How cells become insulin-resistant at the onset of type 2 diabetes is an important question. Endoplasmic reticulum stress has emerged as a factor, but how it affects insulin response has not been settled. In experiments involving a Drosophila model as well as human cells, Zhang et al. present evidence of novel phosphorylation sites on the FOXO transcription factor required for PERK-mediated activation of FOXO. They show that modulation of endogenous PERK activity can restore insulin responsiveness.
Keywords: diabetes, ER stress, unfolded protein response, PERK kinase, Drosophila
Abstract
Endoplasmic reticulum (ER) stress is emerging as a potential contributor to the onset of type 2 diabetes by making cells insulin-resistant. However, our understanding of the mechanisms by which ER stress affects insulin response remains fragmentary. Here we present evidence that the ER stress pathway acts via a conserved signaling mechanism involving the protein kinase PERK to modulate cellular insulin responsiveness. Insulin signaling via AKT reduces activity of FOXO transcription factors. In some cells, PERK can promote insulin responsiveness. However, we found that PERK also acts oppositely via phosphorylation of FOXO to promote FOXO activity. Inhibition of PERK improves cellular insulin responsiveness at the level of FOXO activity. We suggest that the protein kinase PERK may be a promising pharmacological target for ameliorating insulin resistance.
Newly synthesized proteins in the secretory pathway are folded, processed, and assembled in the endoplasmic reticulum (ER). Misfolded proteins are eliminated via the ER-associated degradation (ERAD) pathway to ensure that only correctly folded proteins exit the ER (Nakatsukasa and Brodsky 2008). When substrates exhaust the regulatory capacity of the ERAD pathway, misfolded proteins accumulate, leading to a stress response that halts protein translation and increases the production of molecular chaperones involved in protein folding (Wu and Kaufman 2006). ER stress signaling is mediated through three parallel pathways, including the protein kinases Ire1 and PERK and the transcription factor ATF6 (Malhi and Kaufman 2011). PERK is best known for its ability to inhibit protein biosynthesis through phosphorylation of eIF2α. Whether it has additional regulatory roles in the ER stress pathway remains to be addressed.
ER stress is associated with metabolic disturbance and obesity. ER stress is activated under conditions of cellular nutrient overload, including lipotoxicity (Malhi and Gores 2008). Elevated ER stress is known to cause insulin resistance. However, the molecular pathways through which ER stress impairs cellular insulin response remain incompletely understood. Regulation of the nuclear localization of the Forkhead transcription factor FOXO is a key output of insulin signaling (for review, see Huang and Tindall 2007). Insulin signaling via phosphatidylinositol-3-kinase (PI3K) leads to elevated AKT activity. AKT-mediated phosphorylation of FOXO creates binding sites for 14-3-3 proteins, which promotes cytoplasmic localization of FOXO, thereby lowering its activity in the nucleus. ER stress has been linked to this pathway via Ire1-mediated activation JNK signaling (Urano et al. 2000; Ozcan et al. 2004). JNK phosphorylates the insulin receptor substrate (IRS) proteins and limits activation of PI3K/AKT signaling in response to insulin. Lower AKT activity leads to elevated nuclear FOXO activity. Here we report a parallel mechanism through which the ER stress pathway acting via the protein kinase PERK overrides insulin-induced suppression of the FOXO activity.
Results
ER stress promotes FOXO activity independent of AKT
To identify new regulators of insulin signaling, we performed an RNAi screen for modifiers of FOXO activity (Zhang et al. 2011). Because insulin signaling via AKT leads to inactivation of FOXO family transcription factors, FOXO overexpression can challenge the regulatory capacity of the insulin pathway. This provides a sensitive assay for modulators of pathway activity (Junger et al. 2003; Hietakangas and Cohen 2007). Hrd3 (CG10221), an ERAD protein, was among the FOXO modifiers identified in the RNAi screen. Depletion of Hrd3 enhanced the FOXO overexpression phenotype in the Drosophila eye, further reducing eye size (P < 0.001) (Fig. 1A; quantification in Supplemental Fig. S1A). On its own, Hrd3 depletion had no effect on eye size (Fig. 1B). Efficacy of the RNAi transgene was verified by PCR (Supplemental Fig. S1B). Enhancement of the FOXO overexpression phenotype was also observed when one copy of the endogenous Hrd3 gene was removed, providing a genetically independent confirmation of the interaction (Supplemental Fig. S1C).
Figure 1.
ER stress promotes nuclear localization of FOXO. (A) Photomicrographs of Drosophila adult eyes expressing UAS-FOXO under GMR-Gal4 control. +Hrd3 indicates coexpression of a UAS-Hrd3RNAi transgene together with UAS-FOXO. Eye size was reduced, on average, by 30%, indicating potentiation of the effects of FOXO (n = 9 eyes of each genotype; P < 0.001). Quantification is in Supplemental Figure S1A. (B) Adult eyes expressing GMR-Gal4 alone or together with UAS-Hrd3RNAi. Eye size was unchanged. Histogram: average eye area ± SD; n = 7. (C) S2 cells were treated to deplete Hrd3 transcript by RNAi or with control dsRNA. mRNA levels were measured by quantitative real-time RT–PCR. (Right) RNAi reduced Hrd3 transcript by ∼80%. Bip transcript increased; (**) P < 0.01, Student's t-test. FOXO and Ire1 transcripts were unaffected. Data represent the average of three independent experiments ± SD. (D) Quantification of subcellular localization of a FOXO-GFP transgene expressed in Drosophila S2 cells. Cells were grown in serum-free medium and deprived of insulin prior to treatment. (Left panel) No added insulin. (Second panel) Insulin-treated (10 μg/mL, 30 min). (Third and fourth panels) Insulin-treated 4 d after treatment with dsRNA to deplete Hrd3 or PI3K, as a positive control. (N+CN) Cells with predominantly nuclear localization plus cells with comparable levels in the nucleus and cytoplasm; (C) predominantly cytoplasmic, as described (Zhang et al. 2011). A Fisher's exact test was used to assess the difference in cytoplasmic FOXO comparing insulin-stimulated cells with and without Hrd3 depletion; (***) P < 0.001. (E) Immunoblots of S2 cells treated with insulin and dsRNAs, as indicated. (Top panel) Antibody to S505 phosphorylated Drosophila AKT. (Bottom panel) Antibody to total Drosophila AKT protein. Samples in lanes 1–4 were run on the same gel; intervening lanes were removed between the two sets. (F) Immunoblot to monitor the interaction between FOXO-GFP and 14-3-3ɛ-HA. S2 cells were transfected to express the proteins and treated with insulin with or without tunicamycin to induce ER stress. GFP-tagged FOXO was recovered using GFP trap beads, and blots were probed with anti-GFP and anti-HA to visualize bound 14-3-3.
Hrd3 is a component of the ERAD complex (Carvalho et al. 2006; Smith et al. 2011). Dysfunction of the ERAD complex activates the ER stress response and leads to elevated expression of the ER chaperone BiP (Baumeister et al. 2005). BiP mRNA levels increased in Hrd3-depleted cells (Fig. 1C), confirming that the cells were under ER stress. Hrd3 depletion was then used to probe FOXO activity in S2 cells. FOXO activity is regulated at multiple levels, including nuclear localization (Huang and Tindall 2007). FOXO was predominantly nuclear in cells devoid of growth factors, but upon insulin stimulation, FOXO shifted toward the cytoplasm (Fig. 1D). Depletion of Hrd3 by RNAi limited the insulin-induced increase in cytoplasmic FOXO (P < 0.001) (Fig. 1D). The effect of Hrd3 depletion was comparable with that of inhibiting insulin signaling by depleting PI3K (Fig. 1D). Induction of ER stress using tunicamycin mimicked the effect of Hrd3 depletion and limited the insulin-induced shift of FOXO into the cytoplasm (Supplemental Fig. S1D). Tunicamycin treatment also enhanced the FOXO overexpression phenotype in vivo (P < 0.01) (Supplemental Fig. S1E). These findings suggest that ER stress can increase FOXO activity by promoting nuclear localization of FOXO.
To ask whether the effect of ER stress on FOXO was mediated by regulation of AKT activity, we examined insulin-induced phosphorylation of AKT in Hrd3-depleted S2 cells. No reduction in the level of AKT phosphorylation was observed, compared with control insulin-stimulated cells (Fig. 1E). Furthermore, tunicamycin-induced ER stress did not reduce the amount of FOXO bound to 14-3-3ɛ (Fig. 1F). Thus, in S2 cells, ER stress appears to act on FOXO localization by a mechanism independent of AKT. These findings raised the possibility that ER stress might be able to override the regulation of FOXO activity by insulin signaling.
ER stress acts via PERK to regulate FOXO activity
The protein kinases Ire1 and PERK are activated upon ER stress. Activation of ER stress by depletion of Hrd3 led to an increase in Ire1 levels (Supplemental Fig. S2A). However, we did not observe any effect of Ire1 overexpression on FOXO nuclear export (Supplemental Fig. S2B). In an RNAi screen, depletion of Ire1 was reported to modestly reduce FOXO abundance but not to affect nuclear localization (Mattila et al. 2008). We confirmed that depletion of Ire1 by >50% did not affect insulin-induced relocalization of FOXO to the cytoplasm or FOXO mislocalization caused by depletion of Hrd3 (Supplemental Fig. S2C–E). Furthermore, depleting Ire1 by RNAi or removing one copy of the Ire1 gene did not modify the FOXO overexpression phenotype in vivo (Supplemental Fig. S2F). Thus, it appears unlikely that the effects of ER stress on FOXO are mediated by Ire1 in Drosophila. However, feedback from Ire acting via JNK on IRS might affect FOXO activity in other cellular contexts (Ozcan et al. 2004).
In contrast, depletion of PERK by two independent RNAi lines counteracted the effects of FOXO overexpression, leading to an increase in eye size (Fig. 2A; Supplemental Fig. S3A). PERK depletion had no effect on its own (Fig. 2A). The RNAi transgene gave >80% depletion of the perk transcript (Supplemental Fig. S3B). In S2 cells, PERK depletion prevented the effect of ER stress on FOXO localization (Fig. 2B). Again, PERK depletion had no effect on its own. Next, we made use of the larval fat body to examine the effects of ER stress on endogenous FOXO protein, visualized by antibody labeling. ER stress caused by Hrd3 depletion induced nuclear accumulation of FOXO, and this was prevented by simultaneous depletion of PERK (Fig. 2C). Conversely, PERK overexpression in vivo strongly elevated the mRNA level of 4E-BP (Fig. 2D), a well-established transcriptional target of FOXO. This increase was blunted by reducing FOXO activity (Fig. 2D). These observations suggest that PERK contributes significantly to mediating the effects of ER stress on FOXO activity.
Figure 2.
ER stress acts via PERK to regulate FOXO activity. (A) Adult eyes from animals expressing GMR-Gal4 alone or with UAS-FOXO. (Right panels) Coexpressed UAS-PERKRNAi. Histogram: average eye area ± SD; n ≥ 7. Depletion of PERK offsets the effects of FOXO overexpression, increasing eye area; (**) P < 0.01, Student's t-test. (B) Quantification of subcellular localization of FOXO-GFP in insulin-stimulated S2 cells. Cells were treated with dsRNA to deplete PERK, Hrd3, or both, as indicated. PERK depletion had no effect but counteracted the effect of Hrd3 depletion. A Fisher's exact test was used to assess the difference in cytoplasmic FOXO comparing Hrd3 depletion and codepletion of Hrd3 and PERK; (***) P < 0.001. (C) Immunolabeling of endogenous FOXO protein in larval fat body cells. Anti-FOXO is shown in green. Nuclei were labeled with DAPI (blue). Fat body cells are polyploid. ppl-Gal4 was used to drive UAS-RNAi transgenes for Hrd3 and PERK in fat body cells, as indicated. Hrd3 depletion increased nuclear FOXO (center), compared with the ppl-Gal4 control (left). Simultaneous depletion of PERK reduced nuclear FOXO accumulation. The ratio of nuclear to cytoplasmic FOXO label was quantified using DAPI to define the nucleus. (Note that the larger salivary gland cells at the bottom do not express the Gal4 driver.) Ratios are shown in the histogram at right. (***) P < 0.001. (D) 4E-BP mRNA level measured by quantitative PCR. (Left) RNA from third instar wing discs expressing EP-PERK under Nubbin-Gal4 control. PERK transcript increased 14-fold. (Right) FOXO activity was reduced using the dominant-negative alleles FOXO21/FOXO25. Data represent the average ± SD for three independent experiments; (*) P < 0.05.
GCN2 is a protein kinase related to PERK. GCN2 and PERK have both been shown to mediate the effects of ER stress by phosphorylation of eIF2α in human cells (Hamanaka et al. 2005). However, unlike PERK, depletion of GCN2 did not potentiate the effects of FOXO overexpression in Drosophila.
PERK-mediated FOXO phosphorylation induced by ER stress
Drosophila FOXO-GFP was purified from ER-stressed cells and analyzed by mass spectrometry for phosphorylated peptides (Supplemental Table S1). To study the contribution of these potential phosphorylation sites in mediating the effects of ER stress on FOXO activity, we prepared serine-to-alanine substitution mutants for several of the candidate sites (Supplemental Fig. S4; Supplemental Table S1). None of the mutants tested compromised the ability of insulin stimulation to induce cytoplasmic localization of FOXO. Nuclear localization of the FOXO S243A mutant was comparable with that of unmodified FOXO in response to insulin (Fig. 3A). However, S243A FOXO was refractory to ER stress-induced nuclear localization compared with the control protein (Fig. 3B). Flies expressing FOXO S243A in the eye were insensitive to the effect of Hrd3 depletion, compared with flies expressing intact FOXO (Fig. 3C). To further assess the role of S243 in mediating the effects of PERK on FOXO activity, we coexpressed the two forms of FOXO with PERK in vivo. PERK overexpression had much less effect on eye size when coexpressed with FOXO S243A than with intact FOXO (Fig. 3D). Mutations at the other five sites tested were less effective in reducing the effects of ER stress on FOXO localization or activity (Supplemental Fig. S4). Taken together, these findings suggest that S243 is a key site through which PERK regulates FOXO localization.
Figure 3.
Regulation of FOXO activity by PERK-dependent phosphorylation. (A) Localization FOXO-GFP in S2 cells in response to insulin. (Left) Insulin stimulated with intact FOXO-GFP. (Right) S243A-FOXO-GFP. Data represent the average ± SD for three independent experiments. (B) Localization of S243A mutant FOXO-GFP in response to ER stress. (Left) Control with intact FOXO-GFP. (Middle) Hrd3 depletion shifted FOXO toward the nucleus. (Right) S243A FOXO was refractory to Hrd3 depletion. Data represent the average ± SD for three independent experiments. (***) P < 0.001 comparing C between FOXO and S243A FOXO in Hrd3-depleted cells (Fisher's exact test). (C) Effects of ER stress on eye size in animals expressing UAS-FOXO versus UAS-FOXO-S243A with GMR-Gal4. (Right) Eyes coexpressing UAS-Hrd3RNAi to induce ER stress. Histogram: average area ± SD; n ≥ 9 eyes. Depletion of Hrd3 potentiated the effect of FOXO ([***] P < 0.001) but did not have a significant effect on S243A (NS). The difference between the two +Hrd3 samples was significant (P < 0.001). (D) Effects of PERK overexpression on eye size in animals expressing UAS-FOXO versus UAS-FOXO-S243A with GMR-Gal4. (Left) PERK overexpression reduced the size of eyes expressing UAS-FOXO more than those expressing UAS-FOXO-S243A. Histogram: average area ± SD, n = 10 eyes; (***) P < 0.001. (Right) Immunoblots showing FOXO-GFP protein visualized with anti-GFP. Anti-kinesin was used to monitor loading.
Conservation of PERK/FOXO regulation in human cells
To investigate whether PERK regulates FOXO in human cells, we made use of a luciferase reporter containing the human IRS-2 promoter as a readout of FOXO1 activity (Puig and Tjian 2005). H1299 cells lack endogenous FOXO1 but express FOXO3 (Zhao et al. 2010). Expression of FOXO1 in H1299 cells increased reporter expression. Expression of human PERK alone had little effect, but PERK potentiated the effect of FOXO1 (Fig. 4A). Comparable results were obtained in H1299 cells transfected to express PERK, FOXO3, and a luciferase reporter containing four synthetic FOXO3 sites (4FRE) (Teleman et al. 2008) and in MCF7 cells transfected to express PERK with FOXO1 or FOXO3 (Supplemental Fig. S5A–C).
Figure 4.

Human PERK regulates FOXO activity. (A,B) Luciferase reporter assays for human FOXO1 activity. H1299 cells were transfected to express a FOXO1-responsive luciferase reporter derived from the IRS-2 gene. Cells were cotransfected to express intact human FOXO1 or S298A mutant FOXO1 and PERK as indicated. Data represent the average ± SD for three or more independent experiments. (A) PERK potentiated the effects of FOXO1; (**) P < 0.01, Student's t-test comparing FOXO1 with and without PERK. (B) S298A mutant FOXO1 was refractory to the potentiating effects of PERK; (***) P < 0.001 comparing intact and S298A FOXO1 with PERK, Student's t-test.
To identify potential PERK phosphorylation sites on human FOXO1, we performed an in vitro kinase assay using purified FOXO1 and PERK proteins, followed by mass spectrometry. Five phospho-sites were detected (Supplemental Table S2). Mutation of FOXO1 S261 to alanine had no effect on PERK-induced FOXO1 activity (Supplemental Fig. S5D). The other four sites—S298, S301, S303, and S311—were clustered in a single peptide that corresponds to the region of Drosophila FOXO containing S243.
To assess the contribution of ER stress to phosphorylation of these sites, we performed SILAC (stable isotope labeling by amino acids in cell culture) mass spectrometry on samples of human FOXO1 immunopurified from control MDA-MB231 cells and cells treated with tunicamycin. Six peptides showed increased phosphorylation on serine or threonine residues in the ER-stressed samples (Supplemental Tables S3, S4). Phosphorylation of the peptide containing S298, S301, S303, and S311 was enriched 1.5-fold in FOXO1 from ER-stressed cells. Phosphorylation was observed on S298 alone or on S298 in combination with either S301 or S303. These sites were also phosphorylated by PERK in vitro. The corresponding peptide was also phosphorylated in FOXO1 and FOXO3 in ER-stressed H1299 cells (Supplemental Table S5). We prepared alanine substitution mutants for S298, S301, and S303 to assess the contribution of these sites to mediating the effects of PERK on FOXO1. The S298A mutation reduced the responsiveness of FOXO1 to PERK (Fig. 4B), but mutation of S301 or S303 had little or no effect (Supplemental Fig. S5D). The observation that S298A accounts for approximately half of the effect of PERK on FOXO1 suggests that other phospho-sites also contribute to mediating the effects of ER stress on FOXO1 in vivo.
Previous reports have identified other phosphorylation sites through which FOXO activity can be regulated. Phosphorylation of FOXO1 on S249 by CDK1 has been reported to reduce interaction with 14-3-3, thereby increasing FOXO activity (Yuan et al. 2008). However, CDK2-mediated phosphorylation of FOXO1 at S249 was reported to increase cytoplasmic localization of FOXO and reduce its activity (Huang et al. 2006). Thus, the role of S249 remains unclear. MST1 has also been reported to phosphorylate FOXO1 and FOXO3 on several residues, among which phosphorylation of S207 in FOXO3 was found to reduce interaction with 14-3-3 (Lehtinen et al. 2006). The MST1 target site in FOXO1 is inside the conserved DNA-binding domain and corresponds to S147 in Drosophila FOXO. Mutation of Drosophila FOXO to S147A did not have a significant effect on nuclear accumulation in response to ER stress (Supplemental Fig. S4D).
Opposing activity of AKT and PERK on FOXO
Insulin signaling via PI3K activates AKT and increases phosphorylation of FOXO, creating 14-3-3-binding sites, which promotes cytoplasmic accumulation of FOXO (for review, see Huang and Tindall 2007). Consequently, FOXO activity is expected to be higher under conditions of low insulin pathway activity. Conversely, our findings suggest that PERK potentiates FOXO activity. S298 is not among the FOXO1 sites phosphorylated by AKT, which include T24, S256, and S319 (Greer and Brunet 2005). This raised the possibility that PERK-mediated phosphorylation of FOXO might be able to counteract the effects of AKT, in effect opposing the effects of the insulin pathway.
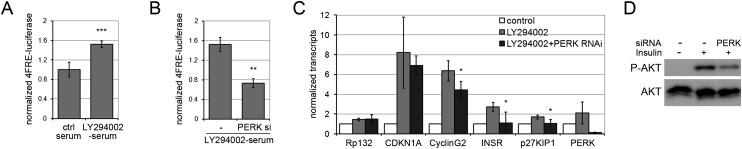
To test this possibility, we used a cellular assay to model insulin resistance and monitored the effects of PERK depletion on FOXO activity. H1299 cells were serum-starved to remove insulin and treated with the PI3K inhibitor LY294002 to further reduce AKT activity. Under these conditions, endogenous FOXO3 activity increased, as monitored using a FOXO3 luciferase reporter (Fig. 5A), while simultaneous depletion of PERK by siRNA treatment lowered FOXO3 activity (Fig. 5B). Next, we examined the effects of reduced PERK activity on endogenous FOXO1 and FOXO3 targets in HEPG2 liver cells (which express both FOXO1 and FOXO3). HEPG2 cells were deprived of serum and treated with the PI3K inhibitor to lower AKT activity. Several FOXO targets showed a consistent pattern of increased expression under these conditions (Medema et al. 2000; Nakae et al. 2003; Martinez-Gac et al. 2004; Puig and Tjian 2005), and this increase too was offset by depletion of PERK (Fig. 5C). Thus, endogenous PERK activity contributes to FOXO activation under conditions in which the inhibitory effects of AKT are removed. These observations support the idea that PERK acts in opposition to AKT in the regulation of FOXO activity.
Figure 5.
Depletion of human PERK limits FOXO activity. (A) Luciferase reporter assays for endogenous FOXO3 activity in H1299 cells. H1299 cells were transfected to express a FOXO3-responsive luciferase reporter containing four synthetic FOXO3-binding sites (4FRE). Cells were serum-starved to reduce insulin signaling and treated with the PI3K inhibitor LY294002 to further reduce AKT activity. Data represent the average ± SD for six independent experiments. (***) P < 0.001. (B) FOXO3 Luciferase assays as in A, except that cells were treated with siRNA to deplete endogenous PERK or were left untreated. Data represent the average ± SD for three independent experiments. (**) P < 0.01. (C) Normalized mRNA levels of FOXO1 targets measured by quantitative PCR. HEPG2 cells were serum-starved to reduce insulin signaling and treated with the PI3K inhibitor LY294002 to further reduce AKT activity. Four of the 14 FOXO targets tested showed a consistent increase in mRNA levels (gray bars) compared with control cells in normal medium (open bars). (Black bars) PERK siRNA treatment. (*) P < 0.05 with/without PERK siRNA treatment. Efficacy of PERK siRNA treatment is shown at the far right. (D) Immunoblot of HepG2 cells treated with insulin and with siRNA to deplete endogenous PERK. (Top panel) Probed with antibody to S473 phosphorylated human AKT. (Bottom panel) Probed with antibody to total AKT protein.
Feedback regulation linking the ER stress and insulin pathways
We noted that the insulin receptor (INSR) was among the FOXO1 targets up-regulated in HEPG2 cells under low AKT activity and that this was offset by depletion of PERK activity (Fig. 5C, INSR). This suggested that PERK, acting via FOXO1, could promote cellular insulin responsiveness by increasing INSR expression. If so, depletion of PERK should lower insulin signaling and AKT activity. We confirmed that this takes place in PERK-depleted HepG2 cells (Fig. 5D).
Discussion
Regulatory interplay between ER stress and the insulin pathway operates at multiple levels, suggesting that fine-tuning this balance is of some importance to cellular metabolism. In this study, we presented evidence that PERK can act directly on FOXO to increase FOXO activity. PERK can also act indirectly via AKT to lower FOXO activity. This relationship has the topology of an incoherent feed-forward motif (Alon 2007). Such motifs provide stability in the output signaling systems, allowing initial activation followed by inhibition to limit the response.
ER stress acts by at least three independent routes to control AKT activity. PERK acts via FOXO to up-regulate INSR levels, which increase AKT activity. PERK has recently been reported to promote AKT activity by acting as a lipid kinase via DAG (Bobrovnikova-Marjon et al. 2012). Regulatory feedback between ER stress and insulin signaling also operates at another level. ER stress has been shown to act via Ire1 and JNK to phosphorylate the IRS and thereby reduce insulin signaling and Akt activation (Ozcan et al. 2004). Thus, at least three distinct mechanisms link activity of the ER stress pathway to control of insulin pathway activity. We note that the SILAC data showed increased phosphorylation of three AKT sites (T24, S256, and S319) in addition to increased phosphorylation of the PERK site S298 in cells subjected to ER stress.
Additionally, AKT feeds back to limit PERK activity. Increased AKT activity has been reported to lower PERK activity (Mounir et al. 2011). We also noted that PERK mRNA levels were increased in cells with low AKT activity (Fig. 5C). Inhibitory feedback from AKT to PERK would reinforce the dampening effect of AKT on the output of the ER stress pathway and also provides a means for insulin or growth factor signaling via AKT to modulate the effects of ER stress mediated by PERK. The extent to which these feedback systems operate may depend on cellular and physiological context.
Genetic models of obesity, such as the ob/ob mouse, as well as diet-induced obesity have been found to induce ER stress (Ozcan et al. 2004). Obesity-induced ER stress, acting via PERK on FOXO, would enhance the effects of obesity-induced insulin resistance, in which Ire1 lowers insulin responsiveness. Obesity-induced ER stress should also synergize with other causes of insulin resistance to exacerbate the cellular metabolic imbalance. Therefore, regulation of FOXO activity emerges as a nodal point on which the effects of obesity on cellular insulin responsiveness converge. Chronic elevation of FOXO activity may contribute to disease progression. In this context, it is noteworthy that reducing FOXO1 activity by removing one copy of the FOXO1 gene has been shown to improve insulin sensitivity in genetic and diet-induced models of insulin resistance in mice (Nakae et al. 2002; Kim et al. 2009).
This study raises the possibility that pharmacological inhibition of PERK could provide a useful therapeutic approach for combating the effects of ER stress in progression to diabetes. Use of chemical chaperones to overcome ER stress has been reported to decrease insulin and glucose levels and improve glucose and insulin tolerance in insulin-resistant ob/ob mice (Ozcan et al. 2006). A similar finding has been reported using the chemical chaperone sodium phenylbutyrate in obese diabetic humans (Xiao et al. 2011). The ER stress pathway may provide a fruitful source of targets for new therapeutics to treat diabetes and metabolic syndrome.
Materials and methods
Fly genetics
Df(3R)Exel9013, Df(3R)BSC636, UAS-RNAi-PERK (BL), and EP-PERK flies were obtained from the Bloomington Stock Center. UAS-RNAi-Hrd3, UAS-RNAi-Ire1, UAS-RNAi-PERK, and UAS-RNAi-Ire1 were from the Vienna Drosophila RNAi Center.
S2 cell culture and treatments
S2 cells were grown at 25°C in SFM (Gibco) supplemented with L-glutamine. dsRNA was prepared using MegascriptT7 (Ambion) with the following templates: Hrd3 #1, nucleotides 1801–2200 of Hrd3 coding sequence (FBpp0083858); Hrd3 #2, nucleotides 2800–3138 of FBtr0084467. If not specified, Hrd3 RNAi refers to sequence #1; PI3K, nucleotides 358–857 of Pi3K92E coding sequence (FBpp0083348); PERK, nucleotides 1045–1506 of FBpp0078417; Ire1, nucleotides 391–883 of FBpp0112161; and GFP, nucleotides 17–633 of EGFP2 were used as control. S2 cells were treated with 37 nM dsRNA. Cells were transfected using Effectene (Qiagen) with pMT-FOXO-GFP, pMT-Gal4, and pUASP-14-3-3ɛ-HA (provided by Pernille Rørth). CuSO4 (0.7 mM) was used to induce FOXO 14-3-3ɛ and Ire1 expression.
Human cell culture and treatments
H1299, MCF7, and HepG2 cells were maintained at 37°C in DMEM supplemented with 10% FBS. Cells were transfected using FuGENE 6 or Lipofectamine 2000 with pcDNA-FOXO1 (Puig and Tjian 2005), pcDNA-FOXO3 (generated by Aurelio Teleman from Addgene1787), pCMV-PERK (SC117132 from OriGene), and pEGFP-N1. pEGFP-N1-FOXO3 was generated at XhoI and BamHI sites using pcDNA-FOXO3 as a template. PERK siRNA (Qiagen, SI02223718) was transfected using HiPerFect reagent. For SILAC mass spectrometry experiments, MDA-MB231cells were maintained in SILAC-DMEM (Thermo Fisher Scientific) supplemented with 10% dialyzed FBS (Thermo Fisher Scientific) and penicillin/streptomycin for at least six cell divisions in order to achieve uniform incorporation. Light medium was supplemented with 0.8 mM L-lysine:HCl and 0.4 mM L-arginine:HCl (Sigma). Heavy medium was supplemented with 0.8 mM L-lysine:2HCl (U-13C6 and U-15N2) and 0.4 mM L-arginine:HCl (U-13C6 and U-15N4) (Cambridge Isotope). Cells were transfected with pcDNA-FOXO1 using TurboFect to express FOXO1-V5 for 2 d. Cells in light medium were left untreated, and cells in heavy medium were treated with tunicamycin for 4 h. FOXO1-V5 was purified using Protein-G Sepharose beads (GE) and anti-V5 (Invitrogen) in RIPA buffer supplemented with protease inhibitor and phosphatase inhibitor (Roche).
Antibodies
Antibody to Drosophila FOXO was from Puig et al. (2003). Antibodies to phospho-S505-AKT (Drosophila), phospho-S473-AKT (human), AKT, and Myc were from Cell Signaling Technology. Anti-GFP was from Invitrogen (A6455). HA antibody was from Roche. Anti-kinesin was from Cytoskeleton. For immunoblotting, samples were homogenized in SDS sample buffer, boiled, and resolved by SDS-PAGE before transfer to nitrocellulose membranes for antibody labeling.
In vitro kinase assay
Purified PERK was purchased from Invitrogen (PV5106). Kinase assays were conducted according to the manufacturer's protocol in 1× kinase buffer (PV3189) for 1 h at 25°C. His-tagged FOXO1 was expressed in Escherichia coli and purified by nickel chelate chromatography.
Mass spectrometry
SDS-PAGE and in-gel digestion
Immunoprecipitated samples were run on an SDS-PAGE using a NuPAGE 4%–12% Bis-Tris gel (Invitrogen). The FOXO bands were excised followed by in-gel digestion (Shevchenko et al. 2006), with minor modifications. Gel pieces were washed with 50 μL of 50 mM ammonium bicarbonate and destained with 50 μL of 50% acetonitrile/25 mM ammonium bicarbonate for 10 min. Reduction was carried out by covering the gel pieces with 10 mM DTT for 30 min at 56°C; alkylation was carried out with 55 mM iodoacetamide for 15 min in the dark at room temperature. Fifty microliters of 50 mM ammonium bicarbonate was used for washing, and 50 μL of 100% acetonitrile was used for shrinking twice for 10 min. Thirty microliters of 13 ng/μL sequencing-grade trypsin (Promega) was added to each well for 30 min at 4°C before 25 mM ammonium bicarbonate was added to cover the gel pieces. Samples were incubated for 3 h at 37°C. Supernatants containing peptides were cleared by centrifugation. Twenty microliters of 5% formic acid was added to each well followed by 20 μL of 100% acetonitrile for peptide extraction. Both steps were repeated.
Liquid chromatography/mass spectrometry analysis
Vacuum-dried sample was reconstituted in 0.1% formic acid and analyzed using nanoHPLC coupled to an LTQ Orbitrap classic, LTQ Orbitrap XL, Q-Exactive, or LTQ Velos (Thermo Fisher Scientific). Peptides were trapped onto a C18 precolumn and separated on an analytical column using a 2- to 4-h gradient ranging from 2% to 40% acetonitrile/0.1% formic acid, followed by a 5-min gradient ranging from 40% to 80% acetonitrile/0.1% formic acid. For Q-Exactive, survey full-scan mass spectrometry spectra (m/z 310–2000) were acquired with a resolution of r = 70,000, an AGC target of 1 × 106, and a maximum injection time of 2 msec. The 10 most intense peptide ions in each survey scan with an intensity threshold of 40,000, underfill ratio of 1%, and charge state ≥2 were isolated sequentially with an isolation window of 2 Th to a target value of 20,000 and a maximum injection time of 50 msec and fragmented in the high-energy collision cell by high-energy collision dissociation using a normalized collision energy of 27%. The tandem mass spectrometry was acquired with a resolution of 17,500 and a starting mass of m/z 100. A dynamic exclusion was applied using an exclusion duration of 10 sec. For LTQ Orbitrap, survey full-scan mass spectrometry spectra (m/z 310–1400) were acquired with a resolution of r = 60,000 at m/z 400, an AGC target of 1 × 106, and a maximum injection time of 1000 msec. The 10 most intense peptide ions in each survey scan with an ion intensity of >2000 counts and a charge state ≥2 were isolated sequentially to a target value of 5000 and fragmented in the linear ion trap by collision-induced dissociation (CID) using a normalized collision energy of 35%. A dynamic exclusion was applied using a maximum exclusion list of 500 with one repeat count, repeat duration of 45 sec, and exclusion duration of 30 sec. For LTQ Velos, we used mass spectrometry scans ranging from 310 to 1400 m/z, AGC target 3 × 104, and maximum injection time of 10 msec. The 20 most intense ions with an ion intensity >1000 and a charge state excluding 1 were sequentially isolated to a maximum AGC target value of 1 × 104 for a maximum of 100 msec and fragmented by either CID using a normalized collision energy of 30% or electron transfer dissociation (ETD). For ETD, the following parameters were used: reagent ion source temp of 160°C, reagent ion source emission current of 50 μA, reagent ion source electron energy of −70.00 V, reagent ion source CI pressure of 20 psi, reagent vial 1 ion time of 50, reagent vial 1 AGC target of 1 × 105, reagent vial 2 ion time of 50, reagent vial 2 AGC target of 1 × 105, and supplemental activation energy of 15. For both ETD and CID, a dynamic exclusion list was applied using an exclusion list size of 500, one repeat count, repeat duration of 45 sec, and exclusion duration of 30 sec, as well as a mass width of 1.0 low and 1.5 high. Expiration count was disabled.
Data processing and database search
Raw file processing for unlabeled samples was carried out using Mascot Daemon (version 2.3.2, Matrix Science). A data import filter for precursor masses from 700 to 4000 Da with a minimum scans per group of 1 and a minimum peak count of 10 was used. Mascot search was performed using the UniProt Drosophila database (released on_2010_09, 34043 entries) or IPI Human database (ipi.HUMAN.v3.68.decoy.fasta or ipi.HUMAN.v3.86.decoy.fasta). For the SILAC-labeled samples, the raw files were processed using MaxQuant version 1.2.0.18 using the default parameters. An Andromeda search was carried out using the IPI Human database (ipi.HUMAN.v3.86.decoy.fasta). In both the Mascot and Andromeda searches, trypsin was specified as the enzyme, and there were two allowed missed cleavages. Carbamidomethyl (C) was set as a static modification, while the dynamic modifications were acetyl (protein N-term), oxidation (M), and phosphorylation (S/T/Y). The tolerance for the precursor masses was 7 parts per million (ppm), and the tolerance for fragments was 0.5 Da for samples analyzed on LTQ Orbitrap. Tolerance for the precursor masses was 2 Da, and the tolerance for fragments was 0.8 Da for samples analyzed on LTQ Velos. The tolerance for fragments was 20 ppm for samples analyzed on Q-Exactive.
Statistical analysis
Statistical analysis used the Fisher's exact test (for categorical data) and Student's t-test (for normally distributed data). All data are presented as average ± SD. P < 0.05 was considered significant.
Acknowledgments
We thank Claire Lee Foon Swa and Suat Peng Neo for technical assistance with mass spectrometry and for providing MDA-MB231 SILAC cells.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.201731.112.
References
- Alon U 2007. Network motifs: Theory and experimental approaches. Nat Rev Genet 8: 450–461 [DOI] [PubMed] [Google Scholar]
- Baumeister P, Luo S, Skarnes WC, Sui G, Seto E, Shi Y, Lee AS 2005. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: Activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol Cell Biol 25: 4529–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrovnikova-Marjon E, Pytel D, Riese MJ, Vaites LP, Singh N, Koretzky GA, Witze ES, Diehl JA 2012. PERK utilizes intrinsic lipid kinase activity to generate phosphatidic acid, mediate Akt activation, and promote adipocyte differentiation. Mol Cell Biol 32: 2268–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA 2006. Distinct ubiquitin–ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126: 361–373 [DOI] [PubMed] [Google Scholar]
- Greer EL, Brunet A 2005. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 24: 7410–7425 [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, Bennett BS, Cullinan SB, Diehl JA 2005. PERK and GCN2 contribute to eIF2α phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol Biol Cell 16: 5493–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Cohen SM 2007. Re-evaluating AKT regulation: Role of TOR complex 2 in tissue growth. Genes Dev 21: 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Tindall DJ 2007. Dynamic FoxO transcription factors. J Cell Sci 120: 2479–2487 [DOI] [PubMed] [Google Scholar]
- Huang H, Regan KM, Lou Z, Chen J, Tindall DJ 2006. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science 314: 294–297 [DOI] [PubMed] [Google Scholar]
- Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, Greenberg ME, Hafen E 2003. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Li P, Huntley J, Chang JP, Arden KC, Olefsky JM 2009. FoxO1 haploinsufficiency protects against high-fat diet-induced insulin resistance with enhanced peroxisome proliferator-activated receptor γ activation in adipose tissue. Diabetes 58: 1275–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, et al. 2006. A conserved MST–FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125: 987–1001 [DOI] [PubMed] [Google Scholar]
- Malhi H, Gores GJ 2008. Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Semin Liver Dis 28: 360–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi H, Kaufman RJ 2011. Endoplasmic reticulum stress in liver disease. J Hepatol 54: 795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gac L, Marques M, Garcia Z, Campanero MR, Carrera AC 2004. Control of cyclin G2 mRNA expression by forkhead transcription factors: Novel mechanism for cell cycle control by phosphoinositide 3-kinase and forkhead. Mol Cell Biol 24: 2181–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila J, Kallijarvi J, Puig O 2008. RNAi screening for kinases and phosphatases identifies FoxO regulators. Proc Natl Acad Sci 105: 14873–14878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema RH, Kops GJ, Bos JL, Burgering BM 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404: 782–787 [DOI] [PubMed] [Google Scholar]
- Mounir Z, Krishnamoorthy JL, Wang S, Papadopoulou B, Campbell S, Muller WJ, Hatzoglou M, Koromilas AE 2011. Akt determines cell fate through inhibition of the PERK-eIF2α phosphorylation pathway. Sci Signal 4: ra62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae J, Biggs WH III, Kitamura T, Cavenee WK, Wright CV, Arden KC, Accili D 2002. Regulation of insulin action and pancreatic β-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet 32: 245–253 [DOI] [PubMed] [Google Scholar]
- Nakae J, Kitamura T, Kitamura Y, Biggs WH III, Arden KC, Accili D 2003. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell 4: 119–129 [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K, Brodsky JL 2008. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9: 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS 2004. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306: 457–461 [DOI] [PubMed] [Google Scholar]
- Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, Hotamisligil GS 2006. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 313: 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Tjian R 2005. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev 19: 2435–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Marr MT, Ruhf ML, Tjian R 2003. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes Dev 17: 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS 2011. Road to ruin: Targeting proteins for degradation in the endoplasmic reticulum. Science 334: 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM 2008. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab 7: 21–32 [DOI] [PubMed] [Google Scholar]
- Urano F, Bertolotti A, Ron D 2000. IRE1 and efferent signaling from the endoplasmic reticulum. J Cell Sci 113: 3697–3702 [DOI] [PubMed] [Google Scholar]
- Wu J, Kaufman RJ 2006. From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ 13: 374–384 [DOI] [PubMed] [Google Scholar]
- Xiao C, Giacca A, Lewis GF 2011. Sodium phenylbutyrate, a drug with known capacity to reduce endoplasmic reticulum stress, partially alleviates lipid-induced insulin resistance and β-cell dysfunction in humans. Diabetes 60: 918–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Becker EB, Merlo P, Yamada T, DiBacco S, Konishi Y, Schaefer EM, Bonni A 2008. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science 319: 1665–1668 [DOI] [PubMed] [Google Scholar]
- Zhang W, Thompson BJ, Hietakangas V, Cohen SM 2011. MAPK/ERK signaling regulates insulin sensitivity to control glucose metabolism in Drosophila. PLoS Genet 7: e1002429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, Zhu WG 2010. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol 12: 665–675 [DOI] [PubMed] [Google Scholar]