Abstract
A major obstacle in the treatment of human immunodeficiency virus type 1 (HIV-1) is a sub-population of latently infected CD4+ T lymphocytes. The cellular and viral mechanisms regulating HIV-1 latency are not completely understood, and a promising technique for probing the regulation of HIV-1 latency is single-cell time-lapse microscopy. Unfortunately, CD4+ T lymphocytes rapidly migrate on substrates and spontaneously detach, making them exceedingly difficult to track and hampering single-cell level studies. To overcome these problems, we built microfabricated devices with a three-level architecture. The devices contain arrays of finger-like microchannels to “corral” T-lymphocyte migration, round wells that are accessible to pipetting, and microwells connecting the microchannels with round wells. T lymphocytes that are loaded into a well first settle into the microwells and then to microchannels by gravity. Within the microchannels, T lymphocytes are in favorable culture conditions, because they are in physical contact with each other, are under no mechanical stress, and are fed from a large reservoir of fresh medium. Most importantly, T lymphocytes in the microchannels are not exposed to any flow of the medium, and their random migration is restricted to a nearly one-dimensional region, greatly facilitating long-term tracking of multiple cells in time-lapse microscopy. The devices have up to nine separate round wells, making it possible to test up to nine different cell lines or medium conditions in a single experiment. Activated primary CD4+ T lymphocytes, resting primary CD4+ T lymphocytes, and THP-1 monocytic leukemia cells loaded into the devices maintained viability over multiple days.
The devices were used to track the fluorescence level of individual primary CD4+ T lymphocytes expressing green fluorescent protein (GFP) for up to 60 hours and to quantify single-cell gene-expression kinetics of four different HIV-1 variants in primary human CD4+ T lymphocytes. The kinetics of GFP expression from the lentiviruses in the primary CD4+ T lymphocytes agree with previous measurements of these lentiviral vectors in the immortalized Jurkat T lymphocyte cell line. These devices offer a simple, robust approach to long-term single-cell studies of environmentally sensitive primary lymphocytes.
INTRODUCTION
Time-lapse microscopy is a powerful quantitative technique in modern biomedical science and is increasingly used to probe dynamic processes in individual cells1. Time-lapse fluorescence microscopy has been critical to the study of stochastic cell-fate decisions by enabling single-cell analysis and mapping of the gene-regulatory circuits that control bacterial sporulation2, bacteriophage-λ lysis-vs.-lysogeny3, and development of antibiotic persistence4.
A similar cell-fate decision occurs in human immunodeficiency virus type 1 (HIV-1), the causative agent of AIDS, as the virus undergoes a stochastic transition between active replication and proviral latency (a long-lived viral dormancy state)5–7. In seroconverted patients, HIV-1 establishes a small reservoir of these latently infected cells. This reservoir is considered the largest obstacle thwarting eradication of HIV-1 from infected patients since interruption of highly active antiretroviral therapy (HAART) allows for viral rebound from these reservoirs. Hence, mapping the genetic circuitry regulating HIV-1 latency is crucial to developing new therapies8. Time-lapse fluorescence microscopy led to progress in mapping the HIV-1 latency circuit, making it possible to quantify single-cell viral-expression kinetics in immortalized tissue culture cell lines, such as Jurkat cells9–11. However, a major remaining challenge is to map the latency circuitry within primary CD4+ T lymphocytes isolated from patients, as it is these cells that are the physiologically relevant target for HIV-1 replication and latency in vivo12.
Single-cell time-lapse assays require tracking of individual cells for many hours or even days, and to achieve statistical significance, large numbers of cells must be tracked in parallel, necessitating some degree of cell immobilization. High experimental throughput is relatively straightforward to achieve with adherent cells, which move relatively slowly, when plated at the bottom of a cell-culture dish. However, for non-adherent or weakly adherent cells, such as T lymphocytes, which can be displaced by uncontrolled flow of the culture medium caused by convection or motion of the microscope stage, time-lapse microscopy becomes significantly more difficult. Uncontrolled displacements of non-adherent and weakly adherent cells can be prevented by using substrates with microwells 13–20, microfluidic devices with semi-permeable barriers (weirs), which also enable time-controlled exchange of the medium21–24, or adhesive coatings, which have been successfully applied to immobilize Jurkat T cells for up to 30 hours 6, 9, 11.
However, long-term tracking of primary human CD4+ T lymphocytes presents a set of specific challenges. When resting, CD4+ T lymphocytes are non-adherent suspension cells. Nevertheless, to support robust HIV-1 infection, viral replication, and reactivation from latency, primary CD4+ T lymphocytes must be activated, which is typically achieved by stimulation with small molecules (e.g., cytokines or anti-CD3 receptor cross-linking)5. As a result of this activation, T lymphocytes become weakly adherent, spontaneously attaching to and detaching from the substrate, and gain a capacity to rapidly migrate along the substrate while attached25, 26. Importantly, the adhesion molecules, which reliably immobilize cultured T lymphocytes, prove to be inadequate for activated primary T lymphocytes. With all the adhesive coatings we tested (including collagen, fibronectin, and Cell-Tak™), primary human T lymphocytes could only be immobilized and attached to the substrate for 2–3 hours at best (unpublished results). Furthermore, unlike strongly adherent cells, activated T lymphocytes do not form confluent monolayers and do not acquire easily distinguishable shapes, making their motion difficult to track over a two-dimensional region of a substrate. While time-lapse microscopy of HIV-1 gene-expression in infected lymphocytes has been demonstrated in regular cover glass-bottom dishes27, very few cells could be tracked and the duration of tracking was only ~8 hours27, much shorter than the 40 hours duration of the life cycle of HIV-1 in an infected cell9.
To immobilize lymphocytes or confine their movement, several microwell and microfluidic techniques have been applied. Arrays of microwells engraved on the surface of polydimethylsiloxane (PDMS) elastomer and sealed by glass slides have been used to analyze molecules secreted by individual lymphocytes15–18, 28. Arrays of sealed microwells have also enabled tracking of lymphocytes in short-term experiments (up to several hours)20, 21, 29–32. However, a sealed microwell limits the access of cells to fresh medium, and a low cell density in a microwell, while postponing depletion of the medium, limits cell-cell interactions that are physiologically important for T lymphocytes; considerable cell loss occurred when tracking of T lymphocytes in sealed microwells was extended to 2 days20. Microfluidic devices have also been used to capture and count human T lymphocytes on substrates with various coatings28. A microfluidic device with bucket-like (weir) structures has been used for capture and imaging of non-adherent T cells by exposing them to a steady hydrodynamic drag14, 33, 34. However, in these studies maintaining good viability of primary T cells required careful adjustment of the flow rate14, 24. Even at an optimal flow rate, the cell viability at 24 hours was ~68% (and much lower at flow rates that were two times faster or slower)14. This presents a major challenge for experiments on HIV-1 latency that require time-lapse microscopy over intervals longer than the HIV-1 life cycle, which is ~40 hours9. Furthermore, while in vivo lymphocytes are exposed to hydrodynamic stress when circulating in the blood plasma, 98% of lymphocytes reside in tissue such as lymph nodes and circulating cells migrate to lymph nodes within 30 minutes35, 36. Therefore, for multi-day time-lapse microscopy of primary T lymphocytes, a static environment may provide a better emulation of the in vivo conditions than continuous perfusion.
To circumvent these technical problems, we developed, built, and tested microfabricated devices that enable easy loading of primary T lymphocytes into arrays of narrow and shallow microchannels that are dead-ended (Fig. 1). These finger-shaped microchannels restrict spontaneous migration of lymphocytes to a nearly one-dimensional region, greatly facilitating cell tracking. The finger-shaped microchannels are located at the bottom of large round wells that are accessible to a pipette for cell loading. A single microfabricated device can have either four or nine separate round ‘loading’ wells and this multi-well design makes it possible to test four to nine different cell lines (or medium conditions) in parallel. Cells in these devices maintained viability over multiple days and an array of cell types were tested: activated primary CD4+ T lymphocytes, resting primary CD4+ T lymphocytes, and THP-1 monocyte-macrophage cells. As a further test of the devices, we performed long-term time-lapse microscopy of T lymphocytes that were infected with HIV-1-derived lentiviruses that expressed green fluorescent protein (GFP). The devices enabled tracking of GFP expression in a large number of individual T lymphocytes for ~60 hours, 20 hours longer than the HIV-1 intracellular life-cycle.
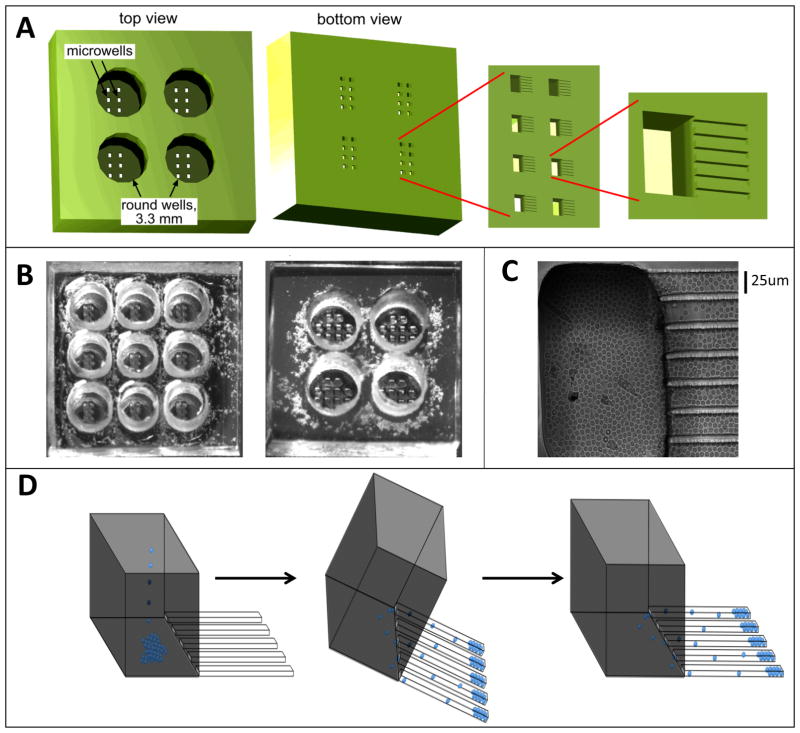
Figure 1. The microfabricated device with finger-like microchannels.
(A) Schematic of a PDMS chip with a 2×2 array of 3.3 mm in diameter, 3-mm-deep round wells. At the bottom of each round well there is a 2×4 array of 100×200-μm rectangular openings in a 100-μm-thick layer of PDMS, forming microwells. At the bottom of each microwell there are finger-like horizontal extensions, 25×25 μm in cross-section and ~100–200 μm in length, forming cell-imaging channels. (B). Photographs of PDMS chips with 3×3 and 2×2 arrays of round wells. The microwells are visible at the bottom of the round wells. (C) Brightfield image of resting primary CD4+ T lymphocytes loaded into a microwell and the adjacent finger-like channels. (D) Schematic of loading of cells into finger-like channels. First, cells are loaded into a round well and allowed to settle onto the bottoms of the microwells by gravity. Then, the device is tilted by ~45° with the finger-like channels pointing downward, making cells slide into the finger-like channels. Third, the normal orientation of the device is restored and it is carefully placed on the microscope stage.
EXPERIMENTAL
Design and operation of the devices
The finger-like microchannels, which are the structures that ultimately “collect” the T lymphocytes for long-term imaging, are 25×25 μm in cross -section and ~100–200 μm long. Seven parallel finger-like microchannels form a cluster connected to a single microwell. Each microwell is 200 μm in width, 100 μm in length, and 100 μm in depth (Fig. 1A). The bottom of the microchannels and microwell is formed by a regular #1.5 microscope cover glass, making cells in the microchannels accessible to high-resolution microscopy. The top of the microwell opens to the bottom of a round well, which is 3.3 mm in diameter and ~6 mm deep. The total volume of a round well is ~50 μL, and medium can be readily loaded into it and aspirated from it with a micropipette. A single round well has an array of 8 rectangular microwells at the bottom, and the microfabricated chips have round wells in either 3×3 (Fig. 1B, left) or 2×2 (Fig. 1B, right) square arrays, with each well connected to a total of 56 finger-like microchannels at the bottom.
The three-level hierarchal structure of the devices facilitates loading of T lymphocytes into the dead-ended finger-like channels, where the low ceiling and small width limit the motion of cells in the vertical and lateral directions, hindering their ability to crawl past each other (Fig. 1C). To load T lymphocytes in the finger-like channels, they are pipetted directly into a round well and allowed to settle by gravity, first settling to the bottom of the round well and then to the bottoms of the microwells (Fig. 1D, left). At this point (usually after 10–15 minutes), cells at the bottom of the round well are removed by repeated aspiration and dispensing of the medium with a micropipette. Importantly, the aspiration and dispensing do not agitate cells at the bottom of the microwells, since these cells are shielded from flow of the medium by the microwell walls37. Next, the device is tilted at 45° in the direction of the finger-like channels (so that they point downward), and cells are allowed to slide into the finger-like channels by gravity (Fig. 1D, middle). Because of the small width of the microwells (100 μm), the sliding of cells from the bottom of the microwell into the finger-like channels only takes several seconds. The horizontal orientation of the device is then restored (Fig. 1D, right), leaving cells in the finger-like channels.
The total footprint of the finger-like channels is nearly the same as the footprint of the microwell, making it simple to estimate the concentration of cells in the suspension loaded into the well that would result in an appropriate number of cells in the finger-like channels. Because the depth of the round wells is much greater that the diameter of a T lymphocyte (6 mm vs. 12 μm), a dense monolayer of cells at the microwell bottoms can be created using a relatively low number of cells in the suspension (~1·106 cells per mL). Therefore complications that might arise from creating an excessively concentrated cellular suspension by centrifugation are avoided. Furthermore, loading of cells into the finger-like channels is nearly as simple as loading of cells into the regular multi-well plates and much simpler that loading of cells into microfluidic devices. Only ~50,000 cells (and ~50 μL of medium) are required to fill all finger-like channels within a single round well.
The volume of a round well is ~1000 times greater than the cumulative volume of all microwells (and finger-like channels) connected to it. Therefore, a round well represents a large reservoir of fresh medium for cells in microwells and finger-like channels. Furthermore, due to the small depth of the microwells and small length of the finger-like channels, the molecular diffusion between a round well and finger-like channels leads to efficient equilibration of the medium content around cells with the medium in the round well (Supplementary Information).
A characteristic time of the diffusion can be estimated as τ = L2/(2D), where L = 250 μm is the cumulative distance from the bottom of a round well to the end of a finger-like channel, and D is the coefficient of diffusion. For small molecules, with D=5·10−5 cm2/s, the value of τ is ~60 s, indicating that, while the density of cells in the finger-like channels and at the bottom of the microwells may be very high (which is usually the case), the medium around cells is likely to be nearly identical to the fresh medium in the well (which does not have any cells in it). Moreover, the short medium equilibration time by diffusion implies that if the medium in the round well is exchanged, cells in the finger-like channels are rapidly exposed to the new medium, even without any active flow through the microwells and finger-like channels37. Close proximity of the round wells (and the finger-like channels connected to them) limits the motion of the microscope stage to <12 mm along one dimension even with the 9-well device, making it possible to use high-resolution oil-immersion objectives and facilitating rapid scanning and high repeatability of the positioning.
Fabrication and loading of the devices
Each microfluidic device consists of a microfabricated PDMS chip and a #1.5 microscope cover glass sealing the chip. The four- and nine-well PDMS chips have footprints of 14×14 and 19×19 mm, respectively. Each chip is assembled of two parts. The first part is 6-mm thick with 3.3-mm diameter through-holes in an either 2×2 or 3×3 array; the second part is 100 μm thick and has 200-μm long grooves with 25×25 μm cross-section engraved on its surface (forming the finger-like channels) and 100×200 μm through-holes (forming the microwells; Fig. 1a). The 6-mm-thick parts are cast of PDMS (Sylgard 184 by Dow Corning) using a master mold machined with a solid printer. To cast the 100-μm-thick parts, a lithographically fabricated master mold is produced by spin-coating a silicon wafer with a 25 μm layer of a UV-curable epoxy (SU8-2015 by MicroChem), exposing it to UV-light through a photomask, spin-coating the wafer with a second layer of SU8 to a total thickness of 100 μm, exposing it to UV light through another photomask, and developing the wafer.
The master mold is spin-coated with an ~100-μm layer of PDMS pre-polymer, so that the upper surfaces of the 100-μm relief features remain exposed (PDMS-free)38, and the PDMS is cured by baking in an 80 °C oven. The 6-mm-thick PDMS parts are bonded to the 100-μm-thick PDMS layer on the mold by treating their surfaces with oxygen plasma. The 100-μm layer is cut around the perimeter of the 6-mm-thick parts and the two-part monolith PDMS chips are separated from the mold. Each chip is reversibly bonded to a #1.5 microscope cover glass by overnight baking in an 80 °C oven forming a complete microfabricated device.
Before loading cells into a device, the microwells and finger-like channels are filled with a pH 7.5 PBS buffer. To this end, the device is treated with oxygen plasma to make its surface hydrophilic, the round wells are filled with the buffer using a micropipette, the device is placed into a plastic bottle with the buffer, and the bottle is pressurized to ~5 psi for 10 minutes. This procedure results in bubble-free filling of the finger-like channels, likely due to a combination of the buffer wicking into voids with hydrophilic walls and the excessive pressure in the bottle pushing the residual bubbles from the microwells and finger-like channels into the bulk of the porous PDMS chip.
Microscopy setup, cells and reagents
Experiments were performed on a Zeiss Axiovert inverted fluorescence microscope equipped with a Yokagawa spinning disc, 488-nm laser excitation light source, a CoolSNAP HQ2 14-bit camera from Photometrics, a computer controlled motorized stage, and environmental enclosure, maintaining a temperature of 37°C and a humidified atmosphere with 5% CO2. In time-lapse experiments, images were captured every 10 minutes, using a 40x oil, 1.3NA objective, 300-ms exposure time, 10% power on a 50-mW 488-nm solid-state laser, and analyzed as described9.
CD4+ T lymphocytes were isolated from patient blood by a negative selection method as described39. Before infection experiments, cells were activated from the resting state for 48 hours with Dynabeads® Human T-Activator conjugated with anti-CD3/CD28 antibodies from Invitrogen. Primary CD4+ T lymphocytes were cultured in RPMI 1640 supplemented with 5% Human Serum AB from GIBCO and 1% penicillin-streptomycin and kept at 1×106 cells/mL. THP-1 monocytes were cultured in RPMI 1640 with 10% fetal calf serum, 1% penicillin-streptomycin, and 0.05 mM 2-mercaptoethanol and cultured at 2×105 – 8×105 cells/mL.
Cell viability was tested with Yo-Pro®-1 Iodide (Life Technologies™) as described14. After isolation from peripheral blood (see above), primary cells were either placed in standard culture dishes or within the microwell device. At 24 and 48 hours, ~100,000 cells from culture dishes were placed on a glass bottom slide and stained with Yo-Pro®-1. Within 10 minutes after Yo-Pro®-1 addition, four images of cells in randomly selected areas were taken under brightfield and fluorescence illumination (with a FITC filter set), using a 10x 0.3NA objective. The total number of cells was 1800 at the 24 hours and 760 at the 48 hours. The percentage of dead cells was calculated by dividing the number of stained cells, as detected under the fluorescence illumination, by the total number of cells counted in the brightfield images. To evaluate the percentage of dead cells in the microwell device, Yo-Pro®-1 was applied to two different microwells at 24 and 48 hours and the numbers of dead cells and all cells were counted in finger-like channels connected to four different microwells at each time point. The total number of cells was 270 at the 24 hours and 609 at the 48 hours. All p-values were calculated using the Student’s two-tailed t-test.
Four lentiviral constructs (LTR-GFP, LTR-GFP-Tat, HIVΔGag-GFP, and full-length HIVΔEnv-GFP)11 were packaged in 293FT cells as described11. Viral preps were clarified and ultra-centrifuged at 18,000 rpm at 18°C for 1.5 hours, then re-suspended in 200 μL of medium together with primary CD4+ T lymphocytes. Infections were performed at a high multiplicity of infection (MOI) in a 96-well plate, and the suspension of cells and viruses was loaded into the microwell device. Activated primary cells cultured by standard techniques were infected with the LTR-GFP-Tat construct and GFP fluorescence of 10,000 live cells was measured at 48 hours post infection by flow cytometry on a FACSCalibur™ DxP8 instrument.
RESULTS
Loading of the microwell device normally resulted in ~25 cells in each finger-like channel and less than a monolayer of cells at the bottoms of rectangular wells (cf. Fig. 1C and 2C). To test for possible depletion of nutrients and accumulation of metabolites in the finger-like channels due to the high density of cells, we performed time-dependent numerical simulations in COMSOL™ (see Supplementary Information). The simulations indicated that medium conditions in the finger-like channels were substantially more favorable than those in a monolayer cell culture in a dish (or multi-well plate). Because of relatively small total number of cells in the finger-like channels and large volume of medium in the circular wells, the depletion of nutrients (both low-molecular (Fig. S1A and S1C) and macromolecular (Fig. S1B and S1D)) in the finger-like channels was less severe than in the dish (see Supplemental Information). Whereas, according to the simulations, concentrations of metabolites secreted by cells (especially those with low diffusivity) considerably varied along the finger-like channels, the average metabolite concentrations were several times lower than in the monolayer culture in a dish (Fig. S1, compare S1A to S1C, and S1B to S1D). Moreover, the oxygen permeability of PDMS is >5 times higher than water40, so medium in the finger-like channels was expected to be better aerated than medium at the bottom of the dish.
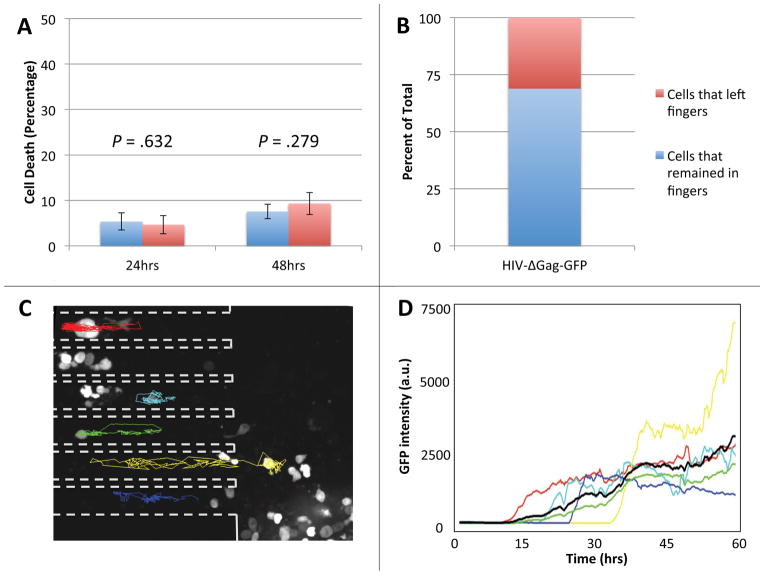
Figure 2. Primary human CD4+ T lymphocytes in the microfabricated device maintain viability and can be reliably tracked.
(A) Cells placed within the finger-like channels of the device or in standard culture dishes were stained with Yo-Pro®-1 Iodide cell death stain after 24 and 48 hours. In the device, 4.66 ± 2.01% of cells stained positive after 24 hours, a statistically insignificant difference from the 5.36 ± 1.91% positively stained cells in culture (p = .632). After 48 hours, there was also no statistically significant difference in the number of dead cells in the device, 9.32 ± 2.45, or in bulk culture, 7.55 ± 1.60 (p = .279). Both p-values were calculated using the Student’s two-tailed t-test. (B) The percent of infected cells that remained within the finger-like channels of the device (69%, blue portion) compared to the number of infected cells that migrated out of the finger-like channels (31%, red portion) over the duration of a 60-hour experiment. (C) Fluorescence micrograph of activated primary human CD4+ T lymphocytes infected with a GFP-expressing HIV-1 virus at 26 hours post infection. Dashed outlines mark the sidewalls of the finger-like channels and colored tracks represent migration trajectories of individual cells over the course of a 60-hour imaging experiment. Random migration of cells in the finger-like channels is largely one-dimensional, making it possible to reliably track cells over the entire 60 hours of the time-lapse experiment. Cells at the bottom of the microwell (large area to the right of the dashed outlines) move about freely in two dimensions and are difficult to track. (D) The intensity of fluorescence of GFP in a cell vs. time for the six infected cells tracked in panel B. The average fluorescence signal (black line) shows that cells start expressing GFP at approximately 10 hours after infection.
The simulations suggested that cell viability would be maintained in the device since there would be no depletion of nutrients and sufficient oxygenation. We then experimentally checked the viability of resting primary CD4+ T lymphocytes with Yo-Pro®-1 cell-viability stain. Resting primary CD4+ T lymphocytes were either cultured in a T-flask using conventional bulk cell-culture techniques (Fig. 2A, blue columns) or placed within the finger-like channels of the device (Fig. 2A, red columns). After 24 and 48 hours in the device cells were just as healthy as cells cultured by standard techniques (Fig. 2A). To test if individual cells could be tracked over extended periods of time, we performed fluorescence time-lapse microscopy of CD4+ T lymphocytes infected with an HIV-1-derived lentivirus expressing GFP. A minority (less than 30%) of cells initially infected within the finger-like channels crawled out of their finger-like channel into the larger microwell area over the duration of the experiment (Fig. 2B). The cells that remained in the finger-like channels of the device were readily tracked since the 2-dimensional movement is restricted in the finger-like channels (Fig. 2C, traces). In contrast, cells at the bottom of the microwells (Fig. 2C, right of dashed lines) freely moved in two dimensions, making them difficult to track over extended time intervals. The time of detection of fluorescence (indicating the onset of GFP expression) varied between cells (Fig. 2D), reflecting inherent variability in the timing of their infection. In agreement with previous reports in Jurkat cells10, the GFP expression of infected cells rapidly increased during the first 10–15 hours after the onset and then reached a plateau (Fig. 2D).
We next compared the expression profiles for diverse HIV-1-derived lentiviral vectors encoding successively fewer HIV-1 regulatory and structural components and all encoding GFP as a reporter. The minimal synthetic lentiviral vectors serve as are important controls since the complexity of their gene-expression circuitry has been reduced to tractable levels and comparing the behavior of these minimal circuits to the behavior of the full-length virus facilitates logical mapping of viral gene-regulation circuitry9–11, 41. Four previously reported viral constructs11 (i.e., LTR-GFP, LTR-GFP-Tat, HIV-ΔGag-GFP, and full-length HIV-ΔEnv-GFP) were used to infect activated primary human CD4+ T lymphocytes in a nine-well device (Fig. 3A). Each infection was performed in duplicate in adjacent wells. The single remaining well was retained as an uninfected control to check cell viability and autofluorescence over the course of the experiment (Fig. 3A). Cellular autofluorescence does not influence the GFP signal since autofluorescence is 30-fold lower than GFP fluorescence even for the very dimmest cell tracked (autofluorescence is 100-fold lower than the average GFP fluorescence from LTR-GFP-Tat expressing cells). A total of 59 cells were tracked for 60 hours, with at least 10 cells tracked for each lentiviral infection. Consistent with the results of the previous experiment (Fig. 2), cells within the finger-like channels could be reliably tracked, whereas tracking of cells at the bottom of the microwells was difficult due to their rapid two-dimensional migration (SI Movie 1). Cells that were infected within the finger-like channels tended to remain inside the finger-like channels for the duration of the experiment (SI Movie 1).
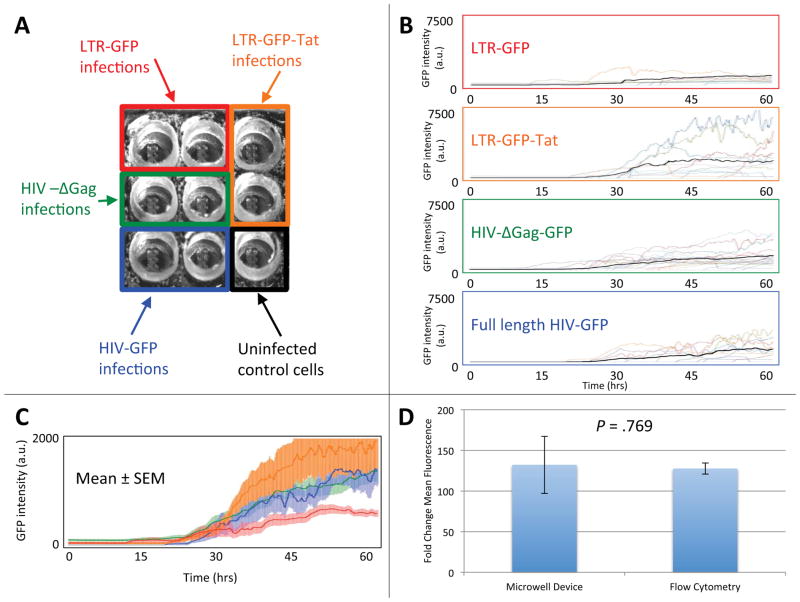
Figure 3. Single-cell gene-expression kinetics measured for primary human CD4+ T lymphocytes infected with four different HIV-1 viral constructs in a single experiment.
(A) Experimental setup for the infection of activated primary CD4+ T lymphocytes in a 3×3 well chip. Four reported HIV-1 lentiviral constructs (LTR-GFP, LTR-GFP-Tat, HIV-ΔGag-GFP, and full-length construct HIV-ΔEnv-GFP11) were used to infect activated primary human CD4+ T lymphocytes. CD4+ T lymphocytes infected with each of the four constructs were loaded into two nearby wells for time-lapse imaging. One well was left for uninfected control cells. (B) Single-cell GFP intensity (arbitrary units) time traces over a 60-hour period for the four lentiviral constructs. Fluorescence signal (reflecting the level of GFP expression) is shown for at least 10 individual cells for each construct. Different cells are represented by time traces of different colors. Mean fluorescence intensity per cell is shown as bold black line. (C) The average time-lapse trace for each construct with error bars representing the standard error of the mean. The LTR-GFP (red with red error bars) average is significantly lower in rate and level of GFP expression than the LTR-GFP-Tat (orange with orange error bars), HIV-ΔGag-GFP (green with green error bars), and full-length HIV-ΔEnv-GFP (blue with blue error bars) average traces. (D) Activated lymphocytes were infected with the LTR-GFP-Tat virus and measurements were taken 48 hours post infection by flow cytometry. There is no statistically significant difference in the fold change in mean fluorescence for cells 48 hours post infection as measured by flow cytometry, 131 ± 7, or by microscopy in the device, 127 ± 35 (p = .769 as calculated by Student’s two-tailed t-test). Error bars represent the standard error of the mean.
Cells infected with the LTR-GFP construct consistently exhibited the lowest fluorescence levels (i.e., expressed the lowest levels of GFP; Fig. 3B, top), which is likely because this vector only encodes the relatively weak HIV-1 long-terminal repeat (LTR) promoter element and no other HIV-1 transactivating genes. Cells infected with the LTR-GFP-Tat had the strongest GFP expression (Fig. 3B, second from top), most likely due to the HIV-1 positive-feedback loop in which the transactivator of transcription (Tat) increases expression from the HIV-1 LTR6, 9, 11, 42. When averaged over an interval from 40 to 50 hours post infection, in which the fluorescence was changing in time relatively slowly, the levels of fluorescence per cell for cells infected with LTR-GFP, LTR-GFP-Tat, HIV-ΔGag-GFP, and full-length HIV-ΔEnv-GFP were, respectively, 542 ± 117, 1605 ± 532, 1062 ± 156, and 932 ± 207. These values indicated that the GFP expression from LTR-GFP was significantly lower than from the other three constructs, whereas the GFP-intensity differences between LTR-GFP-Tat, HIV-ΔGag-GFP, and full-length HIV-ΔEnv-GFP were statistically insignificant. The rate and level of GFP expression from the three lentiviruses encoding the Tat positive-feedback loop (LTR-GFP-Tat, HIV-ΔGag-GFP, and full-length HIVΔEnv-GFP) were significantly higher than LTR-GFP throughout the duration of the experiment (Fig. 3C), in agreement with previous measurements of these lentiviral vectors in the immortalized Jurkat T lymphocyte cell line9, 10. To compare the behavior and function of the cells in the microwell device with standard bulk cell culture, cells in standard bulk culture were infected with the LTR-GFP-Tat lentiviral vector and fluorescence was measured by flow cytometry 48 hours after infection (Fig. 3D). The increase in GFP expression over 48 hours in bulk culture is ~130-fold over background autofluorescence, which is equivalent to the increase in GFP expression for cells residing in the finger-like microchannels 48 hours post infection (Fig. 3D),
DISCUSSION AND CONCLUSIONS
The results of our pilot experiments demonstrate the utility of the proposed microwell devices that contain finger-like imaging channels, microwells, and pipette-accessible ‘loading’ wells. These devices allow for tracking the fluorescence of a large number of individual primary human CD4+ T lymphocytes over a time interval significantly exceeding the 40-hour life-cycle of HIV-1, thus providing a substantial improvement over previous primary-cell studies in either culture dishes27 or continuously perfused microfluidic devices14. The proposed devices combine an important benefit of multi-well plates, the ease of cell loading, with a benefit of microfluidic devices, the confinement of cells to microfabricated channels with a diameter comparable with that of the cells. CD4+ T lymphocytes within the finger-like channels have access to a large reservoir of fresh medium in the round wells, and are in physical contact with each other, thereby allowing them to maintain their viability for multiple days. In addition, the device allows rapid and simple media exchange without displacement of cells. The results of the analysis of cell trajectories from the finger-like channels vs. bottoms of the microwells indicate that the confinement of activated T lymphocytes to a nearly one-dimensional region in a finger-like channel is sufficient for reliable cell tracking. Moreover, the finger-like channels appear to provide a nearly optimal degree of cell confinement for long-term imaging, because more aggressive confinement and immobilization of T lymphocytes (e.g., by placing them in small closed wells or pushing them against a semi-permeable barrier) might create less physiological conditions or even compromise T-lymphocyte viability.
The proposed devices have the potential to enable quantitative studies of kinetics of HIV-1 infection and reactivation from latency with single-cell resolution. The devices can be readily expanded to include a larger number of the round wells (each with microwells and finger-like channels at the bottom), making it possible to test a larger number of medium conditions and different cell lines in a single experiment. The proposed devices can also be modified and adapted to experiments on other non-adherent cells such as other leukocytes, hematopoietic stem cells, or yeast.
Supplementary Material
Acknowledgments
We are grateful to the Center for AIDS Research (CFAR) of San Diego, Weinberger and Groisman Laboratory for helpful discussions. This work was supported by the NIH Director’s New Innovator Award DP2OD00667 (to LSW) and NIH awards AI090935 and GM083395 (LSW). BSR acknowledges support from the NIH Molecular Biophysics Training Grant (Grant GM08326) and is supported by the NSF graduate research fellowship program (GRFP). AG acknowledges support from NIH award GM084332. EG acknowledges support from NIH award HL078784.
References
- 1.Locke JC, Elowitz MB. Nat Rev Microbiol. 2009;7:383–392. doi: 10.1038/nrmicro2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eldar A, Chary VK, Xenopoulos P, Fontes ME, Loson OC, Dworkin J, Piggot PJ, Elowitz MB. Nature. 2009;460:510–514. doi: 10.1038/nature08150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng L, Skinner SO, Zong C, Sippy J, Feiss M, Golding I. Cell. 2010;141:682–691. doi: 10.1016/j.cell.2010.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balaban N. Curr Opin Genet Dev. 2011 doi: 10.1016/j.gde.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. Nat Rev Microbiol. 2007;5:95–106. doi: 10.1038/nrmicro1580. [DOI] [PubMed] [Google Scholar]
- 6.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Singh A, Weinberger LS. Curr Opin Microbiol. 2009;12:460–466. doi: 10.1016/j.mib.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 9.Weinberger LS, Dar RD, Simpson ML. Nat Genet. 2008;40:466–470. doi: 10.1038/ng.116. [DOI] [PubMed] [Google Scholar]
- 10.Weinberger LS, Shenk T. PLoS Biol. 2007;5:e9. doi: 10.1371/journal.pbio.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Razooky BS, Weinberger LS. Methods. 2011;53:68–77. doi: 10.1016/j.ymeth.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siliciano RF, Greene WC. Cold Spring Harb Perspect Med. 2011;1:a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li PC, de Camprieu L, Cai J, Sangar M. Lab Chip. 2004;4:174–180. doi: 10.1039/b400770k. [DOI] [PubMed] [Google Scholar]
- 14.Faley S, Seale K, Hughey J, Schaffer DK, VanCompernolle S, McKinney B, Baudenbacher F, Unutmaz D, Wikswo JP. Lab Chip. 2008;8:1700–1712. doi: 10.1039/b719799c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogunniyi AO, Story CM, Papa E, Guillen E, Love JC. Nat Protoc. 2009;4:767–782. doi: 10.1038/nprot.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love JC, Ronan JL, Grotenbreg GM, van der Veen AG, Ploegh HL. Nat Biotechnol. 2006;24:703–707. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- 17.Han Q, Bradshaw EM, Nilsson B, Hafler DA, Love JC. Lab Chip. 2010;10:1391–1400. doi: 10.1039/b926849a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong Y, Ogunniyi AO, Love JC. Lab Chip. 2010;10:2334–2337. doi: 10.1039/c004847j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khademhosseini A, Yeh J, Jon S, Eng G, Suh KY, Burdick JA, Langer R. Lab Chip. 2004;4:425–430. doi: 10.1039/b404842c. [DOI] [PubMed] [Google Scholar]
- 20.Guldevall K, Vanherberghen B, Frisk T, Hurtig J, Christakou AE, Manneberg O, Lindstrom S, Andersson-Svahn H, Wiklund M, Onfelt B. PLoS One. 2010;5:e15453. doi: 10.1371/journal.pone.0015453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiffenbauer YS, Kalma Y, Trubniykov E, Gal-Garber O, Weisz L, Halamish A, Sister M, Berke G. Lab Chip. 2009;9:2965–2972. doi: 10.1039/b904778f. [DOI] [PubMed] [Google Scholar]
- 22.Yang X, Forouzan O, Burns JM, Shevkoplyas SS. Lab Chip. 2011 doi: 10.1039/c1lc20293f. [DOI] [PubMed] [Google Scholar]
- 23.Deutsch M, Deutsch A, Shirihai O, Hurevich I, Afrimzon E, Shafran Y, Zurgil N. Lab Chip. 2006;6:995–1000. doi: 10.1039/b603961h. [DOI] [PubMed] [Google Scholar]
- 24.Kobel S, Valero A, Latt J, Renaud P, Lutolf M. Lab Chip. 2010;10:857–863. doi: 10.1039/b918055a. [DOI] [PubMed] [Google Scholar]
- 25.Volkov Y, Long A, Kelleher D. J Immunol. 1998;161:6487–6495. [PubMed] [Google Scholar]
- 26.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 27.Saez-Cirion A, Nicola MA, Pancino G, Shorte SL. Biotechnol J. 2006;1:682–689. doi: 10.1002/biot.200600045. [DOI] [PubMed] [Google Scholar]
- 28.Thorslund S, Larsson R, Bergquist J, Nikolajeff F, Sanchez J. Biomed Microdevices. 2008;10:851–857. doi: 10.1007/s10544-008-9199-y. [DOI] [PubMed] [Google Scholar]
- 29.Yamamura S, Kishi H, Tokimitsu Y, Kondo S, Honda R, Rao SR, Omori M, Tamiya E, Muraguchi A. Anal Chem. 2005;77:8050–8056. doi: 10.1021/ac0515632. [DOI] [PubMed] [Google Scholar]
- 30.Lindstrom S, Mori K, Ohashi T, Andersson-Svahn H. Electrophoresis. 2009;30:4166–4171. doi: 10.1002/elps.200900572. [DOI] [PubMed] [Google Scholar]
- 31.Tokimitsu Y, Kishi H, Kondo S, Honda R, Tajiri K, Motoki K, Ozawa T, Kadowaki S, Obata T, Fujiki S, Tateno C, Takaishi H, Chayama K, Yoshizato K, Tamiya E, Sugiyama T, Muraguchi A. Cytometry A. 2007;71:1003–1010. doi: 10.1002/cyto.a.20478. [DOI] [PubMed] [Google Scholar]
- 32.Ozawa T, Kinoshita K, Kadowaki S, Tajiri K, Kondo S, Honda R, Ikemoto M, Piao L, Morisato A, Fukurotani K, Kishi H, Muraguchi A. Lab Chip. 2009;9:158–163. doi: 10.1039/b810438g. [DOI] [PubMed] [Google Scholar]
- 33.Di Carlo D, Aghdam N, Lee LP. Anal Chem. 2006;78:4925–4930. doi: 10.1021/ac060541s. [DOI] [PubMed] [Google Scholar]
- 34.Di Carlo D, Wu LY, Lee LP. Lab Chip. 2006;6:1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 35.von Andrian UH, Mempel TR. Nat Rev Immunol. 2003;3:867–878. doi: 10.1038/nri1222. [DOI] [PubMed] [Google Scholar]
- 36.Chen JJ, Huang JC, Shirtliff M, Briscoe E, Ali S, Cesani F, Paar D, Cloyd MW. J Leukoc Biol. 2002;72:271–278. [PubMed] [Google Scholar]
- 37.Carvalho A, Olson SK, Gutierrez E, Zhang K, Noble LB, Zanin E, Desai A, Groisman A, Oegema K. PLoS One. 2011;6:e24656. doi: 10.1371/journal.pone.0024656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kartalov EP, Walker C, Taylor CR, Anderson WF, Scherer A. Proc Natl Acad Sci U S A. 2006;103:12280–12284. doi: 10.1073/pnas.0602890103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terry VH, Johnston IC, Spina CA. Virology. 2009;388:294–304. doi: 10.1016/j.virol.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polinkovsky M, Gutierrez E, Levchenko A, Groisman A. Lab Chip. 2009;9:1073–1084. doi: 10.1039/b816191g. [DOI] [PubMed] [Google Scholar]
- 41.Franz K, Singh A, Weinberger LS. Methods Enzymol. 2011;497:603–622. doi: 10.1016/B978-0-12-385075-1.00026-3. [DOI] [PubMed] [Google Scholar]
- 42.Singh A, Razooky B, Cox CD, Simpson ML, Weinberger LS. Biophys J. 2010;98:L32–34. doi: 10.1016/j.bpj.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.