohnsack et al. 2009). However, at present, we cannot exclude that Prp43 recognizes several target structures in the ILS and that the disassembly of the ILS takes place in several, successive remodeling steps. It is clear that the elucidation of the molecular details of the dissociation of the ILS will require the characterization of the structure of Prp43 and its target. We anticipate that our purified disassembly system will pave the way for these and similar studies in the future.
Materials and methods
Splicing reconstitution and disassembly assays
To obtain matrix-bound post-catalytic spliceosomes, typically, 2 pmol of BactΔPrp2 (5 nM) complexes bound to the amylose matrix were supplemented with 20 pmol (50 nM; an ∼10-fold molar excess) of recombinant proteins (Prp2, Spp2, Cwc25, Prp16, Slu7, and Prp18), and the reaction volume was adjusted to 400 μL with GK75 buffer. After incubating for 5 min on ice, 40 μL of 10× “rescue” solution (200 mM KPO4 at pH 7.4, 10 mM MgCl2, 20 mM ATP, 20% PEG 8000) was added to the reaction. After thorough mixing, the reaction was incubated for 45 min at 23°C. To purify post-catalytic spliceosomes, beads were subsequently washed three times with 10 column volumes of GK75 buffer, and the complexes were eluted with 400 μL of 12 mM maltose in GK75 buffer. The eluate was loaded onto a 3.8-mL linear 10%–30% (v/v) glycerol gradient containing GK75 buffer. Gradients were centrifuged at 60,000 rpm for 2 h in a Sorvall TH660 rotor, and 175-μL fractions were harvested from the top of the gradient and analyzed by Cherenkov counting. Peak fractions were then analyzed by MS. To obtain ILSs, post-catalytic spliceosomes bound to the matrix were reconstituted as described above. Then, 20 pmol of recombinant Prp22 protein was added, and the volume was adjusted to 400 μL with 1× “rescue” solution prepared in GK75 buffer. After thorough mixing, the reaction was incubated for 10 min at 23°C. The supernatant was collected, GK75 buffer was added to the matrix to a final volume of 400 μL and then gently mixed, and the supernatant was once again collected after centrifugation at 2000 rpm for 1 min. Four-hundred microliters of the pooled supernatants (containing the released ILS) was subjected to glycerol gradient centrifugation exactly as described above. For RNA analysis, each gradient fraction was subjected to Proteinase K digestion, and the RNAs were extracted by PCI, precipitated by ethanol, and then analyzed by 8% denaturing PAGE followed by autoradiography. To obtain sufficient amounts of the ILS for MS analysis, after Prp22 action, the KCl concentration of the sample was adjusted to 250 mM, and the supernatant was collected as described above. GK0 buffer containing 0 mM KCl was then added to the matrix to obtain the ILS remaining on the beads, and the supernatant was combined with the 250 mM supernatant such that the end concentration of KCl was 75 mM. These stages lasted only 4 min. The sample was then subjected to glycerol gradient centrifugation exactly as described above, and peak fractions were pooled and analyzed by MS. To obtain dissociated ILS, 20 pmol of recombinant Prp43, Ntr1, and Ntr2 was added to the ILS in solution, and the volume was adjusted to 400 μL with 1× “rescue” solution prepared in GK75 buffer. After thorough mixing, the reaction was incubated for 10 min at 23°C and then subjected to glycerol gradient centrifugation exactly as described above. Every two fractions were pooled and then analyzed by MS. For ATP depletion, purified ILSs were supplemented in solution with 2 mM glucose and 2 U of recombinant hexokinase and incubated for 15 min at 23°C.
Acknowledgments
We thank T. Conrad for excellent technical assistance, M. Raabe and U. Plessmann for help with the MS analysis, S.M. Jovin for help with the experiment shown in Supplemental Figure S5, and S.C. Cheng (Academia Sinica) and T. Rocha de Moura for providing antibodies. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 860 to R. L.) and a fellowship from the GGNB (DFG Grant GSC 226/1 to H. C.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.207779.112.
References
- Arenas JE, Abelson JN 1997. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci 94: 11798–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa I, Haque N, Fiorini F, Barrandon C, Tomasetto C, Blanchette M, Le Hir H 2012. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat Struct Mol Biol 19: 983–990 [DOI] [PubMed] [Google Scholar]
- Bell M, Schreiner S, Damianov A, Reddy R, Bindereif A 2002. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J 21: 2724–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D 2009. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell 36: 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon KL, Auchynnikava T, Edwalds-Gilbert G, Barrass JD, Droop AP, Dez C, Beggs JD 2006. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol Cell Biol 26: 6016–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brys A, Schwer B 1996. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA 2: 707–717 [PMC free article] [PubMed] [Google Scholar]
- Chan SP, Cheng SC 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem 280: 31190–31199 [DOI] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282 [DOI] [PubMed] [Google Scholar]
- Chiu YF, Liu YC, Chiang TW, Yeh TC, Tseng CK, Wu NY, Cheng SC 2009. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol Cell Biol 29: 5671–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M, Arenas J, Abelson J 1991. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349: 487–493 [DOI] [PubMed] [Google Scholar]
- Cordin O, Hahn D, Beggs JD 2012. Structure, function and regulation of spliceosomal RNA helicases. Curr Opin Cell Biol 24: 431–438 [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B 2004. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J 23: 4847–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 36: 593–608 [DOI] [PubMed] [Google Scholar]
- Gottschalk A, Neubauer G, Banroques J, Mann M, Lührmann R, Fabrizio P 1999. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J 18: 4535–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger RJ, Barrass JD, Jacquier A, Rain JC, Beggs JD 2009. Physical and genetic interactions of yeast Cwc21p, an ortholog of human SRm300/SRRM2, suggest a role at the catalytic center of the spliceosome. RNA 15: 2161–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz DS 2012. The mechanism of the second step of pre-mRNA splicing. Wiley Interdiscip Rev RNA 3: 331–350 [DOI] [PubMed] [Google Scholar]
- Laggerbauer B, Achsel T, Lührmann R 1998. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci 95: 4188–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardelli RM, Thompson JX, Yates JR 3rd, Stevens SW 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208 [DOI] [PubMed] [Google Scholar]
- Martin A, Schneider S, Schwer B 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem 277: 17743–17750 [DOI] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Staley JP 2006. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol 13: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrail JC, Krause A, O'Keefe RT 2009. The RNA binding protein Cwc2 interacts directly with the U6 snRNA to link the nineteen complex to the spliceosome during pre-mRNA splicing. Nucleic Acids Res 37: 4205–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW. 1998. RNA-RNA interactions in nuclear pre-mRNA splicing. In RNA structure and function (ed. M Grundber-Manago and RW Simons), pp. 279–307. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Ohi MD, Gould KL 2002. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8: 798–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohrt T, Prior M, Dannenberg J, Odenwälder P, Dybkov O, Rasche N, Schmitzová J, Gregor I, Fabrizio P, Enderlein J, et al. 2012. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA 18: 1244–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Lynn B, Rymond BC 2006. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc Natl Acad Sci 103: 13700–13705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Paul S, Zhang L, Chen M, Durbin N, Harrison SM, Rymond BC 2009. Spp382p interacts with multiple yeast splicing factors, including possible regulators of Prp43 DExD/H-box protein function. Genetics 183: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader SD, Guthrie C 2002. A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA 8: 1378–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol 8: 847–855 [DOI] [PubMed] [Google Scholar]
- Rasche N, Dybkov O, Schmitzová J, Akyildiz B, Fabrizio P, Lührmann R 2012. Cwc2 and its human homologue RBM22 promote an active conformation of the spliceosome catalytic centre. EMBO J 31: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy J, Kim K, Maddock JR, Anthony JG, Woolford JL Jr 1995. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1: 375–390 [PMC free article] [PubMed] [Google Scholar]
- Santos KF, Jovin SM, Weber G, Pena V, Lührmann R, Wahl MC 2012. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci 109: 17418–17423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B 2008. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell 30: 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Gross CH 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J 17: 2086–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ 2004. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol Cell Biol 24: 10101–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small EC, Leggett SR, Winans AA, Staley JP 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell 23: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 3: 55–64 [DOI] [PubMed] [Google Scholar]
- Steckelberg AL, Boehm V, Gromadzka AM, Gehring NH 2012. CWC22 connects pre-mRNA splicing and exon junction complex assembly. Cell Rep 2: 454–461 [DOI] [PubMed] [Google Scholar]
- Stevens SW, Barta I, Ge HY, Moore RE, Young MK, Lee TD, Abelson J 2001. Biochemical and genetic analyses of the U5, U6, and U4/U6 × U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA 7: 1543–1553 [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Aronova A, Schwer B 2007. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev 21: 2312–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC 2005. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 19: 2991–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RT, Tseng CK, Lee PJ, Chen HC, Fu RH, Chang KJ, Yeh FL, Cheng SC 2007. Dynamic interactions of Ntr1–Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol Cell Biol 27: 8027–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 16: 1237–1243 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R 2011. Spliceosome structure and function. In RNA worlds (ed. RF Gesteland, et al.), pp. 181–203. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Yeh TC, Liu HL, Chung CS, Wu NY, Liu YC, Cheng SC 2011. Splicing factor Cwc22 is required for the function of Prp2 and for the spliceosome to escape from a futile pathway. Mol Cell Biol 31: 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]