Abstract
Background
The gene for chromodomain helicase/ATPase DNA binding protein 1-like (CHD1L) was recently identified as a target oncogene within the 1q21 amplicon, which occurs in 46% to 86% of primary hepatocellular carcinoma (HCC) cases. However, the prognostic significance of CHD1L in HCC remains uncertain. In this study, we investigated the roles of CHD1L in the prognosis of HCC.
Methods
We investigated the expressions of CHD1L in tumor tissue microarrays of 281 primary HCC patients who underwent surgical resection using immunohistochemistry. Prognostic factors of HCC were examined by univariate and multivariate analyses. The median follow-up period was 75.6 months.
Results
CHD1L expression was observed in 48 of the 281 HCCs (17.1%). CHD1L expression was associated with a younger age (p=0.033), higher Edmondson grade (p=0.019), microvascular invasion (p<0.001), major portal vein invasion (p=0.037), higher American Joint Committee on Cancer T stage (p=0.001), lower albumin level (p=0.047), and higher α-fetoprotein level (p=0.002). Multivariate analyses revealed that CHD1L expression (p=0.027), Edmondson grade III (p=0.034), and higher Barcelona Clinic Liver Cancer stage (p<0.001) were independent predictors of shorter disease-free survival.
Conclusions
CHD1L expression might be a prognostic marker of shorter disease-free survival in HCC patients after surgical resection.
Keywords: CHD1L; Carcinoma, hepatocellular; Survival
The prognosis of hepatocellular carcinoma (HCC) remains poor mainly because of high recurrence rates even after surgical resection.1 Tumor recurrence complicates more than 70% of cases at five years.2 Many signaling pathways and molecules are involved in HCC carcinogenesis. Molecules involved in cancer progression might serve as markers for early detection of recurrence and metastasis after surgical resection.3 Given the high rates of HCC recurrence, a critical need exists to identify patients at high-risk of recurrence and to develop more effective and targeted treatment strategies to improve patient outcomes.
The chromodomain helicase/ATPase DNA binding protein 1-like gene (CHD1L; previously called amplified in liver cancer 1 gene) was recently identified as a target oncogene within the 1q21 amplicon,4 which occurs in 46% to 86% of primary HCC cases.5,6 CHD1L belongs to the sucrose nonfermenting 2 (SNF2)-like subfamily of the SNF2 family.7 SNF2 proteins stabilize or perturb protein-DNA interactions by using the energy released by their DNA-dependent ATPase activity and play important roles in transcriptional regulation, maintenance of chromosome integrity, and DNA repair.8 Ma et al.4 reported that amplification of CHD1L at the genomic level and overexpression of CHD1L at the protein level were detected in 50.6% (86/170) and 52.4% (163/311) of informative HCC tissues, respectively. Ma et al.4 also reported that CHD1L-transfected cells possessed a strong oncogenic ability.4 Recent studies showed that CHD1L expression was significantly associated with venous infiltration, microsatellite tumor nodule formation, advanced tumor stage, poor disease-free survival (DFS), and poor overall survival.9,10 However, the prognostic significance of CHD1L expression in HCC remains uncertain. In this study, we investigated the expression of CHD1L to evaluate the prognostic roles in HCC patients with long-term follow-up and extensive information on clinicopathologic characteristics.
MATERIALS AND METHODS
Patients and histopathology
A total of 281 primary HCC tissues were collected from patients who were treated with surgical resection at Samsung Medical Center, Seoul, Korea from July 2000 to May 2006 (age, 52.6 years; range, 17 to 76 years; 232 males and 49 females). Surgical resection margins were free of tumor. Two hundred and eleven (75.1%) patients were infected with hepatitis B and 30 (10.7%) with hepatitis C. No viral marker was recognized in 40 (14.2%) patients. None of the patients received preoperative chemotherapy. This study was approved by the Institutional Review Board of Samsung Medical Center. Tissue samples were fixed in 10% formaldehyde and embedded in paraffin for histopathologic examination. Histopathologic features of HCCs examined were tumor size, histological differentiation, microvascular invasion, major portal vein invasion, intrahepatic metastasis, multicentric occurrence, and non-tumor liver pathology. Differentiation was graded histologically using Edmondson and Steiner's criteria.11 Microvascular invasion was considered present when at least one or more endothelial cells or the tunica media of the vessel surrounded a neoplastic cell group. Intrahepatic metastasis and multicentric occurrence were defined according to previously reported criteria.12 Stage was determined according to both the American Joint Committee on Cancer (AJCC)13 and the Barcelona Clinic Liver Cancer (BCLC) staging classification.14
We followed up all patients every three months after surgery until December 31, 2010. Serum α-fetoprotein levels were evaluated and three phase dynamic computed tomography scans were performed. When tumor recurrence was suspected, precise diagnostic imaging was performed using magnetic resonance imaging. DFS was defined from the date of resection until the detection of both intrahepatic and extrahepatic recurrence. We chose HCC-related mortality (disease-specific death) as the clinical endpoint for survival analysis, defined as: 1) tumor occupying more than 80% of the liver, 2) portal venous tumor thrombus (PVTT) proximal to the second bifurcation, 3) obstructive jaundice due to the tumor, 4) distant metastases, or 5) variceal hemorrhage with PVTT proximal to the first bifurcation.15 The median follow-up period was 75.6 months (range, 1.7 to 126.9 months). Tumor recurrence was detected in 185 patients (65.8%), and 99 (35.2%) died of HCC. Thirty of the 129 deaths in this study were due to non-HCC causes.
Tissues with dysplastic nodule (DN), a precancerous lesion of HCC (n=28) were included, and DNs were subdivided into low-grade DN and high-grade DN according to the International Working Party guideline.16
Preparation of tissue microarrays and immunohistochemistry
All histologic sections were examined by two pathologists (CK.P. and S.A.) and representative tumor areas free from necrosis or hemorrhage were pre-marked in formalin-fixed paraffin-embedded blocks. Two 2.0-mm-diameter tissue cores were obtained from donor blocks and arranged in recipient paraffin blocks. Two cores of normal liver tissue from 12 patients with metastatic colonic carcinoma of the liver were included in each array block. Each tissue array block contained up to 60 tissue cores.
Horseradish peroxidase staining was used to visualize antigens on consecutive 4-µm thick sections. Sections were deparaffinized, hydrated, and immersed in peroxidase-blocking solution (Dako, Glostrup, Denmark) to inhibit endogenous peroxidase. For antigen retrieval, microwave pretreatment was performed with a 0.01 mol/L citrate buffer (pH 6.0) for 30 minutes. Sections were incubated overnight at 4℃ with mouse monoclonal antibody to CHD1L (1:50, ab51324, Abcam Inc., Cambridge, MA, USA). Sections were then incubated in Dako-REAL EnVision/HRP rabbit/mouse detection reagent (Dako) for 20 minutes at room temperature. Staining was visualized using the diaminobenzidine color substrate. The slides were counterstained with Mayer's hematoxylin. The positive control (human normal kidney) showed intense nuclear CHD1L expression in epithelial cells of convoluted tubules while no immnoreactivity was observed in tissue sections used as negative controls where the primary antibody was replaced by an isotype-matched irrelevant antibody. To validate the concordance between tissue microarrays and whole tumor sections, we used immunohistochemistry to detect the expression of CHD1L in 40 corresponding whole tumor sections randomly chosen from the 281 cases.
Evaluation of immunohistochemical staining
To assess the positivity of immunostaining, only nuclear staining was regarded as positive. We used a scoring method based on intensity and proportion of stained cells following a previous report.17 The percentage of positive tumor cells was determined semi-quantitatively and each sample was scored on a scale of 0-4 (0, <1%; 1, 1-25%; 2, 26-50%; 3, 51-75%; 4, 76-100%). Staining intensity was determined as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). The immunoreactive score of each tumor was calculated by the sum of these two parameters. The total score was graded as negative (total score; 0-1) or positive (total score; 2-7). All stained sections were assessed by two independent pathologists (CK.P. and J.H.) without knowledge of clinicopathologic features, and any differences in interpretation were resolved by consensus. Duplicate tissue cores for each tumor showed high levels of homogeneity for both intensity and proportion of stained cells. In cases of differences between duplicate tissue cores, the higher score was taken as the total score.
Statistical analysis
Statistical analyses were performed using SPSS software (SPSS Inc., Chicago, IL, USA). Chi-square and Fisher's exact tests were used for comparisons of categorical variables. Kaplan-Meier plots and log-rank tests were used for survival analysis. Univariate and multivariate analyses were based on the Cox proportional hazards regression model. p-values less than 0.05 were regarded as statistically significant.
RESULTS
CHD1L protein expression in HCC
In HCC, immunoreactivity for CHD1L was observed only in the nuclei of cancer cells. No immunoreactivity was found in hepatocytes, bile ducts, or stromal cells in normal liver tissues. CHD1L protein expression was observed in 48 (17.1%) of the 281 HCCs (Fig. 1A). CHD1L expression was significantly associated with a younger age (p=0.033), higher Edmondson grade (p=0.019), microvascular invasion (p<0.001), major portal vein invasion (p=0.037), higher AJCC T stage (p=0.001), lower albumin level (p=0.047), and higher α-fetoprotein level (p=0.002) (Table 1). All 28 DNs showed no nuclear immunoreactivity for CHD1L (Fig. 1B).
Fig. 1.
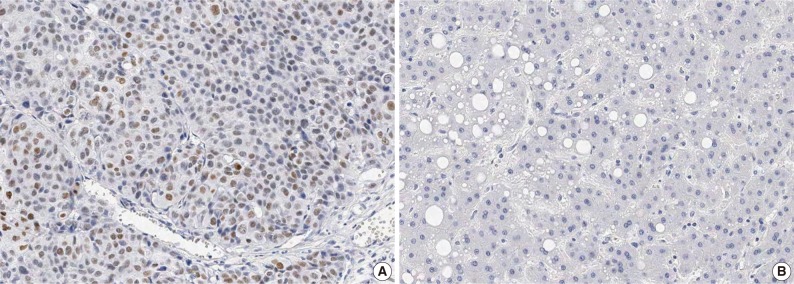
Immunostaining of CHD1L showing (A) nuclear expression in hepatocellular carcinoma and (B) no nuclear expression in dysplastic nodule (horseradish peroxidase stain).
Table 1.
Correlation of CHD1L expression with clinicopathologic features in 281 hepatocellular carcinomas

Values are presented as number (%).
AJCC, American Joint Committee on Cancer; BCLC, Barcelona Clinic Liver Cancer; AFP, α-fetoprotein; HBV, hepatitis B virus; HCV, hepatitis C virus.
Survival analysis
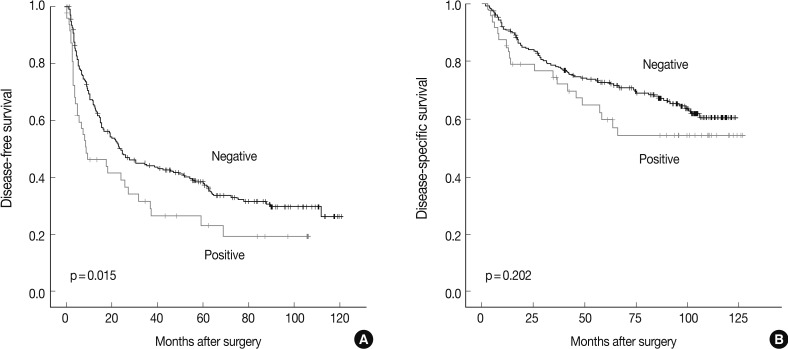
The DFS and disease-specific survival (DSS) rates for the 281 HCCs were 42.0% and 77.6% at 3 years, 35.9% and 70.6% at 5 years, 29.4% and 66.2% at 7 years, and 27.8% and 59.7% at 9 years, respectively. On univariate analyses, larger tumor size, Edmondson grade III, microvascular invasion, major portal vein invasion, intrahepatic metastasis, higher AJCC T stage, higher BCLC stage, lower albumin level, and higher α-fetoprotein level showed unfavorable influences on DFS and DSS (Table 2). The 5-year DFS rate of the CHD1L-positive group was significantly lower than that of the CHD1L-negative group (23.1% vs. 38.4%, p=0.015) (Fig. 2A). The median DFS of CHD1L-positive group and CHD1L-negative group were 59.7 months and 72.8 months, respectively. However, CHD1L expression was not a significant prognostic factor for DSS (p=0.202) (Fig. 2B). The 5-year DSS rate of the CHD1L-positive group and CHD1L-negative group were 59.7% and 72.8%, respectively.
Table 2.
Univariate analyses of disease-free survival and disease-specific survival in 281 hepatocellular carcinomas
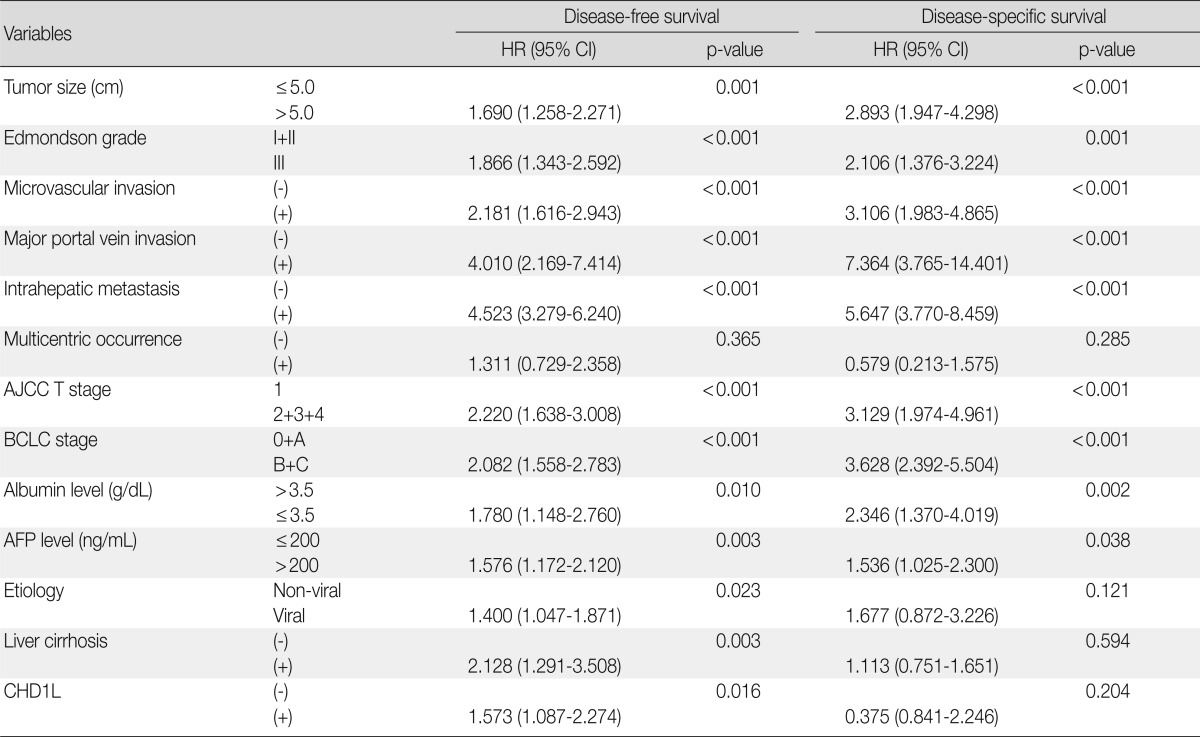
HR, hazard ratio; CI, confidence interval; AJCC, American Joint Committee on Cancer; BCLC, Barcelona Clinic Liver Cancer; AFP, α-fetoprotein.
Fig. 2.
Kaplan-Meier analysis of (A) disease-free survival and (B) disease-specific survival for CHD1L expression in 281 hepatocellular carcinomas.
As tumor size, microvascular invasion, major portal vein invasion, intrahepatic metastasis, multicentric occurrence, AJCC stage, and albumin serum level were associated with BCLC stage, we did not enter these into multiple analyses with the indices to avoid potential bias. An evaluation of the significant weight of serum α-fetoprotein level was not performed due to missing data (n=270). On multivariate analyses, Edmondson grade III (p=0.034), higher BCLC stage (p<0.001), and CHD1L expression (p=0.027) were independent predictors of shorter DFS. CHD1L-positive patients were more likely to suffer from recurrence than CHD1L-negative patients (hazard ratio, 1.534). Higher BCLC stage was an independent predictor of shorter DSS (p<0.001). However, CHD1L expression was not an independent predictor for DSS (p=0.155) (Table 3).
Table 3.
Multivariate analyses of disease-free survival and disease-specific survival in 281 hepatocellular carcinomas
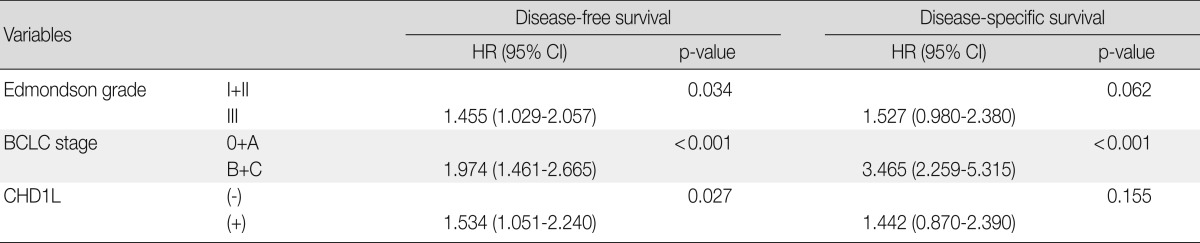
HR, hazard ratio; CI, confidence interval; BCLC, Barcelona Clinic Liver Cancer.
DISCUSSION
During HCC progression, regional chromosomal gain is a major mechanism in the activation of proto-oncogenes. A 1q gain is one of the most frequently detected alterations in HCC and has been suggested as an early genomic alteration during HCC progression.18 Amplification of 1q21 has been associated with HCC metastasis.19 CHD1L, a 1q21 target gene, is amplified and overexpressed in HCC cases.4 The molecular mechanism of CHD1L in HCC tumorigenesis has been associated with its role in promoting cell proliferation.4,20 Both in vitro and in vivo functional studies found that CHD1L upregulated ARHGEF9 transcription, which subsequently increased Cdc42 activity, causing filopodia formation, epithelial-mesenchymal transition, and finally HCC invasion and metastasis.9 Amplification of 1q has also been frequently detected in other human cancers, including biliary tract cancer,21 gastric cancer,22 and breast cancer,23 which suggests that CHD1L plays an important role in cancer progression in many human cancers.
In the present study, no nuclear immunoreactivity for CHD1L was observed in normal liver or DN tissues. CHD1L expression might not be an early event in HCC carcinogenesis. CHD1L expression in HCC was significantly associated with microvascular invasion, major portal vein invasion, and higher AJCC T stage. These findings were consistent with previous reports.9,10 Chen et al.9 reported that increased expression of CHD1L was detected in 34 of 50 (68%) metastatic HCCs compared with their paired primary HCCs. We collected a large number of clinical HCC tissues with long-term follow-up data and extensive information on clinicopathologic characteristics. Thus, we were able to determine the roles of CHD1L on patient survival independent of other prognostic factors of clinical outcome. As a result, CHD1L was an independent predictor of shorter DFS. A recent study showed that the DFS rate significantly decreased in the CHD1L-positive group compared with the CHD1L-negative group.9 CHD1L expression status might be correlated with progression and poor prognosis of HCC. In this study, CHD1L expression was not an independent predictor for DSS. Chen et al.10 reported that CHD1L expression was significantly associated with worse overall survival of HCC patients. Different clinical variables (e.g., overall survival vs. DSS), case number, and racial differences may have resulted in the discrepancy between this previous study10 and ours.
Our findings indicate that CHD1L is a marker for poor prognosis of HCC after surgical resection, and could help clinicians identify patients at high-risk of recurrence and enable them to administer adjuvant therapy after surgery. CHD1L could be used as an immunohistochemical biomarker to detect patients with a high-risk of recurrence. Moreover, undetectable expression of CHD1L in normal liver tissues suggests that targeting CHD1L for HCC therapy may not damage liver tissue.
This study demonstrates for the first time that CHD1L expression is an independent predictor of shorter DFS in HCC patients after surgical resection. Further studies are needed to identify its roles in the progression and prognosis of HCC.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Llovet JM, Schwartz M, Mazzaferro V. Resection and liver transplantation for hepatocellular carcinoma. Semin Liver Dis. 2005;25:181–200. doi: 10.1055/s-2005-871198. [DOI] [PubMed] [Google Scholar]
- 2.Poon RT. Prevention of recurrence after resection of hepatocellular carcinoma: a daunting challenge. Hepatology. 2011;54:757–759. doi: 10.1002/hep.24569. [DOI] [PubMed] [Google Scholar]
- 3.Ho MC, Lin JJ, Chen CN, et al. A gene expression profile for vascular invasion can predict the recurrence after resection of hepatocellular carcinoma: a microarray approach. Ann Surg Oncol. 2006;13:1474–1484. doi: 10.1245/s10434-006-9057-1. [DOI] [PubMed] [Google Scholar]
- 4.Ma NF, Hu L, Fung JM, et al. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology. 2008;47:503–510. doi: 10.1002/hep.22072. [DOI] [PubMed] [Google Scholar]
- 5.Sakakura C, Hagiwara A, Taniguchi H, et al. Chromosomal aberrations in human hepatocellular carcinomas associated with hepatitis C virus infection detected by comparative genomic hybridization. Br J Cancer. 1999;80:2034–2039. doi: 10.1038/sj.bjc.6690638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang J, Kim NG, Piao Z, et al. Assessment of chromosomal losses and gains in hepatocellular carcinoma. Cancer Lett. 2002;182:193–202. doi: 10.1016/s0304-3835(02)00083-6. [DOI] [PubMed] [Google Scholar]
- 7.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Chan THM, Yuan YF, et al. CHD1L promotes hepatocellular carcinoma progression and metastasis in mice and is associated with these processes in human patients. J Clin Invest. 2010;120:1178–1191. doi: 10.1172/JCI40665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Yuan YF, Li Y, et al. Clinical significance of CHD1L in hepatocellular carcinoma and therapeutic potentials of virus-mediated CHD1L depletion. Gut. 2011;60:534–543. doi: 10.1136/gut.2010.224071. [DOI] [PubMed] [Google Scholar]
- 11.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 12.Liver Cancer Study Group of Japan. The general rules for the clinical and pathological study of primary liver cancer. 2nd ed. Tokyo: Kanehara; 2003. p. 38. [DOI] [PubMed] [Google Scholar]
- 13.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A., 3rd . AJCC cancer staging manual. 7th ed. Chicago: Springer; 2010. [Google Scholar]
- 14.Llovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 15.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Working Party. Terminology of nodular hepatocellular lesions. Hepatology. 1995;22:983–993. doi: 10.1016/0270-9139(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 17.Gao J, Song Z, Chen Y, et al. Deregulated expression of Notch receptors in human hepatocellular carcinoma. Dig Liver Dis. 2008;40:114–121. doi: 10.1016/j.dld.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Wong N, Lai P, Lee SW, et al. Assessment of genetic changes in hepatocellular carcinoma by comparative genomic hybridization analysis: relationship to disease stage, tumor size, and cirrhosis. Am J Pathol. 1999;154:37–43. doi: 10.1016/S0002-9440(10)65248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong N, Chan A, Lee SW, et al. Positional mapping for amplified DNA sequences on 1q21-q22 in hepatocellular carcinoma indicates candidate genes over-expression. J Hepatol. 2003;38:298–306. doi: 10.1016/s0168-8278(02)00412-9. [DOI] [PubMed] [Google Scholar]
- 20.Chen M, Huang JD, Hu L, et al. Transgenic CHD1L expression in mouse induces spontaneous tumors. PLoS One. 2009;4:e6727. doi: 10.1371/journal.pone.0006727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller G, Socci ND, Dhall D, et al. Genome wide analysis and clinical correlation of chromosomal and transcriptional mutations in cancers of the biliary tract. J Exp Clin Cancer Res. 2009;28:62. doi: 10.1186/1756-9966-28-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gümüs-Akay G, Unal AE, Elhan AH, et al. DNA copy number changes in gastric adenocarcinomas: high resolution-comparative genomic hybridization study in Turkey. Arch Med Res. 2009;40:551–560. doi: 10.1016/j.arcmed.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Tsuji K, Kawauchi S, Saito S, et al. Breast cancer cell lines carry cell line-specific genomic alterations that are distinct from aberrations in breast cancer tissues: comparison of the CGH profiles between cancer cell lines and primary cancer tissues. BMC Cancer. 2010;10:15. doi: 10.1186/1471-2407-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]