Abstract
Current evidence highlights the importance of developing a healthy intestinal microbiota in the neonate. Many aspects that promote health or disease are related to the homeostasis of these intestinal microbiota. Their delicate equilibrium could be strongly influenced by the intervention that physicians perform as part of the medical care of the neonate, especially preterm infants. As awareness of the importance of the development and maintenance of these intestinal flora increase and newer molecular techniques are developed, it will be possible to provide better care of infants with interventions that will have long-lasting effects.
Keywords: Necrotizing enterocolitis, Microbiome, Intestinal microbiota, Noncultured based techniques, Sequencing
Key points
-
•
The intestinal microbiota normally exist in a commensal or symbiotic relationship with the host, but in the neonate, and especially in the premature infant, this relationship needs to develop.
-
•
Alterations of the intestinal microbiota may predispose the preterm infant to the development of necrotizing enterocolitis.
-
•
Composition and quality of the intestinal microbiota depend on many factors, such as mode of birth, type of feeding, and use of antibiotics. There are many opportunities for physicians to provide interventions that could improve the adequate colonization of the neonatal gut.
-
•
Culture-based techniques are highly limited, but the development of new molecular technologies to study previously unidentified organisms has enhanced our understanding of the microbial environment that may predispose to necrotizing enterocolitis.
Introduction
Necrotizing enterocolitis (NEC) is an enigmatic disease that has been recognized for more than a century, but with the advent of neonatal intensive care, it has become one of the most common and devastating diseases in neonates.1, 2, 3 Among the reasons why NEC has been so difficult to understand and eradicate is that what has been termed NEC is certainly more than 1 disease with multiple causes.2 For example, when an infant born at term who has a hypoplastic left ventricle presents with pneumatosis intestinalis at 2 days of age, the cause and pathophysiology of this baby’s NEC is likely different from the 25-week gestation preterm who presents with pneumatosis intestinalis at 5 weeks of age. The first is more likely related to ischemic injury caused by hemodynamic insufficiency and hypoxic-ischemic injury4 rather than a coalescence of factors that result in intestinal inflammation and injury largely as a result of intestinal immaturity, as in the preterm infant. In the latter case, sometimes referred to as classic NEC,2 the interactions of a predisposing genetic background, an immature intestinal barrier, and a microbial environment that is conducive to the development of NEC are believed to play an interactive and critical role in pathogenesis.2, 3, 5
The linkage of NEC to bacterial colonization was recognized by Santulli and colleagues6 more than 3 decades ago. Additional observations showing clusters of cases, outbreaks in institutions, the finding of pneumatosis intestinalis, which likely represents submucosal gas produced by bacterial fermentation, and the common findings of bacteremia and endotoxinemia in affected neonates support a microbial role in the pathogenesis of this disease.5 Numerous bacteria have been related to NEC, but none of them has been found to fulfill Koch’s postulates, because they are commonly found among patients without NEC.7 Viruses have also been implicated in the pathogenesis of NEC, and coronavirus within fecal samples and resected intestinal segments were reported in patients with NEC,8 but their role in the causation of the disease has not been substantiated.
Recently, the Human Microbiome Project has enabled development of novel technologies that should be highly instrumental in helping understand the contribution of the intestinal microbial ecology to the pathogenesis of NEC.9 The realization that culture-based techniques are limited in delineating the vast array of microbes present in the human intestine along with the development of new technologies to study these previously unidentified organisms has enhanced our optimism for better describing the microbial environment that may predispose to classic NEC. Furthermore, an appreciation of the role of commensal microbes in protection of the intestine and the disease that may occur when the balance of these commensals is disrupted is opening a new era of thought in the understanding of NEC as well as diseases ranging from obesity to autism. Efforts by the National Institutes of Child Health and Human Development have also paved the way for new studies in NEC.10 This review focuses on the role of the intestinal microbiota in the pathogenesis of classic NEC.
Complementing traditional culture-based techniques
The traditional view of the human is that they are composed of 10 trillion cells, which are the product of approximately 23,000 genes. However, in various niches of the human body, there reside several species of microbes and microbial genes that vastly outnumber those of the human host. In the past few years, emerging technologies derived largely from the Human Genome Project have been applied to evaluating the intestinal microbiota, and new discoveries using these techniques have prompted new initiatives such as the Human Microbiome Roadmap, designed to evaluate the role of the intestinal microbiome in health and disease.
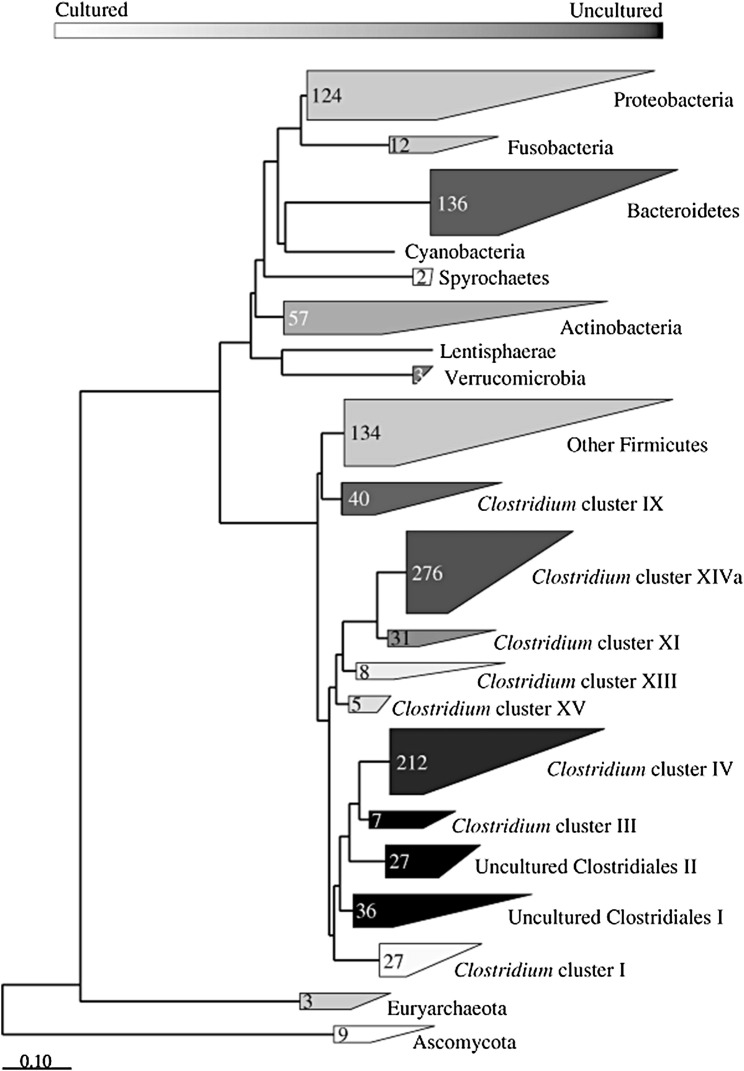
Until the beginning of the last decade, culture-based techniques were the mainstay of evaluating intestinal microbes. However, most bacterial cells seen microscopically in feces cannot be cultured in the laboratory (Fig. 1 ).11 Recently developed high-throughput molecular techniques analyze microbial DNA and RNA. There are 2 general approaches, both of which comprise several variants. One commonly used general approach is to use the 16S rRNA gene12 and the other is a more complex metagenomic approach, in which community DNA is subject to shotgun (whole-genome) sequencing.13 A comprehensive review of these techniques is beyond the scope of this article, but some of the more commonly used techniques are summarized in Box 1 .
Fig. 1.
Intestinal microbiota remain mostly uncultured by traditional culture-based methods. Black fills indicate phylotypes detected in cultivation-independent studies and white indicates species detected in cultivation-based studies.
(From Rajilic-Stojanovic M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol 2007;9:2125–36; with permission.)
Box 1. Nonculture techniques of intestinal microbiota identification.
16s rRNA sequencing V3/V4 and V4/V5 regions
Shotgun approach: Illumina (Illumina, Inc, San Diego, CA, USA) or 454 Titanium (454 Life Sciences Corporation, Branford, CT, USA) for longer sequence reads
Fingerprinting: DGGE/TGGE: separates individual rRNA genes and provides a fingerprint of the complexity of the intestinal microbiota
T-RFLP: rapid comparative analysis and very sensitive
FISH: best for enumeration of species in the intestinal tract
PhyloChip (Affymetrix Corporation, Lawrence Berkeley Lab (LBNL), San Francisco, CA, USA): DNA microarray for multiple bacterial identification
Both methods include the extraction of community DNA or RNA from feces or other samples of interest. The 16S rRNA is a part of the ribosomal RNA. It is commonly used for phylogenetic studies, because some regions of the gene encoding it are highly conserved and can act as primer binding sites, whereas other regions containing phylogenetic information are highly variable and enable their use for taxonomic classification. Using this approach, polymerase chain reaction (PCR) amplicons are sequenced using novel high-throughput pyrosequencing technology. Based on the degree of nucleotide similarity (usually between 95% and 99%), sequences can be binned into operational taxonomic units that form the basis for comparisons of microbiota composition and diversity between samples.
Although most of the initial efforts for taxonomic evaluation of the microbiome involved use of the 16S rRNA, more recent work has focused on sequencing of the genomes of the entire community using shotgun techniques that sequence the entire genome. Using this approach, Venter and colleagues14 identified a vast number of microbes that had not previously been recognized to exist in the oceans. Over the past few years, similar techniques are being applied to the human intestinal microbiome, and with future refinement of the technology, bioinformatics, statistical analyses, and decreases in cost of the analyses should yield important new information about the microbes present in the intestine and how they relate to health and disease.
Adjuncts to microbiome analysis: functional characterization
Even if the taxonomy of a particular microbial community is identified, the functional expression as it relates to physiology and interaction with the host is not clarified. Simple identification of individual microbes or microbial genotypes often does not clarify their phenotypic expression. Thus, in addition to microbiome identification-based technologies, it is important to clarify the function of the microbial communities of interest within a particular niche. It thus is of interest to identify the mRNA and protein expression of the genes as well as the metabolites that result before and after interaction of the microbial gene products with the host. Additional “-omic” disciplines are thus being applied to augment studies of the microbiome (Box 2 ). Metatranscriptomics, metaproteinomics, and metabolomics identify gene expression products (mRNA), proteins, and metabolites resulting from the genes within a complex microbial community, such as that found in fecal sample.
Box 2. Studies of functional expression of the intestinal microbiome.
Metagenomics (profiling intestinal microbiota, DNA): comparison with known functional expression of similar sequences
Metabolomics: metabolic profiles (metabolites) associated with microbiota
Metaproteomics: catalytic potential of microbiota (proteins)
Metatranscriptomics: microbiota responses to environmental changes (RNA)
Several metatranscriptome and metaproteome studies describing the human intestinal microbiota have confirmed the importance of bacterial functions related to carbohydrate metabolism in the colon. Similar results were seen in a transcriptional analysis of fecal samples from a monozygotic, obese twin pair15 and metatranscriptomic analysis of fecal samples from 10 healthy volunteers.16 Metatranscriptomic data from the less studied small intestinal microbiota showed enrichment in sugar phosphotransferase (PTS) and other carbohydrate transport systems, as well as energy, central metabolic, and amino acid conversion pathways compared with the metagenome.17 This finding suggests rapid uptake and fermentation of available simple sugars by the small intestinal microbiota, compared with the degradation of more complex carbohydrates by the bacteria in the colon. The importance of carbohydrate metabolism is also evident from the enormous amount of carbohydrate-active enzymes (CAZymes) present in the gut microbiome. Another recent study using fecal samples from studies of adult humans found that most assigned transcripts belonged to the metabolism cluster (26% of all sequences), underlining that even at the end of the intestinal tract the microbiota are still very active.18
Metaproteomics is the study of proteins collectively expressed within microbial communities that usually use mass spectrometry-based analyses to detect proteins associated with the microbiota identified within a given niche. These studies usually also use a systems-biology approach to evaluate the likely functions of the proteins produced by the microbiota. A recent study used a nontargeted, shotgun mass spectrometry-based whole-community proteomics, or metaproteomics, approach for the first deep proteome measurements of thousands of proteins in human fecal samples.19 The resulting metaproteomes had a distribution that was unexpected relative to the metagenome, with more proteins for translation, energy production, and carbohydrate metabolism when compared with what was earlier predicted from metagenomics. Human proteins, including antimicrobial peptides, were also identified, providing a nontargeted glimpse of the host response to the microbiota. Several unknown proteins represented previously undescribed microbial pathways or host immune responses, revealing a novel complex interplay between the human host and its associated microbes.
The intestinal microbiota are involved in the regulation of multiple host metabolic pathways, giving rise to interactive host-microbiota metabolic, signaling, and immune-inflammatory processes connecting the intestine, liver, muscle, and brain.
These interactions begin at birth and likely even during fetal life. The microbiota shape the development of the immune system, and the immune system in turn shapes the composition of the microbiota through a cross-talk between the microbes and the host immune system. The signaling processes, together with direct chemical interactions between the microbe and host, act on multiple organs such as the gut, liver, muscle, and brain.
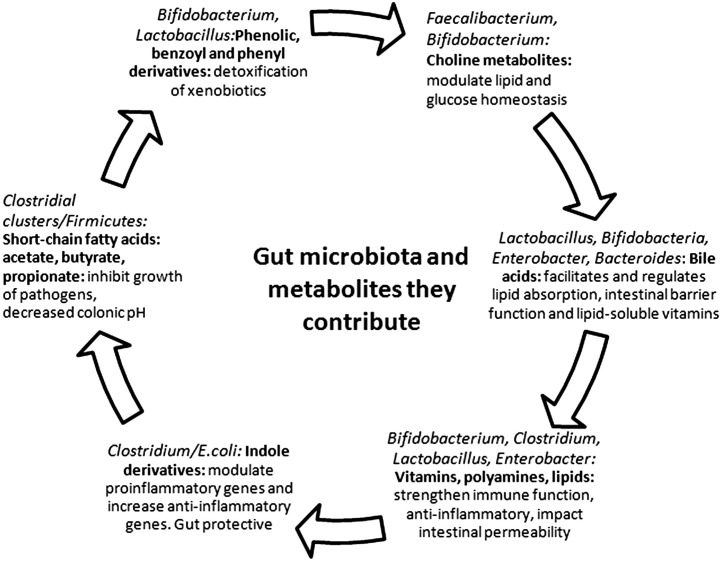
Fig. 2 summarizes some of the gut bacteria and the metabolites they contribute. Notable among these metabolites are the short-chain fatty acids (acetate, propionate, and butyrate), choline, bile acids, vitamin K, and polyamines, all believed to be important for optimal functioning of the gastrointestinal tract as well as the entire human. The production of these metabolites by microbes contributes to the host metabolic phenotype and hence to disease risk. Despite the putative importance of these metabolic functions, few studies are available as they pertain to NEC in the neonate.
Fig. 2.
Intestinal microbiota and the potential biologic functions and metabolites.
Alterations of the intestinal microbiota in NEC
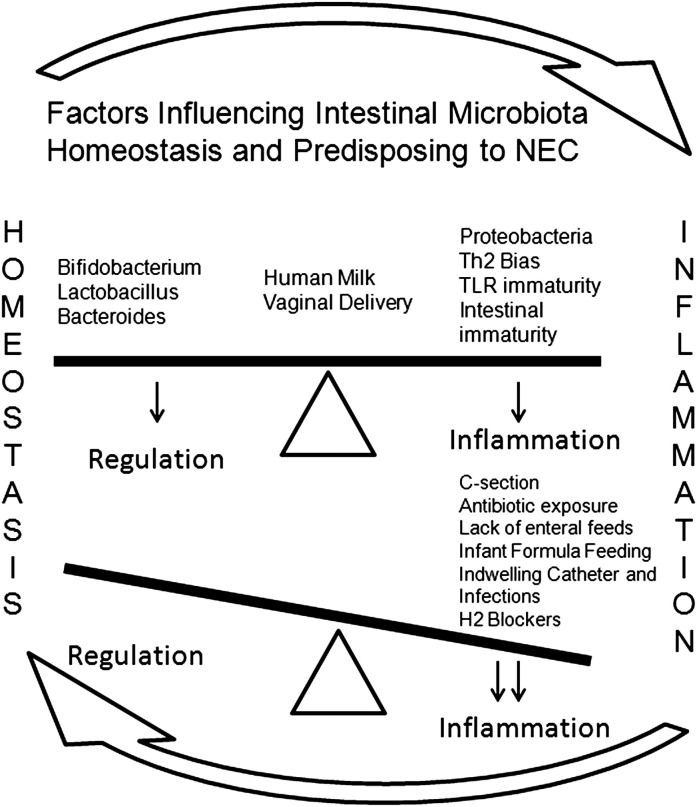
The intestinal microbiota normally exist in a commensal or symbiotic relationship with the host,20 but in the neonate and especially in the premature infant, this relationship needs to develop, and many factors define its delicate equilibrium with the capability to modulate immune responses and promote health (Fig. 3 ).21 As mentioned earlier, a specific pathogen that fulfills Koch’s postulates for the cause of NEC has not been found. Whether the use of new-generation sequencing technologies will help in this search is unknown, although some preliminary studies are offering clues.
Fig. 3.
Factors influencing intestinal microbiota homeostasis and predisposing to NEC. The intestinal microbiota are in perfect equilibrium with bacteria that regulate and others that have the potential to cause inflammation. Disruption of this equilibrium (dysbiosis) for different factors leads to inflammation.
In a study by Wang and colleagues,22 fecal samples from 20 preterm infants, 10 with NEC and 10 matched controls (including 4 twin pairs), were obtained from patients in a single-site level III neonatal intensive care unit. Bacterial DNA was subjected to terminal restriction fragment length polymorphism analysis and library sequencing of the 16S rRNA gene. The distribution of samples from patients with NEC distinctly clustered separately from controls. Intestinal bacterial colonization in all preterm infants was notable for low diversity. Patients with NEC had even less diversity, an increase in abundance of Gammaproteobacteria, a decrease in other bacteria species, and had received a higher mean number of previous days of antibiotics. These results suggested a relationship with previous use of antibiotics in patients with NEC. Whether the differences in clustering were because the samples were obtained at the time of NEC when these babies were receiving antibiotics is not clear.
In another study by Mai and colleagues,23 starting with the first stool and continuing until discharge, weekly stool specimens were collected prospectively from infants with gestational ages 32 completed weeks or less or birth weights 1250 g or less. High-throughput 16S rRNA sequencing was used to compare the diversity of microbiota and the prevalence of specific bacterial signatures in 9 NEC infants and in 9 matched controls. A bloom (34% increase) of Proteobacteria and a decrease (32%) in Firmicutes in NEC cases between 1 week and less than 72 hours was detected. No significant change was identified in the controls. Several molecular signatures were identified to be increased in NEC cases 1 week before and within 72 hours of NEC development. One of the bacterial signatures detected more frequently in NEC cases (P<.01) matched closest to γ-Proteobacteria (similar to the Wang study22). Although this sequence grouped to the well-studied Enterobacteriaceae family, it did not match any sequence in GenBank by more than 97%. These observations suggest that abnormal patterns of microbiota and potentially a novel pathogen contribute to the cause of NEC.
Factors affecting the intestinal microbiota
Prenatal
Thus, at this juncture, it seems that the microbiota of babies who subsequently develop NEC are different from those who do not. It will be important to dissect the factors that contribute to these differences, which may begin in fetal life (see Fig. 3). Recent PCR-based studies estimate the prevalence of microbial invasion of the amniotic cavity to be 30% to 50% higher than that detected by cultivation-based methods.24 These studies have shown that cultivation-resistant anaerobes belonging to the family Fusobacteriaceae (particularly Sneathia sanguinegens and Leptotrichia spp) are also commonly found in amniotic fluid. Other diverse microbes detected by PCR of amniotic fluid include as-yet uncultivated and uncharacterized species. A causal relationship between diverse microbes, as detected by PCR, and preterm birth is supported by types of association (eg, space, time, and dose) proposed as alternatives to Koch's postulates for inferring causality from molecular findings. Whether this colonization affects the gastrointestinal tract of the fetus is not known, but the fact that the fetus swallows large quantities of amniotic fluid during the last trimester of pregnancy and the recent finding that microbial DNA is present in meconium suggest that the fetal intestine is exposed to the amniotic fluid microbes. The high sensitivity of the fetal intestine to inflammatory mediators such as lipopolysaccharide (LPS) suggests that in utero microbes may trigger intestinal inflammation in utero25 Furthermore, it can be speculated that exposure of the intestinal tract to toll-like receptor (TLR) agonists such as LPS, flagellin, or microbial CpG in low quantities may also make the intestine tolerant to further inflammatory stimuli.26
Postnatal
Postnatally, many factors can influence intestinal bacterial colonization as well as the responses to colonization. Some studies have shown that mode of delivery (vaginal vs cesarean section), type of milk (human vs formula), and use of antibiotics influence the intestinal flora.27, 28, 29 Those infants born via cesarean section, fed formula milk, and exposed to antibiotics have a decrease in diversity of intestinal microbiota and abnormal patterns of colonization, with suppression of healthy bacteria such as Lactobacillus and bifidobacteria.
Cesarean Section Versus Vaginal Delivery
Epidemiologic data showing a relationship between increased rate of cesarean section and risk for development of subsequent diseases was recently reviewed.30 In the United States, the rate of cesarean delivery has increased 48% since 1996, reaching a level of 31.8% in 2007. This trend is reflected in many parts of the world, with the most populous country in the world, China, approaching 50% and some private clinics in Brazil approaching 80%. Concurrent with the trend of increasing deliveries by cesarean section, there has been an epidemic of both autoimmune diseases, such as type 1 diabetes, Crohn disease, and multiple sclerosis, and allergic diseases, such as asthma, allergic rhinitis, and atopic dermatitis. The occurrence of these diseases is higher in more affluent, Western, industrialized countries. As reviewed in Ref,30 several previous studies have shown differences in microbial colonization after cesarean section and vaginal deliveries. A more recent study,31 in which nonculture-based sequencing technology was used, examined the early stages of the colonization of the body by microbes. Babies born vaginally were colonized predominantly by Lactobacillus, whereas babies delivered by cesarean section were colonized by a mixture of potentially pathogenic bacteria typically found on the skin and in hospitals, such as Staphylococcus and Acinetobacter, suggesting that babies born by cesarean delivery were colonized with skin flora in lieu of the traditionally vaginal type of bacterium.32 The data relating these changes to later outcomes such as celiac disease, allergies, and atopy are compelling, but a relationship of cesarean section versus vaginal delivery, intestinal colonization, and the development of NEC has not been found.
Human Milk Versus Infant Formula
Animal and human studies have shown that human milk decreases the incidence of NEC.33, 34, 35 Beneficial factors of breast milk include immunoglobulins, cytokines, lactoferrin, lysozyme, and growth factors, but more importantly, healthy bacteria are promoted by human milk oligosaccharides (HMO).27, 36 HMOs contain a lactose core and act as prebiotics stimulating growth of Bifidobacterium species.37, 38 At birth, the rapid colonization of intestinal flora develops in the newborn first with aerobic or facultative anaerobic bacteria such as enterobacteria, enterococci, and staphylococci, and then during growth, they consume oxygen, allowing the proliferation of anaerobic bacteria such as Bacteroides, Bifidobacterium, and Clostridium species.39, 40 In the formula-fed infant, this transition tends to not occur that way and the newborn intestinal flora differs in its pattern of colonization with respect to breast-fed infants with predominance of gram-negatives and fewer anaerobes.41, 42 If they are grouped by phyla, breast-fed infants have predominance of Firmicutes mainly Lactobacillus, Bacteroides and Actinobacteria (Bifidobacterium) compared with formula-fed infants, who have predominance of Proteobacteria such as Escherichia coli and Firmicutes, some whom have pathogenic characteristics such as Clostridium and Staphylococcus. In 1 study, it was speculated that the abnormal pattern of colonization early at birth with delayed bloom of more pathogenic bacteria late in preterm infants could potentially explain those patients who develop NEC.23 Sullivan and colleagues33 in 1 multicenter study using donor human milk showed that an exclusively human milk-based diet is associated with significantly lower rates of NEC and surgical NEC when compared with a mother’s milk-based diet that also includes bovine milk-based products. The reduction in NEC using strategies of only human milk was of 50% for clinical NEC and almost 90% for surgical NEC. The number needed to treat to prevent 1 case of NEC was estimated to be 10. In a recent policy statement by the American Academy of Pediatrics regarding breastfeeding and the use of human milk, the recommendation was to offer donor milk to all preterm infants in whom mother’s own milk is unavailable.43
Antibiotic Exposure
The use of antibiotics is widespread in the neonatal intensive care unit (NICU). Often, their use is justified, but sometimes, it is the product of fear of infections in the preterm infant. Nevertheless, the use of antibiotics in neonates and especially in the preterm infant has unintended and sometimes devastating consequences. Antibiotic exposure may reduce the diversity of intestinal microbiota, delay the colonization of beneficial bacteria, and predispose preterm neonates to NEC. Previous studies44, 45, 46 have shown that duration of antibiotic exposure is associated with an increased risk of NEC among neonates without previous sepsis. One study47 found that overgrowth of pathogenic species is increased after 3 days of antimicrobial exposure. In the United States, most mothers giving birth prematurely are treated with antibiotics and many very-low-birth-weight infants are treated with a course of broad-spectrum antibiotics such as ampicillin and gentamicin. Studies28 have shown the detrimental effects of antibiotics to the intestinal flora, even after 1 dose of antibiotics with alterations in the intestinal microbiota that could take years to recover. One study48 showed a decrease in diversity, a predominance of less desirable bacteria and highly resistant clones with abundance of specific resistance genes as a result of antibiotic exposure that did not recover after 2 years after exposure. Cotten and colleagues49 found that each empiric treatment day with antibiotic was associated with increased odds of death, NEC, and the composite measure of NEC or death. These investigators concluded that prolonged initial empiric antibiotic therapy may be associated with increased risk of NEC or death and should be used with caution.
Extreme caution should then be exerted at the moment of deciding to start antibiotics in preterm infants. Long-lasting consequences and life-threatening situations could result from the indiscriminate use of antibiotics.
Intestinal microbiota alteration induces intestinal inflammation and cytokines
Immature intestinal response and alterations in the intestinal microbiota predispose the intestine of neonates to inflammation and to a cascade of proinflammatory and counterinflammatory cytokines response. Proinflammatory cytokines such as tumor necrosis factor (TNF), interleukin 1 (IL-1), IL-6, IL-8, endothelin 1, and platelet-activating factor are all increased in patients that develop NEC.50, 51, 52
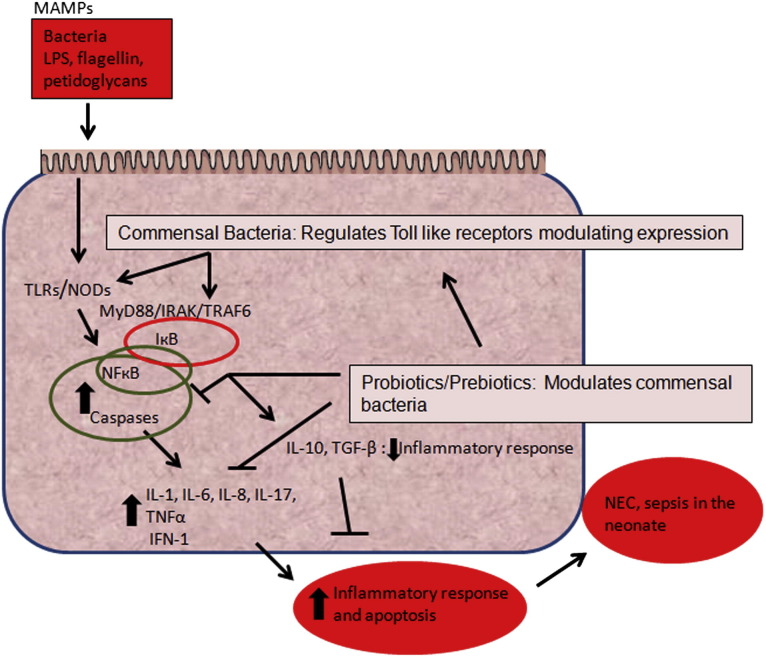
The intestinal flora in the neonate have important functions in metabolism, nutrition, immune system, and defense against pathogens.53, 54 The immune system is developmentally regulated; therefore, in preterm babies, this immune system is still immature. Normally, it recognizes and fights harmful bacteria but does not react against healthy species of bacteria.55, 56 To avoid triggering pathologic immune responses, commensal bacteria must not express key virulence factors. For example, LPS is pentacylated in Bacteroides species, which are abundant in the intestinal flora of breast-fed, healthy infants. This modification makes lipid A in commensal Bacteroides a poor agonist of TLRs.57 Conversely, lipid A of Proteobacteria is a potent agonist of TLR-4 and more likely to elicit an immune response because of its hexacylated characteristics.58, 59 The intestinal mucosa has mechanisms of recognition of bacterial products via highly specialized pattern recognition receptors called TLRs, which recognize specific microbial-associated molecular patterns (MAMPs).60 Each MAMP has its own specific TLR.61 For example, LPS, which acts as the endotoxin for gram-negative bacteria, is recognized by TLR-4. Flagellin is recognized by TLR-5, whereas peptidoglycan and lipotechoic acid from gram-positive bacteria are detected by TLR-2. TLR expression seems to be regulated by patterns of intestinal colonization, and therefore, abnormal patterns of colonization can trigger inappropriate responses. These MAMPs activate the specific TLR that leads to the activation of nuclear factor κ B (NF-κB) and caspases, which in turn activate transcription genes and induce cytokines such as IL-1, IL-6, IL-8, TNF-α, and interferon (IFN-1) (Fig. 4 ). Commensal healthy bacteria are believed to regulate this intestinal inflammatory response.60, 62 For example, studies have shown that Lactobacillus protects against cytokine-mediated cell injury.63 Bifidobacterium, Bacteroides thetaiotaomicron, and Streptococcus thermophilus are examples of other bacteria that play a role in cytoprotection of the enterocyte and intestinal mucosa.64 Many of these bacteria have been studied as therapeutic agents in the treatment of NEC, with promising results. Several studies, including a recent meta-analysis of the literature,65, 66 showed that the incidence of NEC was decreased in infants treated with probiotics containing different combination of these beneficial bacteria. Probiotics seems to help maintain a normal intestinal flora in neonates, thereby decreasing the incidence of NEC. Current evidence led to a recommendation level B in 2011 of effectiveness (recommendation is based on positive-controlled studies, but 1 negative study did not support the primary outcome).67 Others68, 69 have suggested that dead microbes may be as effective as live microbes in modulating excessive inflammatory stimuli (see Fig. 4).
Fig. 4.
Simplified TLR signaling leading to NF-κB activation, inflammatory response, and possible NEC. Surface enterocytes can recognize MAMPs via TLRs. Each of these receptors recognizes a specific bacterial product. For example, TLR-2 recognizes products of gram-positive bacteria, TLR-4 recognizes LPS from gram-negative bacteria, and TLR-5 recognizes flagellin. Cell stimulation signals recruitment of MyD88, IRAK, and TRAF6, then triggers activation of IκB. NF-κB activates the transcription of genes, including cytokines and chemokines. IκB: inhibitor of kappa B.
There are many opportunities for the physicians taking care of neonates to provide interventions that improve the adequate colonization of the neonatal gut and to minimize the deleterious but often unavoidable effects of medical treatment.
Intestinal microbiota bacterial translocation and other risk factors for NEC
Bacterial translocation has been suggested as one of the mechanisms for developing NEC.70, 71 Bacterial translocation is not restricted only to the invasion of intestinal bacteria but can also include bacterial toxins or antigens that damage the intestinal epithelia and enter the circulation, resulting in a systemic inflammatory response.72 During inflammation, production of nitric oxide alters expression and localization of the tight junction. Disruption of the tight junction zonulin proteins ZO-1, ZO-2, ZO-3, and occludin leads to intestinal permeability and bacterial translocation.73 Few studies have shown the relationship between bacterial translocation and NEC. In 1 study74 of bloodstream infections (BSIs) in very-low-birth-weight infants, BSI was more common after diagnosis of NEC in very-low-birth-weight infants who developed intestinal failure (60%) compared with those with surgical NEC without intestinal failure (42%) and those with medical NEC (20%). Coagulase-negative Staphylococcus and Klebsiella were the most frequently identified microorganisms. Alteration in the intestinal permeability was believed to be one of the mechanisms of BSI. Causation is difficult to establish. Nonculture-based analyses of intestinal microbiota have given us some indication of the alterations in diversity and intestinal flora composition of those infants who develop NEC.
However, what causes bacterial translocation? Bacterial translocation is believed to be secondary to abnormal intestinal colonization. Risk factors for bacterial translocation are discussed elsewhere in this article. Other factors have been associated with increased risk of NEC probably secondary to bacterial translocation. Histamine 2 (H2) blockers, which are sometimes used in NICUs, have been shown to increase the risk of sepsis and meningitis.75 It seems that increasing the gastric pH in preterm infants predisposes the neonate to bacterial translocation and infection.76 Guillet and colleagues75 showed in their study that those infants less than 1500 g treated with H2 blocker were more likely to develop NEC. In a study performed several years ago, Carrion and Egan77 reported that by maintaining an acidic gastric pH less than 3.0, the incidence of NEC decreased and higher gastric enteric bacterial colony counts were strongly correlated with gastric pH level of more than 4.
References
- 1.Obladen M. Necrotizing enterocolitis–150 years of fruitless search for the cause. Neonatology. 2009;96(4):203–210. doi: 10.1159/000215590. [DOI] [PubMed] [Google Scholar]
- 2.Neu J., Walker W.A. Necrotizing enterocolitis. N Engl J Med. 2011;364(3):255–264. doi: 10.1056/NEJMra1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin Patricia W., Stoll Barbara J. Necrotising enterocolitis. Lancet. 2006;368(9543):1271–1283. doi: 10.1016/S0140-6736(06)69525-1. [DOI] [PubMed] [Google Scholar]
- 4.Young C.M., Kingma S.D., Neu J. Ischemia-reperfusion and neonatal intestinal injury. J Pediatr. 2011;158(2 Suppl):e25–e28. doi: 10.1016/j.jpeds.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Claud E.C., Walker W.A. Hypothesis: inappropriate colonization of the premature intestine can cause neonatal necrotizing enterocolitis. FASEB J. 2001;15(8):1398–1403. doi: 10.1096/fj.00-0833hyp. [DOI] [PubMed] [Google Scholar]
- 6.Sántulli T.V., Schullinger J.N., Heird W.C. Acute necrotizing enterocolitis in infancy: a review of 64 cases. Pediatrics. 1975;55(3):376–387. [PubMed] [Google Scholar]
- 7.Falkow S. Molecular Koch's postulates applied to bacterial pathogenicity–a personal recollection 15 years later. Nat Rev Microbiol. 2004;2(1):67–72. doi: 10.1038/nrmicro799. [DOI] [PubMed] [Google Scholar]
- 8.Resta S., Luby J.P., Rosenfeld C.R. Isolation and propagation of a human enteric coronavirus. Science. 1985;229(4717):978–981. doi: 10.1126/science.2992091. [DOI] [PubMed] [Google Scholar]
- 9.Neu J., Mshvildadze M., Mai V. A roadmap for understanding and preventing necrotizing enterocolitis. Curr Gastroenterol Rep. 2008;10(5):450–457. doi: 10.1007/s11894-008-0084-x. [DOI] [PubMed] [Google Scholar]
- 10.Grave G.D., Nelson S.A., Walker W.A. New therapies and preventive approaches for necrotizing enterocolitis: report of a research planning workshop. Pediatr Res. 2007;62(4):510–514. doi: 10.1203/PDR.0b013e318142580a. [DOI] [PubMed] [Google Scholar]
- 11.Ben-Amor K., Heilig H., Smidt H. Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl Environ Microbiol. 2005;71(8):4679–4689. doi: 10.1128/AEM.71.8.4679-4689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leblond-Bourget N., Philippe H., Mangin I. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int J Syst Bacteriol. 1996;46(1):102–111. doi: 10.1099/00207713-46-1-102. [DOI] [PubMed] [Google Scholar]
- 13.Rondon M.R., August P.R., Bettermann A.D. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol. 2000;66(6):2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venter J.C., Remington K., Heidelberg J.F. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304(5667):66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 15.Turnbaugh P.J., Quince C., Faith J.J. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A. 2010;107(16):7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosalbes M.J., Durban A., Pignatelli M. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One. 2011;6(3):e17447. doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zoetendal E.G., Raes J., van den Bogert B. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6(7):1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Booijink C.C., Boekhorst J., Zoetendal E.G. Metatranscriptome analysis of the human fecal microbiota reveals subject-specific expression profiles, with genes encoding proteins involved in carbohydrate metabolism being dominantly expressed. Appl Environ Microbiol. 2010;76(16):5533–5540. doi: 10.1128/AEM.00502-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verberkmoes N.C., Russell A.L., Shah M. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3(2):179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 20.Hooper L.V., Gordon J.I. Commensal host-bacterial relationships in the gut. Science. 2001;292(5519):1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 21.Forsythe P., Bienenstock J. Immunomodulation by commensal and probiotic bacteria. Immunol Invest. 2010;39(4–5):429–448. doi: 10.3109/08820131003667978. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Hoenig J.D., Malin K.J. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3(8):944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mai V., Young C.M., Ukhanova M. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS One. 2011;6(6):e20647. doi: 10.1371/journal.pone.0020647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiGiulio D.B., Gervasi M.T., Romero R. Microbial invasion of the amniotic cavity in pregnancies with small-for-gestational-age fetuses. J Perinat Med. 2010;38(5):495–502. doi: 10.1515/JPM.2010.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nanthakumar N.N., Fusunyan R.D., Sanderson I. Inflammation in the developing human intestine: a possible pathophysiologic contribution to necrotizing enterocolitis. Proc Natl Acad Sci U S A. 2000;97(11):6043–6048. doi: 10.1073/pnas.97.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medzhitov R., Schneider D.S., Soares M.P. Disease tolerance as a defense strategy. Science. 2012;335(6071):936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harmsen H.J., Wildeboer-Veloo A.C., Raangs G.C. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30(1):61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 28.Jernberg C., Lofmark S., Edlund C. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1(1):56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 29.Gronlund M.M., Lehtonen O.P., Eerola E. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr. 1999;28(1):19–25. doi: 10.1097/00005176-199901000-00007. [DOI] [PubMed] [Google Scholar]
- 30.Neu J., Rushing J. Cesarean versus vaginal delivery: long-term infant outcomes and the hygiene hypothesis. Clin Perinatol. 2011;38(2):321–331. doi: 10.1016/j.clp.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominguez-Bello M.G., Blaser M.J., Ley R.E. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140(6):1713–1719. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominguez-Bello M.G., Costello E.K., Contreras M. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan S., Schanler R.J., Kim J.H. An exclusively human milk-based diet is associated with a lower rate of necrotizing enterocolitis than a diet of human milk and bovine milk-based products. J Pediatr. 2010;156(4):562–567.e1. doi: 10.1016/j.jpeds.2009.10.040. [DOI] [PubMed] [Google Scholar]
- 34.Barlow B., Santulli T.V., Heird W.C. An experimental study of acute neonatal enterocolitis–the importance of breast milk. J Pediatr Surg. 1974;9(5):587–595. doi: 10.1016/0022-3468(74)90093-1. [DOI] [PubMed] [Google Scholar]
- 35.Lucas A., Cole T.J. Breast milk and neonatal necrotising enterocolitis. Lancet. 1990;336(8730):1519–1523. doi: 10.1016/0140-6736(90)93304-8. [DOI] [PubMed] [Google Scholar]
- 36.Hanson L.A., Korotkova M., Telemo E. Breast-feeding, infant formulas, and the immune system. Ann Allergy Asthma Immunol. 2003;90(6 Suppl 3):59–63. doi: 10.1016/s1081-1206(10)61662-6. [DOI] [PubMed] [Google Scholar]
- 37.Ward R.E., Ninonuevo M., Mills D.A. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72(6):4497–4499. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ninonuevo M.R., Park Y., Yin H. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54(20):7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 39.Adlerberth I., Lindberg E., Aberg N. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res. 2006;59(1):96–101. doi: 10.1203/01.pdr.0000191137.12774.b2. [DOI] [PubMed] [Google Scholar]
- 40.Bjorkstrom M.V., Hall L., Soderlund S. Intestinal flora in very low-birth weight infants. Acta Paediatr. 2009;98(11):1762–1767. doi: 10.1111/j.1651-2227.2009.01471.x. [DOI] [PubMed] [Google Scholar]
- 41.Le Huerou-Luron I., Blat S., Boudry G. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev. 2010;23(1):23–36. doi: 10.1017/S0954422410000065. [DOI] [PubMed] [Google Scholar]
- 42.Penders J., Thijs C., Vink C. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 43.Section on Breastfeeding Breastfeeding and the use of human milk. Pediatrics. 2012;129(3):e827–e841. doi: 10.1542/peds.2011-3552. [DOI] [PubMed] [Google Scholar]
- 44.Kuppala V.S., Meinzen-Derr J., Morrow A.L. Prolonged initial empirical antibiotic treatment is associated with adverse outcomes in premature infants. J Pediatr. 2011;159(5):720–725. doi: 10.1016/j.jpeds.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexander V.N., Northrup V., Bizzarro M.J. Antibiotic exposure in the newborn intensive care unit and the risk of necrotizing enterocolitis. J Pediatr. 2011;159(3):392–397. doi: 10.1016/j.jpeds.2011.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weintraub A.S., Ferrara L., Deluca L. Antenatal antibiotic exposure in preterm infants with necrotizing enterocolitis. J Perinatol. 2012;32(9):705–709. doi: 10.1038/jp.2011.180. [DOI] [PubMed] [Google Scholar]
- 47.Goldmann D.A., Leclair J., Macone A. Bacterial colonization of neonates admitted to an intensive care environment. J Pediatr. 1978;93(2):288–293. doi: 10.1016/s0022-3476(78)80523-x. [DOI] [PubMed] [Google Scholar]
- 48.Jakobsson H.E., Jernberg C., Andersson A.F. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5(3):e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cotten C.M., Taylor S., Stoll B. Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics. 2009;123(1):58–66. doi: 10.1542/peds.2007-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caplan M.S., MacKendrick W. Necrotizing enterocolitis: a review of pathogenetic mechanisms and implications for prevention. Pediatr Pathol. 1993;13(3):357–369. doi: 10.3109/15513819309048223. [DOI] [PubMed] [Google Scholar]
- 51.Caplan M.S., Sun X.M., Hseuh W. Role of platelet activating factor and tumor necrosis factor-alpha in neonatal necrotizing enterocolitis. J Pediatr. 1990;116(6):960–964. doi: 10.1016/s0022-3476(05)80661-4. [DOI] [PubMed] [Google Scholar]
- 52.Edelson M.B., Bagwell C.E., Rozycki H.J. Circulating pro- and counterinflammatory cytokine levels and severity in necrotizing enterocolitis. Pediatrics. 1999;103(4 Pt 1):766–771. doi: 10.1542/peds.103.4.766. [DOI] [PubMed] [Google Scholar]
- 53.Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murgas Torrazza R., Neu J. The developing intestinal microbiome and its relationship to health and disease in the neonate. J Perinatol. 2011;31(Suppl 1):S29–S34. doi: 10.1038/jp.2010.172. [DOI] [PubMed] [Google Scholar]
- 55.Rook G.A. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: Darwinian medicine and the ‘hygiene’ or ‘old friends’ hypothesis. Clin Exp Immunol. 2010;160(1):70–79. doi: 10.1111/j.1365-2249.2010.04133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Macpherson A.J., Harris N.L. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4(6):478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 57.Coats S.R., Do C.T., Karimi-Naser L.M. Antagonistic lipopolysaccharides block E. coli lipopolysaccharide function at human TLR4 via interaction with the human MD-2 lipopolysaccharide binding site. Cell Microbiol. 2007;9(5):1191–1202. doi: 10.1111/j.1462-5822.2006.00859.x. [DOI] [PubMed] [Google Scholar]
- 58.Sansonetti P.J., Medzhitov R. Learning tolerance while fighting ignorance. Cell. 2009;138(3):416–420. doi: 10.1016/j.cell.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 59.Munford R.S., Varley A.W. Shield as signal: lipopolysaccharides and the evolution of immunity to gram-negative bacteria. PLoS Pathog. 2006;2(6):e67. doi: 10.1371/journal.ppat.0020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma R., Young C., Neu J. Molecular modulation of intestinal epithelial barrier: contribution of microbiota. J Biomed Biotechnol. 2010;2010:305879. doi: 10.1155/2010/305879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rhee S.H. Basic and translational understandings of microbial recognition by toll-like receptors in the intestine. J Neurogastroenterol Motil. 2011;17(1):28–34. doi: 10.5056/jnm.2011.17.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caplan M.S. Probiotic and prebiotic supplementation for the prevention of neonatal necrotizing enterocolitis. J Perinatol. 2009;29(Suppl 2):S2–S6. doi: 10.1038/jp.2009.21. [DOI] [PubMed] [Google Scholar]
- 63.Tien M.T., Girardin S.E., Regnault B. Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J Immunol. 2006;176(2):1228–1237. doi: 10.4049/jimmunol.176.2.1228. [DOI] [PubMed] [Google Scholar]
- 64.Otte J.M., Podolsky D.K. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am J Physiol Gastrointest Liver Physiol. 2004;286(4):G613–G626. doi: 10.1152/ajpgi.00341.2003. [DOI] [PubMed] [Google Scholar]
- 65.Lin H.C., Hsu C.H., Chen H.L. Oral probiotics prevent necrotizing enterocolitis in very low birth weight preterm infants: a multicenter, randomized, controlled trial. Pediatrics. 2008;122(4):693–700. doi: 10.1542/peds.2007-3007. [DOI] [PubMed] [Google Scholar]
- 66.Deshpande G., Rao S., Patole S. Updated meta-analysis of probiotics for preventing necrotizing enterocolitis in preterm neonates. Pediatrics. 2010;125(5):921–930. doi: 10.1542/peds.2009-1301. [DOI] [PubMed] [Google Scholar]
- 67.Floch M.H., Walker W.A., Madsen K. Recommendations for probiotic use-2011 update. J Clin Gastroenterol. 2011;45(Suppl):S168–S171. doi: 10.1097/MCG.0b013e318230928b. [DOI] [PubMed] [Google Scholar]
- 68.Zhang L., Li N., Caicedo R. Alive and dead Lactobacillus rhamnosus GG decrease tumor necrosis factor-alpha-induced interleukin-8 production in Caco-2 cells. J Nutr. 2005;135(7):1752–1756. doi: 10.1093/jn/135.7.1752. [DOI] [PubMed] [Google Scholar]
- 69.Zhang L., Li N., des Robert C. Lactobacillus rhamnosus GG decreases lipopolysaccharide-induced systemic inflammation in a gastrostomy-fed infant rat model. J Pediatr Gastroenterol Nutr. 2006;42(5):545–552. doi: 10.1097/01.mpg.0000221905.68781.4a. [DOI] [PubMed] [Google Scholar]
- 70.Sherman M.P. New concepts of microbial translocation in the neonatal intestine: mechanisms and prevention. Clin Perinatol. 2010;37(3):565–579. doi: 10.1016/j.clp.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hunter C.J., Upperman J.S., Ford H.R. Understanding the susceptibility of the premature infant to necrotizing enterocolitis (NEC) Pediatr Res. 2008;63(2):117–123. doi: 10.1203/PDR.0b013e31815ed64c. [DOI] [PubMed] [Google Scholar]
- 72.Gatt M., Reddy B.S., MacFie J. Review article: bacterial translocation in the critically ill–evidence and methods of prevention. Aliment Pharmacol Ther. 2007;25(7):741–757. doi: 10.1111/j.1365-2036.2006.03174.x. [DOI] [PubMed] [Google Scholar]
- 73.Anand R.J., Leaphart C.L., Mollen K.P. The role of the intestinal barrier in the pathogenesis of necrotizing enterocolitis. Shock. 2007;27(2):124–133. doi: 10.1097/01.shk.0000239774.02904.65. [DOI] [PubMed] [Google Scholar]
- 74.Cole C.R., Hansen N.I., Higgins R.D. Bloodstream infections in very low birth weight infants with intestinal failure. J Pediatr. 2012;160(1):54–59.e2. doi: 10.1016/j.jpeds.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guillet R., Stoll B.J., Cotten C.M. Association of H2-blocker therapy and higher incidence of necrotizing enterocolitis in very low birth weight infants. Pediatrics. 2006;117(2):e137–e142. doi: 10.1542/peds.2005-1543. [DOI] [PubMed] [Google Scholar]
- 76.Canani R.B., Terrin G. Gastric acidity inhibitors and the risk of intestinal infections. Curr Opin Gastroenterol. 2010;26(1):31–35. doi: 10.1097/MOG.0b013e328333d781. [DOI] [PubMed] [Google Scholar]
- 77.Carrion V., Egan E.A. Prevention of neonatal necrotizing enterocolitis. J Pediatr Gastroenterol Nutr. 1990;11(3):317–323. doi: 10.1097/00005176-199010000-00006. [DOI] [PubMed] [Google Scholar]