Abstract
Multiple homeodomain transcription factors are crucial for pituitary organogenesis and cellular differentiation. A homeodomain repressor, Msx1, is expressed from the ventral aspect of the developing anterior pituitary and implicated in gonadotrope differentiation. Here, we find that Msx1 represses transcription of lineage-specific pituitary genes such as the common α-glycoprotein subunit (αGSU) and GnRH receptor (GnRHR) promoters in the mouse gonadotrope-derived cell lines, αT3-1 and LβT2. Repression of the mouse GnRHR promoter by Msx1 is mediated through a consensus-binding motif in the downstream activin regulatory element (DARE). Truncation and mutation analyses of the human αGSU promoter map Msx1 repression to a site at −114, located at the junctional regulatory element (JRE). Dlx activators are closely related to the Msx repressors, acting through the same elements, and Dlx3 and Dlx2 act as transcriptional activators for GnRHR and αGSU, respectively. Small interfering RNA knockdown of Msx1 in αT3-1 cells increases endogenous αGSU and GnRHR mRNA expression. Msx1 gene expression reaches its maximal expression at the rostral edge at e13.5. The subsequent decline in Msx1 expression specifically coincides with the onset of expression of both αGSU and GnRHR. The expression levels of both αGSU and GnRHR in Msx1-null mice at e18.5 are higher compared with wild type, further confirming a role for Msx1 in the repression of αGSU and GnRHR. In summary, Msx1 functions as a negative regulator early in pituitary development by repressing the gonadotrope-specific αGSU and GnRHR genes, but a temporal decline in Msx1 expression alleviates this repression allowing induction of GnRHR and αGSU, thus serving to time the onset of gonadotrope-specific gene program.
After closure of Rathke's pouch on embryonic day (e)12 in the mouse, 5 endocrine cell types arise in a specific spatial and temporal pattern giving rise to the anterior pituitary (1, 2). The gonadotrope, which regulates reproduction through the synthesis and secretion of LH and FSH, arises last during pituitary organogenesis. Gonadotrope emergence can be traced throughout pituitary development by the sequential appearance of earlier markers of the lineage, first the common α-subunit of LH and FSH (αGSU) at e11.5, followed by the nuclear receptor steroidogenic factor (SF)1, then GnRH receptor (GnRHR), and finally, the appearance of the LHβ and FSHβ subunits on e16.5 and e17.5, respectively.
Molecular genetic investigation of the regulation of gonadotrope gene expression is greatly facilitated by the use of cultured cell lines that represent differentiated cell types (3–6). The αT1-1 cell line represents a precursor to the gonadotrope-thyrotrope lineages (4) and expresses only one glycoprotein hormone gene, αGSU (7). The immature gonadotrope αT3-1 cell line expresses both αGSU and GnRHR, and the mature gonadotrope LβT2 cell line expresses the 4 gonadotrope-specific genes αGSU, GnRHR, LHβ, and FSHβ (4, 8, 9). The availability of these cultured cell models, complemented by genetically modified mice in vivo, has made it possible to study pituitary specificity of the gonadotropin genes and the molecular basis of gonadotrope maturation during development. Moreover, although transfection of the αGSU promoter provides expression in all 3 cell lines, the GnRHR promoter is only expressed in αT3-1 and LβT2 cells (10). Transfection of the LHβ and FSHβ promoters into αT3-1 cells fails to promote basal or hormone-regulated expression, whereas transfection of these promoters into LβT2 cells reflects both tissue-specific and hormone-regulated expression (8). These unique cell lines serve as model systems for investigating the molecular mechanisms of gonadotrope differentiation, because they express the known tissue-specific regulators of the 4 gonadotrope-specific differentiated target genes, including SF1, Lhx3, Pitx1, Runx, FoxL2, and GATA2 (10–19). These tissue-specific transcription factors play direct roles in regulating the transcription of the gonadotrope-specific target genes, yet the coordinated program of gonadotrope maturation remains to be elucidated (20, 21).
Msx1 is a transcription factor that is expressed in the ventral aspect of the developing anterior pituitary, regulated by bone morphogenetic protein, and implicated in gonadotrope differentiation (2, 22). It is a member of the Antennapedia class of Q50 non-Hox homeodomain transcription factors that regulate gene expression and influence development of craniofacial structures and the anterior forebrain (23, 24). The mammalian Msx gene family consists of 3 genes, each on a different chromosome: Msx1, Msx2, and Msx3. In developing vertebrate embryos, Msx1 and Msx2 are widely expressed in many organs, whereas Msx3 is only expressed in the dorsal neural tube. Neither Msx2 nor Msx3 is expressed during anterior pituitary development (25). In vivo, Msx1 is expressed at sites of epithelial-mesenchymal interaction during embryogenesis (23, 26), and Msx1-null mice are perinatal lethal with defects in craniofacial development, cleft palate, and tooth agenesis (23, 27). Msx1 expression is detected in Rathke's pouch starting on e10.5, but by e12.5, expression is more regionalized in areas destined to become the pars distalis, a region pituitary of the highly enriched in gonadotrope precursors (22).
Dlx homeodomain proteins are closely related to the Msx family and have overlapping but distinct expression patterns during vertebrate development. Both Msx and Dlx genes are required during the early, middle, and late phases of development, where their differential expression mediates patterning, morphogenesis, and histogenesis of tissues. Members of the Msx family are transcriptional repressors, whereas Dlx family members activate transcription (24, 28–32). Msx and Dlx bind to a common DNA motif (C/GTAATTG), and heterodimerization and/or competitive binding results in functional antagonism (30). The ability to oppose each other's transcriptional actions implies a mechanism underlying the complementary or antagonistic functions of Msx and Dlx proteins during development. For example, in the hypothalamus, spatiotemporal expression patterns of the Msx and Dlx family members largely coincide with the migratory route of GnRH neurons and coexpress with GnRH in neurons during embryonic development (33). Msx and Dlx1/2 can bind directly to CAATTA repeated elements in the rat GnRH promoter and enhancer, and in vivo, Msx1-null mice have increased numbers of GnRH neurons, and Dlx1/2-double-null mice have reduced numbers of GnRH neurons in regions where these factors are expressed.
Transducin-like enhancer of split (TLE) homologues control many embryonic and postembryonic processes, such as differentiation, cell specification, embryonic patterning, and apoptosis (34–37). TLE corepressors lack a DNA-binding domain but are tethered to cis-acting regulatory elements via protein-protein interactions, where they repress transcription (38). For example, TLEs interact with Msx1 to repress GnRH transcription in hypothalamic GnRH neurons (39, 40). TLE4 (Grg4) is recruited by repressor Msx1 for a dynamic switch between activation and repression of GnRH transcription in hypothalamic neurons. TLEs also act as corepressors for other transcription factors during early pituitary development (41–43). However, the role of TLEs in gonadotropes remains to be resolved.
Here, we show that Msx1 represses transcription of both αGSU and GnRHR, the early gonadotrope-specific genes, in the immature gonadotrope cell line, αT3-1. Small interfering RNA (siRNA)-mediated knockdown of Msx1 in vitro and Msx1 deletion in vivo both result in increased αGSU and GnRHR mRNA levels, indicating that Msx1 is capable of temporally regulating expression of the αGSU and GnRHR during gonadotrope development by repressing its expression. We hypothesize that the decline in Msx1 expression during anterior pituitary and gonadotrope development alleviates Msx1-induced repression of GnRHR and αGSU expression and, thus, helps time the onset of the gonadotrope-specific gene program.
Materials and Methods
Materials
The wild-type mouse GnRHR and μDARE (BR15) luciferase reporter constructs were generously provided by Dr Colin Clay (Colorado State University, Ft Collins, Colorado). The mouse FSHβ, rat SF1, and rat LHβ luciferase reporter plasmids have been previously described (15, 44, 45). The wild-type human αGSU and its mutants have been previously described or constructed by QuikChange Site-Directed Mutagenesis kit from Agilent Technologies (Santa Clara, California) (46). The Msx1, Dlx, TLE4 expression vectors, and Msx1 plasmid for in vitro translation have also been previously described (28, 33, 39). Oligonucleotides were obtained from IDT (San Diego, California). DNA-modifying enzymes were obtained from New England Biolabs (Beverly, Massachusetts). siRNA and transfection reagents were obtained from Dharmacon (Lafayette, Colorado). Immunohistochemistry reagents were from Vector Laboratories (Burlingame, California). Anti-Msx1 antibody (MMS-261R) was obtained from Covance (Princeton, New Jersey), anti-β-galactosidase (β-gal) antibody (ab9361) was from Abcam (Cambridge, Massachusetts), and anti-αGSU antiserum was available through the National Hormone and Pituitary Program.
Cell culture, transient transfections, and luciferase assays
Cell cultures were maintained in a humidified atmosphere of 5% CO2 in air at 37°C. αT3-1 cells were maintained in high-glucose DMEM containing 10% fetal bovine serum and 100-U/mL penicillin/streptomycin as previously described (47). Transient transfection overexpression assays were performed according to the FuGENE 6 protocol (Promega, Madison, Wisconsin). A total of 100 ng of TK-β-gal expression vector was transfected as internal control for transfection efficiency. Forty-eight hours after transfection, cells were harvested in lysis buffer (100-mm potassium phosphate [pH 7.8] and 0.2% Triton X-100). Luciferase assays were performed as previously described (33), and β-gal assays were performed using the Galacto-Light Plus assay system as directed by the manufacturer (Applied Biosystems, Foster City, California). All transfections were performed in triplicate and repeated at least 4 times, and values are presented as the mean ± SEM. Data were analyzed using JMP with an asterisk indicating significant differences (P < .05) from empty vector control.
RNA isolation and RT-PCR
Total RNA was harvested and isolated from αT1-1, αT3-1, LβT2, and NIH 3T3 cells using TRIzol reagent (Sigma-Aldrich, St. Louis, Missouri). RNA was quantified and treated to remove DNA with Turbo DNA-free from Ambion (Austin, Texas) according to manufacturer's protocol. Purified RNA was then reverse transcribed with SuperScript III First-Strand (Invitrogen, Carlsbad, California), or mock reverse transcribed as a negative control (-RT), to generate cDNA. Resulting cDNA was subject to 35 cycles of PCR using specific Msx, Dlx, and TLE primers previously described (33, 39), in addition to the coding sequence of β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as controls. Resulting PCR reactions were run on 1.2% agarose gels.
Nuclear extracts and EMSA
Nuclear protein extracts were prepared from αT3-1 cells as previously described. αT3-1 cells were scraped in hypotonic buffer (20mM Tris-HCl [pH 7.4], 10mM NaCl, 1mM MgCl2, 10mM NaF, 0.5mM EDTA, and 0.1mM EGTA) with a protease inhibitor cocktail (Sigma, St Louis, Missouri) and 1mM phenylmethylsulfonyl fluoride and allowed to swell on ice. Cells were lysed by passing them through a 25-gauge needle, and the nuclei were collected by centrifugation. Nuclear proteins were extracted on ice for 30 minutes in hypertonic buffer (20mM HEPES [pH 7.9], 20% glycerol, 420mM KCl, 2mM MgCl2, 10mM NaF, 0.5mM EDTA, and 0.1mM EGTA) with protease inhibitor cocktail (Sigma) and 1mM phenylmethylsulfonylfluoride. Protein concentration was determined using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, California). Oligonucleotide probes are shown in the figures. Msx1 was in vitro translated using a reticulocyte lysate kit according to the manufacturer's protocol (48). Oligonucleotide probes were annealed, end labeled using T4 Polynucleotide Kinase (New England Biolabs) and [γ32P]ATP (7000 Ci/mmol; MP Biomedicals, Solon, Ohio), then purified using G25 Probe Quant columns (Amersham, Piscataway, New Jersey). Binding reactions were carried out using 2 μg of nuclear protein and 4 fmol of labeled oligonucleotide in a 10-μL reaction containing 5mM dithiothreitol, 0.025-μg/μL Poly dIdC, and binding buffer (50mM HEPES [pH 7.8], 250mM KCl, 5mM EDTA, and 30% glycerol). For competition assays, 1000-fold excess of double-stranded unlabeled oligonucleotides were annealed and used in binding reactions. After the addition of probe and nuclear protein, binding reactions were incubated 5 minutes before electrophoresis on a 5% nondenaturing polyacrylamide gel in 0.25× Tris-borate, EDTA. For supershift EMSA assays, 2 μL of anti-Msx1 (MMS-261R; Covance) or normal mouse IgG (sc-2025; Santa Cruz Biotechnology, Inc, Santa Cruz, California) were added to reactions before the addition of nuclear protein and incubated for 10 minutes. Gels were run at 250 V for approximately 2 hours, dried under vacuum, and exposed to autoradiographic film overnight.
Msx1 siRNA knockdown
αT3-1 cells were transfected for 48 hours with 100nM ON-TARGET SMARTpool scrambled control, ON-TARGET plus SMARTpool MSX1 siRNA purchased from Dharmacon. DharmaFECT transfection reagent was used according to manufacturer's protocol.
Total RNA was harvested with RNeasy mini kit (QIAGEN, Germantown, Maryland) according to manufacturer's protocol. Two micrograms of purified RNA were then reverse transcribed with SuperScript III First-Strand (Invitrogen) to generate cDNA. Resulting cDNA was subjected to quantitative PCR (qPCR) using intron-spanning primers for Msx1, GnRHR, αGSU, and GAPDH primers (Msx1-forward 5′-CAGAGTCCCCGCTTCTCC-3′ and Msx1-reverse 5′-GTCTTGTGCTTGCGTAGGG-3′, GnRHR-forward 5′-GCCCCTTGCTGTACAAAGC-3′ and GnRHR-reverse 5′-CCGTCTGCTAGGTAGATCATCC-3′, αGSU-forward 5′-GGTTCCAAAGAATATTACCTCG-3′ and αGSU-reverse 5′-GTCATTCTGGTCATGCTGTCC-3′, and GAPDH-forward 5′-ATGGGTGAAGGTCGGTGTGA-3′ and GAPDH-reverse 5′-GCTTCCCGTTGATGACAAGC-3′). Values represent the average of the square means of 4 independent experiments, and all samples are normalized to GAPDH levels. All values are normalized to scrambled siRNA control and are expressed as mean ± SEM. Means were separated by 1-way ANOVA using Dunnet's Method, and an asterisk denotes values significantly different (P < .05) from scrambled siRNA control.
Western blotting
Cells were washed in ice-cold PBS and lysed in radioimmunoprecipitation assay buffer containing 20mM Tris (pH 8.0), 137mM NaCl, 10% glycerol, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate, 0.5% deoxycholate, and 0.2mM phenylmethylsulfonylfluoride. Protein concentration in lysates was determined by Bradford assay before gel loading to ensure equal protein loading; 6× sample buffer (300mM Tris-HCl [pH 6.8], 60% glycerol, 30mM dithiothreitol, and 6% sodium dodecyl sulfate) was added to yield a final concentration of 1×, lysates were heated to 95°C for 5 minutes. Samples were subjected to SDS-PAGE on a 10% gel (acrylamide:bis-acrylamide ratio of 29:1) and electroblotted to Immobilin (Millipore, Billerica, Massachusetts). Membranes were blocked in 5% BSA in Tris-buffered saline. Anti-Msx1 (MMS-261R; Covance) was incubated overnight at a 1:1000 dilution at 4°C. Blots were washed and then incubated with a 1:10 000 dilution of antimouse horseradish peroxidase for 1 hour at room temperature. GAPDH-horseradish peroxidase (ab9842; Abcam) was used at a 1:5000 dilution as loading control. All blots were washed for 30 minutes (3 × 10 min) with Tris-buffered saline after secondary antibody and then visualized by chemiluminescence using Pierce SuperSignal reagents (Pierce, Rockford, Illinois).
Msx1-Null mice and timed matings
The Msx1-null mice were a generous gift from Dr Robert and have been previously described (33, 49). Animals were maintained under a 12-hour light, 12-hour dark cycle and received food and water ad libitum. All experiments were performed under approval from the University of California San Diego Animal Care and Use Committee and in accordance with the National Institutes of Health Animal Care and Use Guidelines. Screening for Msx1 deletion has previously been described (49). Briefly, genomic DNA was extracted from toe biopsies and analyzed for the presence of the insertion of β-gal into the Msx1 gene by PCR amplification using primers specific to Msx1 and β-gal/LacZ: LacZ-forward 5′-CGACACCACGGCCACCGATATTATTTG-3′ and LacZ-reverse 5′-GGTATTCGCTGGTCACTTCGATGGTT-3′, Msx1-forward 5′-GCGGAATTCTCCAGCTCGCTCAGCCTCA-3′ and Msx1-reverse 5′-TTTGGCCTCTGGTCTCCTTCAGCCTCTA-3′. Heterozygous males and females were used for timed mating experiments. Males and females were paired in the afternoon, and the following morning, females were checked for vaginal plugs with that time designated as e0.5. At the desired stage of embryonic development, females were killed and embryos harvested. Biopsies of embryos were taken and genotyped before embryo fixing.
Immunohistochemistry
Embryos were fixed (10% acetic acid, 30% formaldehyde, and 60% ethanol) overnight at 4°C and dehydrated in ethanol/water washes before embedding in paraffin. Fixed embryos were embedded in paraffin by the University of California San Diego, Histology Core. Embedded whole embryos (e11.5, e13.5, and e15.5) or heads (e17.5) were cut into 10-μm sagittal sections with a microtome and floated onto SuperFrost Plus slides (Fisher Scientific, Auburn, Alabama) and dried overnight at room temperature. Before staining, slides were incubated at 60°C for 30 minutes. Slides were deparaffinized in xylene washes, then rehydrated in ethanol/water washes. Antigen unmasking was performed by boiling for 10 minutes in 10mM sodium citrate. After cooling and washing 2 times in water, endogenous peroxidase was quenched by incubating for 10 minutes in 0.3% hydrogen peroxide. After washing in PBS, slides were blocked in PBS with 5% goat serum and 0.3% Triton X-100 for 45 minutes. Slides were then incubated in anti-β-gal antibody or anti-αGSU antisera diluted 1:1000 in blocking solution overnight at 4°C. After washing 3 times in PBS, biotinylated goat antichicken or goat antirabbit IgG (Vector Laboratories) was diluted 1:300 in PBS for 30 minutes, then was washed 3 times in PBS. The Vectastain ABC elite kit (Vector Laboratories) was used per the manufacturer's instructions and incubated for 30 minutes. After washing, the VIP peroxidase (Msx/β-gal) or DAB (αGSU) kit was used for colorometric staining, and sections were counterstained using methyl green (Vector Laboratories). Slides were dehydrated in an ethyl alcohol series and xylenes and coverslips mounted with Vectamount (Vector Laboratories).
qPCR analysis of embryo pituitary at e18.5
Adult Msx1 heterozygous (+/−) mice were set up in timed matings. On e18.5, the pregnant females were euthanized by carbon dioxide inhalation, and the embryos were extracted. Pituitaries were collected and placed individually in tubes on dry ice. The embryos were genotyped from tail biopsies using primers for the β-gal transgene. The pituitaries were pooled into wild-type and Msx1-null groups. Each group contained 5 individual embryonic pituitaries. The groups were prepared during the RNA extraction process. RNA was isolated from the pituitaries using a QIAshredder and RNeasy mini kit (QIAGEN Sciences, Germantown, Maryland), as directed by the manufacturers. Total RNA was reverse transcribed using an iScript cDNA Synthesis kit (Bio-Rad Laboratories), and cDNA was stored at −20°C. For qPCR, cDNA was diluted 1:10 in water. qPCR was performed using an iQ SYBR Green supermix and an IQ5 real-time PCR machine (Bio-Rad Laboratories). Five genes were analyzed for both the wild-type and null groups: GAPDH as an internal control, as well as LHβ, FSHβ, GnRHR, and αGSU.
Statistical analysis
Transient transfection experiments were repeated independently at least 3 times. Data were normalized for transfection efficiency with luciferase activity relative to β-gal. Data were also normalized to the pGL3 plasmid. In the figures, the error bars represent the SEM. Data were analyzed by 1-way ANOVA followed by Tukey honestly significant difference, or 2-way ANOVA to determine synergy using the statistical package JMP version 9.0 (SAS, Cary, North Carolina). For all analyses, the result was considered significant if P ≤ .05. An asterisk indicates values that are significantly different from the control (P < .05).
Results
Msx1 is expressed in immortalized mouse gonadotrope cell lines
In vivo, Msx1 expression is detected in the ventral aspect of the developing anterior pituitary at e11.5, the approximate spatial and temporal location of immature gonadotropes (22, 25, 50). To address the role of Msx1 during gonadotrope development, we first want to determine the expression patterns of Msx family members in the immortalized gonadotrope cell lines. By using validated Msx1-specific primers (33), RT-PCR analysis was performed in 3 mouse gonadotrope-derived cell lines that represent different developmental stages, including progenitor αT1-1, immature αT3-1, and mature LβT2 cells. Mouse NIH 3T3 cells serve as a negative control and do not express any Msx family members. Msx1 expression has been shown to maintain cells in a mitotically active state by preventing their terminal differentiation (23, 50, 51). Among the 3 gonadotrope cell lines, Msx1 mRNA is highly expressed in αT1-1 and LβT2 cell lines but is somewhat lower in αT3-1 gonadotropes (Figure 1A). Msx2 mRNA is weakly detected at early stages of gonadotropes αT1-1 and αT3-1. Msx3 is barely detected in progenitor αT1-1 cells, consistent with in vivo Msx3 expression, which is confined primarily to the developing neural tube (Figure 1A) (50). All RT-PCR results were further validated by quantitative RNA-seq (Whole Transcriptome RNA Sequencing) analysis on gonadotrope cell lines (data not shown). The presence of Msx1 in the immature gonadotrope-derived cell lines suggests that Msx1 may play a role on transcription of gonadotrope-specific genes.
Figure 1.
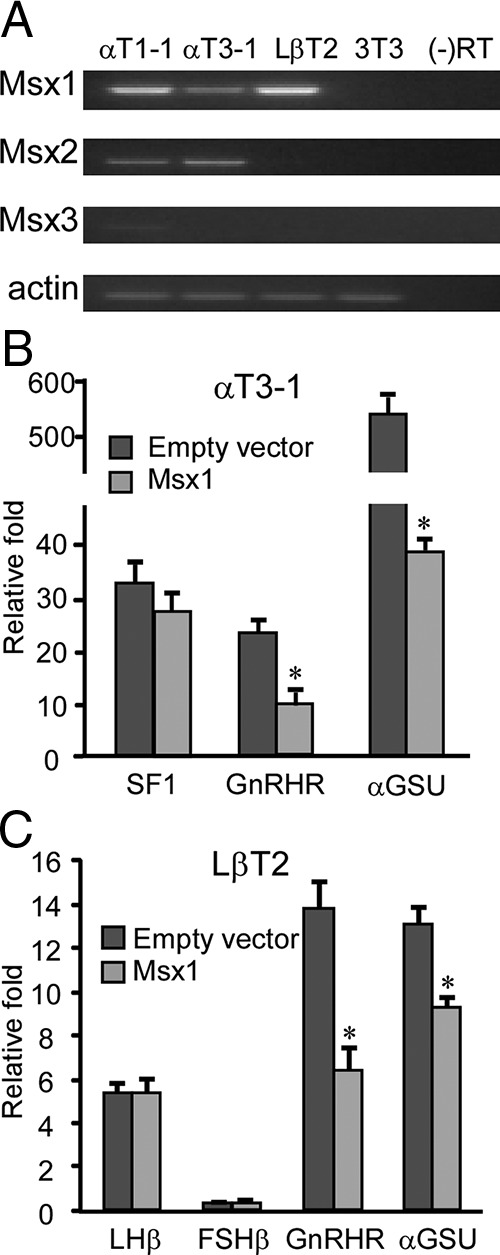
Msx1 represses transcription of GnRHR and αGSU promoters in gonadotropes. (A) Msx mRNA expression in the gonadotrope-derived cell lines. Total RNA was harvested from the mouse gonadotrope-derived cell lines: progenitor αT1-1, immature αT3-1, mature LβT2, and NIH 3T3 cells as control, then reverse transcribed to generate cDNA. β-Actin was used as loading control. (B) The wild-type rat SF1, human αGSU, and mouse GnRHR-luciferase reporters were cotransfected with Msx1 expression vector or an equal mass of empty expression vector into αT3-1 cells. (C) The wild-type FSHβ, LHβ, human αGSU, mouse GnRHR, and pGL3 luciferase reporters were cotransfected with Msx1 expression vector or an equal mass of empty expression vector in LβT2 cells. Abbreviation: RT, reverse transcriptase. An asterisk indicates significant differences (P < .05) from empty vector control.
Overexpression of Msx1 represses activity of αGSU and GnRHR transcription
Sequence analysis shows that the mouse GnRHR gene promoter contains a consensus Msx-binding site (CTAATTG) located in the downstream activin regulatory element (DARE) from −352 to −346, whereas the human αGSU promoter contains several partial Msx motifs. Conversely, the promoters of SF1, an early gonadotrope-specific transcription factor, LHβ, and FSHβ have no putative Msx consensus sites. Therefore, we first tested whether Msx1 represses activity of the αGSU and GnRHR-luciferase reporters in αT3-1 cells. Overexpression of Msx1 causes an approximately 60% and 90% decrease in the GnRHR and αGSU-luciferase reporter activity, respectively, in αT3-1 cells, indicating that Msx1 may play a role in repressing expression of both genes during early gonadotrope differentiation (Figure 1B) (52). This repression is specific, because the rat SF1 promoter is not affected by Msx1 overexpression. Msx1 mRNA is highly expressed in LβT2 cells (Figure 1A), and overexpression of Msx1 also represses transcription of both promoters in this more mature gonadotrope cell line, although Msx1 repression of the αGSU promoter is not as robust as in αT3-1 cells. However, on transfected reporters of LHβ and FSHβ, the 2 late gonadotrope-specific genes expressed only in LβT2 cells (but not αT3-1-1 cells), Msx1 has no inhibitory effect (Figure 1C), although, as shown previously, basal levels of FSHβ-luciferase reporters is very low (12, 13). Interestingly, the decrease in Msx1 expression in the developing pituitary coincides with the onset of expression of GnRHR and αGSU but precedes expression of the LHβ and FSHβ subunits, which do not contain Msx-binding motifs. Thus, Msx1 overexpression represses activity of the gonadotrope-specific αGSU and GnRHR promoters, indicating that Msx1 may play a role in regulating gene expression primarily in early gonadotropes.
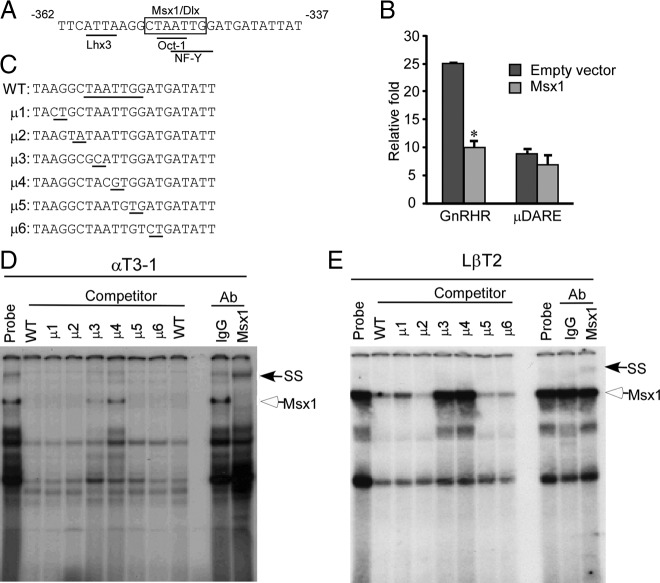
Repression of mouse GnRHR transcription by Msx1 localizes to a consensus Msx-binding site in the DARE element
The DARE element is a composite regulatory element capable of binding Lhx3, Oct-1, and NF-Y (10, 45, 53), including 2 TAAT repeats, the core binding domain for homeodomain proteins and a CCAAT box in the antisense strand. Because the GnRHR promoter contains a consensus Msx-binding site in the DARE element (Figure 2A) (10, 45, 53), we hypothesized that this site mediates Msx1 repression of the mouse GnRHR-luciferase reporter. To test this hypothesis, the wild-type mouse GnRHR-luciferase promoter with or without a 8-base pair (bp) mutation in the DARE element (μDARE) (45) was transiently transfected into αT3-1 cells. Basal transcription of μDARE was lower than that of the wild-type promoter, consistent with the previous findings that the DARE element is responsible for basal transcriptional regulation of GnRHR gene (Figure 2B). Overexpression of Msx1 repressed the wild-type mouse GnRHR-luciferase reporter but failed to repress luciferase activity of the mutant DARE reporter, signifying the critical nature of this DARE element for Msx1-induced repression (Figure 2B). Consequently, Msx1 repression of the mouse GnRHR promoter localizes to the DARE element, which contains a consensus Msx-binding site that may be critical for timing expression of GnRHR expression during gonadotrope development.
Figure 2.
Msx1 represses GnRHR promoter through DARE element. (A) Schematic of DARE element on mouse GnRHR promoter. Box indicates the putative Msx1/Dlx-binding site. The Lhx3, Oct-1, and NF-Y sites on the DARE element are underlined. (B) Msx1 represses GnRHR transcription through DARE element. The wild-type GnRHR and DARE mutant (μDARE) reporters were transiently transfected into αT3-1 cells. The pGL3 empty luciferase reporter was used as control. Cells were cotransfected with Msx1 expression vector or an equal mass of empty expression vector. (C) The oligonucleotides used as the DARE probe and its mutants (μ1–μ6) are listed, and the Msx1-binding element and the 2-bp transversions are underlined. The binding of Msx1 on the GnRHR promoter maps to the DARE element using αT3-1 (D) and LβT2 (E) gonadotrope nuclear extracts. Specific protein/DNA complexes were identified by competition with 1000-fold excess unlabeled oligonucleotides, including homologous oligonucleotides or ones containing a defined Msx1-binding site from the rat GnRH promoter (positive control) or the 2-bp transversions. Confirmation of the presence of Msx1 in specific complexes was determined by inclusion of an antibody directed against Msx1 or mouse IgG as a control. The hollow arrow marks the Msx1 complex, and the arrow marks the supershift of the Msx1 complex after binding with Msx1 antibody. SS represents supershift. Abbreviations: WT, wild type; Ab, antibody.
The consensus Msx-binding site in the DARE element shares the same core TAAT motif with the Oct-1-binding site and 4 of 5 nucleotides with the NF-Y-binding site in the antisense strand (Figure 2A). To investigate whether Msx1 can directly bind to the DARE sequence, αT3-1 nuclear extract was incubated with excess wild-type mouse GnRHR DARE oligonucleotide probes and competed with unlabeled oligonucleotides containing 2-bp transversion mutations, μ1–μ6, that scanned the consensus Msx-binding site (Figure 2C). Interestingly, only the 2-bp mutations that eliminated the TAAT motif of the Msx1 consensus sequence (C/GTAATTG) failed to compete (μ3 and μ4) in both αT3-1 and LβT2 nuclear extracts (Figure 2, D and E). Inclusion of an antibody directed against Msx1 resulted in the formation of a new supershift complex of markedly reduced mobility demonstrating the highly specific binding of Msx1 to the mouse DARE probe in both immature and mature gonadotrope cell lines (Figure 2, D and E). To avoid the overlapping transcription factors, such as Oct-1 and NF-Y, binding on the same region, instead of αT3-1 nuclear extract, we also performed EMSA using the in vitro-translated Msx1, and the results are consistent with αT3-1 nuclear extract showing that Msx1 binding requires the TAAT motif (data not shown). These results lead to the conclusion that transcription of the mouse GnRHR promoter is regulated by Msx1, and this repression maps to a consensus Msx-binding motif in the DARE element.
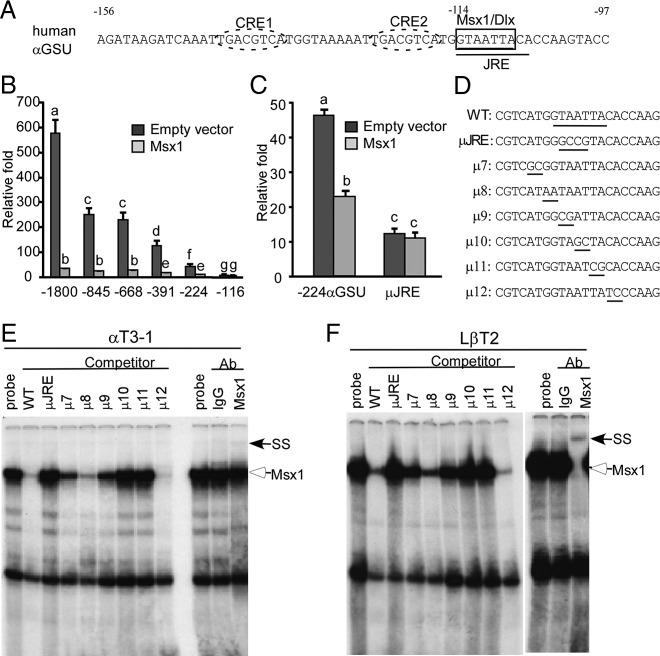
Repression of human αGSU-luciferase promoter activity by Msx1 is mediated by an Msx-binding site overlapping the junctional regulatory element (JRE)
Transcription of the other early gonadotrope-specific gene, αGSU, is also regulated by Msx1 (Figure 1B). In contrast to the GnRHR promoter, the human αGSU promoter has no full consensus Msx1 response element, but it does have multiple partial binding elements that may be responsible for Msx1-regulted repression, including 1 known Dlx3-binding element (Figure 3A) (54). To map the putative Msx1-binding elements in the human αGSU promoter, truncation analysis was used to define the sequences of importance (46). Basal transcription of the truncated αGSU promoters (−845, −668, −391, and −224 αGSU) was lower than that of the wild-type −1800 αGSU promoter, whereas the −116 αGSU promoter completely lost activity. Overexpression of Msx1 with these truncations maps the Msx1-mediated repression within −224 bp of the αGSU mRNA start site (Figure 3B). One putative partial Msx1-binding motif was mapped in this region: a −114 binding site (GTAATTA) that can bind Dlx3 in placental cells via binding at the JRE located from −114 to −106 (Figure 3A) (54). Overexpression of Dlx3 in LβT2 cells inhibits the transcriptional activation of the human αGSU promoter by glucocorticoids, and this activity is also mediated by the −114 JRE site (46). Because Msx and Dlx share the common consensus-binding element (C/GTAATTG), to assess the importance of this putative site for Msx1-mediated repression in αT3-1 cells, this JRE site was mutated within the −224 αGSU promoter truncation (μJRE). Basal transcription of the μJRE mutant was lower than wild-type −224 αGSU, and this mutation of the −114 JRE site also disrupts Msx1-mediated repression (Figure 3C).
Figure 3.
Msx1 represses the αGSU promoter at the −114 JRE within −224 bp of αGSU mRNA start site. (A) Gonadotrope regulatory elements in the proximal human αGSU promoter. Thick boxes indicate the putative Msx1/Dlx-binding elements, the −114 site. The JRE element is underlined. CRE1 and CRE2 are marked with dashed ovals. (B) The full-length human αGSU promoter and its truncated reporters map Msx1 repression inside of −224 bp of the start site on the αGSU promoter. The wild-type −1800 αGSU-luc, and its truncations: −845 αGSU-luc, −668 αGSU-luc, −391 αGSU-luc, −224 αGSU-luc, and −116 αGSU-luc, were transiently transfected into αT3-1 cells with the Msx1 expression vector or an equal mass of empty expression vector. (C) Identification of Msx1-binding sites on the human αGSU promoter. Wild-type −224 αGSU reporter (WT) and mutants (μJRE) were transiently transfected into αT3-1 cells, pGL3 empty luciferase reporters were used as control. Cells were cotransfected with Msx1 or an equal mass of empty expression vector. Groups labeled with different letters are significantly different from each other (P < .05). (D) The oligonucleotides used for the αGSU JRE probe and its mutants (μ7–μ12) are listed, and the Msx1-binding element and 2-bp transversions are underlined. Msx1 binding on the αGSU promoter maps to the −114 site in αT3-1 (E) and LβT2 (F) gonadotropes. The hollow arrow marks the Msx1 complex, and the arrow marks the supershift of the Msx1 complex after binding with Msx1 antibody. SS represents supershift complex. Abbreviation: Ab, antibody.
To investigate whether Msx1 binds the −114 JRE site directly, either αT3-1 or LβT2 nuclear extracts were incubated with excess wild-type oligonucleotide probes and competed with unlabeled oligonucleotides containing 2-bp transversion mutations (μ7–μ12) that scanned the consensus Msx-binding site (Figure 3D). The 2-bp mutations of the −114 JRE site (μ9–μ11) fail to compete, indicating the location of the Msx1-binding site. A new supershift complex of markedly reduced mobility formed with the inclusion of Msx1 antibody, demonstrating the highly specific binding of Msx1 to the −114 JRE site, although it is not completely shifted (Figure 3E). Instead, the Msx1 band in LβT2 nuclear extracts formed supershift complex with Msx1 antibody completely (Figure 3F), maybe due to the higher endogenous Msx1 expression in LβT2 cells (Figure 1A). In summary, these results indicate that the transcription of the human αGSU promoter is repressed by Msx1 homeodomain protein, and this repression maps to the Msx-binding motif on JRE element.
Dlx family members are transcriptional activators for αGSU and GnRHR transcription
Previous studies have shown that the consensus Msx-binding site also serves as the binding site for other homeodomain transcription factors, such as Dlx proteins (23). In contrast to Msx1, Dlx proteins are transcriptional activators primarily expressed in cells undergoing differentiation (29). Our RT-PCR data show that 4 of 6 Dlx family member mRNAs, Dlx1–Dlx4, are expressed at different levels in all 3 gonadotrope cell lines, whereas Dlx5 and Dlx6 show low or no expression (Figure 4A). All RT-PCR results were further validated by quantitative RNA-seq (Whole Transcriptome RNA Sequencing) analysis on gonadotrope cell lines (data not shown).
Figure 4.
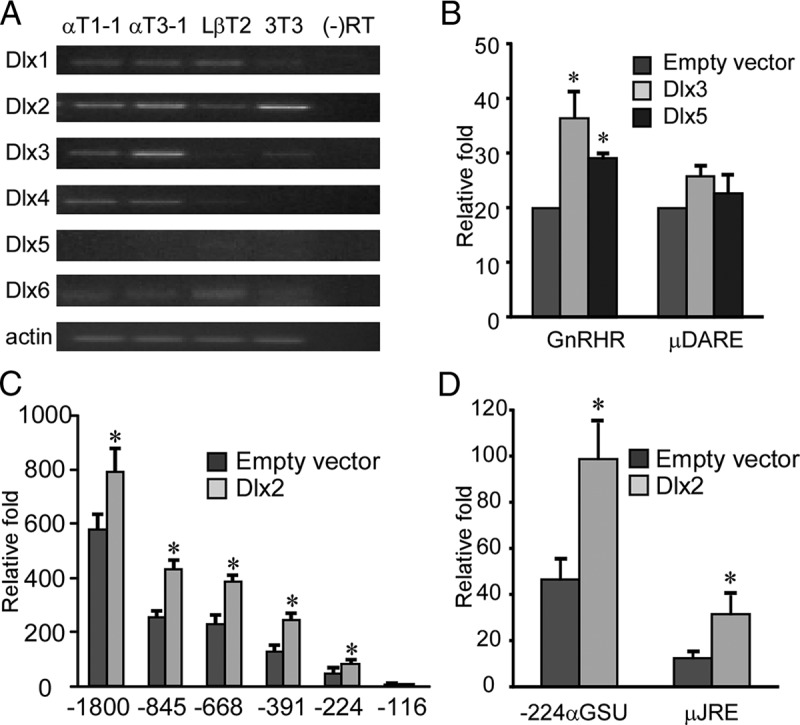
Dlx proteins are activators of both αGSU and GnRHR transcription. (A) Dlx mRNA expression in the gonadotrope-derived cell lines. Total RNA was harvested from gonadotrope-derived cell lines: progenitor αT1-1, immature αT3-1, mature LβT2, and NIH 3T3 cells as control, then reverse transcribed to generate cDNA. β-Actin was used as loading control. Resulting PCR reactions were run on a 1.5% agarose gel. (B) Dlx3 and Dlx5 activate GnRHR transcription through the DARE element. Wild-type GnRHR and DARE mutant (μDARE) reporters were transiently transfected into αT3-1 cells, pGL3 luciferase reporters were used as control. Cells were cotransfected with Dlx expression vector or an equal mass of empty expression vector. (C) Dlx2 activates αGSU transcription, and this effect maps to inside of −224 bp on the αGSU promoter. The wild-type −1800 αGSU-luciferase and its truncations were transiently transfected into the αT3-1 cell line with Dlx2 expression vector or an equal mass of empty expression vector. (D) Identification of Dlx2-binding sites on αGSU promoter. Wild-type −224 αGSU and mutant (μJRE) reporters were transiently transfected into αT3-1 cells, pGL3 luciferase reporters were used as control. Cells were cotransfected with Dlx2 expression vector or an equal mass of empty expression vector. Abbreviation: RT, reverse transcriptase. An asterisk indicates significant differences (P < .05) from empty vector control.
To investigate the transcriptional activities of Dlx proteins during gonadotrope development, we overexpressed several Dlx proteins, including Dlx1, Dlx2, Dlx3, Dlx5, and Dlx6, with either the GnRHR or αGSU promoter. Only Dlx3 and Dlx5 stimulated GnRHR transcription, and this activation also maps to the DARE element (Figure 4B and data not shown). Considering RT-PCR and RNA-seq data showed that there was no Dlx5 mRNA expression in αT3-1 cells (Figure 4A), we conclude that Dlx3 is the only Dlx protein that may stimulate GnRHR transcription in αT3-1 cells. These results lead to the conclusion that transcription of the mouse GnRHR promoter is regulated by both Msx1 and Dlx3; that is, Msx1 represses and Dlx3 activates GnRHR transcription.
Only Dlx2 can induce αGSU transcription in αT3-1 cells, an effect that is also retained within −224 bp of the human αGSU promoter (Figure 4C). Similar to Msx1-mediated repression, the mutant at the −114 site (μJRE) disrupted the Dlx2 activation of αGSU transcription (Figure 4D). In summary, these results indicate that the transcription of the αGSU promoter is regulated by Msx1 and Dlx2 homeodomain proteins functioning as a repressor and an activator, respectively.
TLE, the Msx1 corepressor, is required for regulation of the αGSU and GnRHR genes during gonadotrope differentiation
In the hypothalamus, Msx1 physically interacts with the corepressor TLE4 to augment repression of GnRH expression in GnRH neurons (39). Our hypothesis is that Msx1 can recruit TLE4 as a corepressor for regulation of both the GnRHR and αGSU promoters in pituitary as well. Investigation of TLE mRNA expression in gonadotropes using RT-PCR (Figure 5A) and quantitative RNA-seq analysis (data not shown) on gonadotrope cell lines reveals the expression patterns of 6 TLE mRNAs in the gonadotrope cell lines, and TLE4 mRNA shows a similar expression pattern as Msx1 (Figures 1A and 5A). To explore whether TLE4 functions as an Msx1 corepressor for early expression of the αGSU and GnRHR genes, we cotransfected TLE4 and Msx1 with either the GnRHR (Figure 5B) or αGSU promoter (Figure 5C) into αT3-1 cells. The results show that the overexpression of TLE4 augments Msx1-mediated repression of both αGSU and GnRHR promoters. However, due to the existence of the endogenous Msx1 and TLE4 expression in αT3-1 cells, the corepression of Msx1 and TLE4 on both the GnRHR and αGSU is not dramatic.
Figure 5.
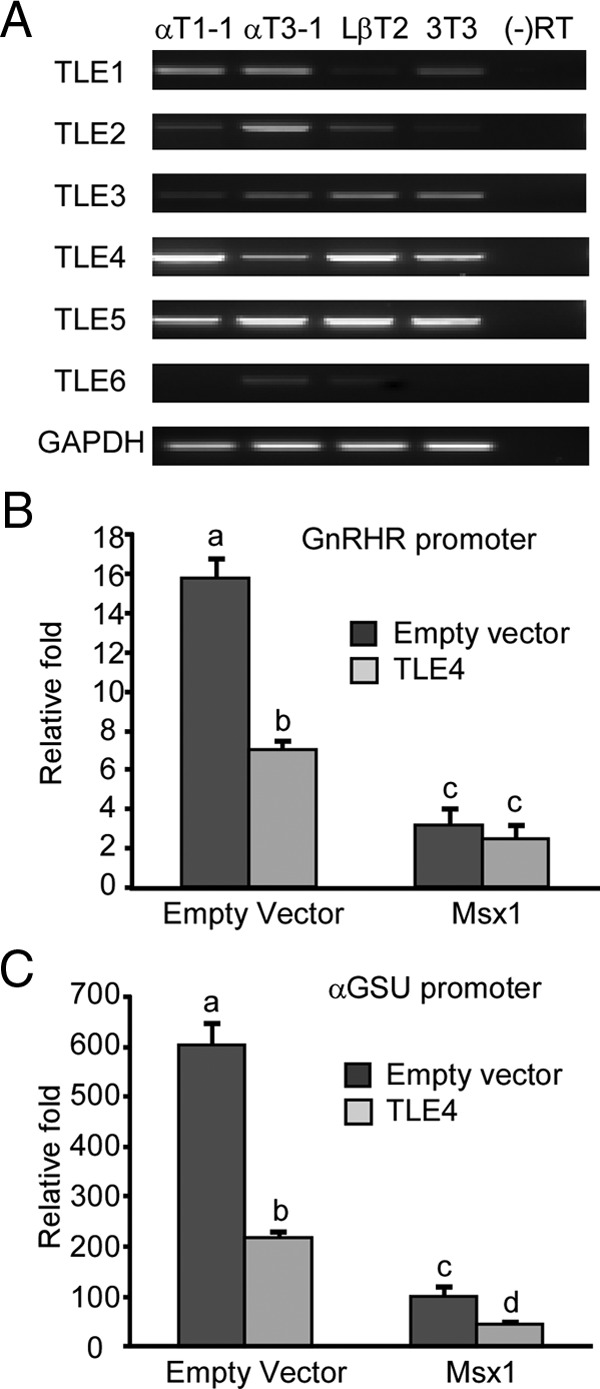
Msx1 recruits TLE4 as a corepressor to repress αGSU and GnRHR transcription. (A) TLE expression in the gonadotrope-derived cell lines. Total RNAs were harvested from progenitor αT1-1, immature αT3-1, mature LβT2, and fibroblast NIH 3T3 cells, then reverse transcribed to generate cDNA. GAPDH was used as loading control. (B and C) TLE4 is involved in Msx1-mediated GnRHR (B) and αGSU (C) repression. pGL3 empty luciferase reporters were used as control. Cells were cotransfected with Msx1 expression vector or an equal mass of empty expression vector, with or without TLE4 expression vector. Groups labeled with different letters are significantly different from each other (P < .05). Abbreviation: RT, reverse transcriptase.
siRNA-mediated knockdown of Msx1 in αT3-1 cells results in increased GnRHR and αGSU mRNA levels
Overexpression of Msx1 results in dramatic reduction in activity of both αGSU- and GnRHR-luciferase reporters. To investigate the actions of endogenous Msx1 in αT3-1 cells, we used siRNA-mediated knockdown of Msx1, hypothesizing that it would attenuate the Msx1 repressive action on αGSU and GnRHR expression, resulting in an increase in both αGSU and GnRHR mRNA. To test this hypothesis, αT3-1 cells were transfected with a SMARTpool of either Msx1 siRNA constructs or a nontargeting scrambled siRNA, which serves as a negative control. Transfection of Msx1 siRNA results in an approximately 80% reduction in the levels of Msx1 mRNA (Figure 6A) and decreased Msx1 protein expression (Figure 6B) in αT3-1 cells compared with a scrambled control (P < .05). Consequently, knockdown of Msx1 results in a significant increase in endogenous αGSU mRNA and a modest, although significant, increase in GnRHR mRNA (Figure 6A). Thus, Msx1 acts to suppress expression of αGSU and GnRHR in αT3-1 gonadotrope lineage cells.
Figure 6.
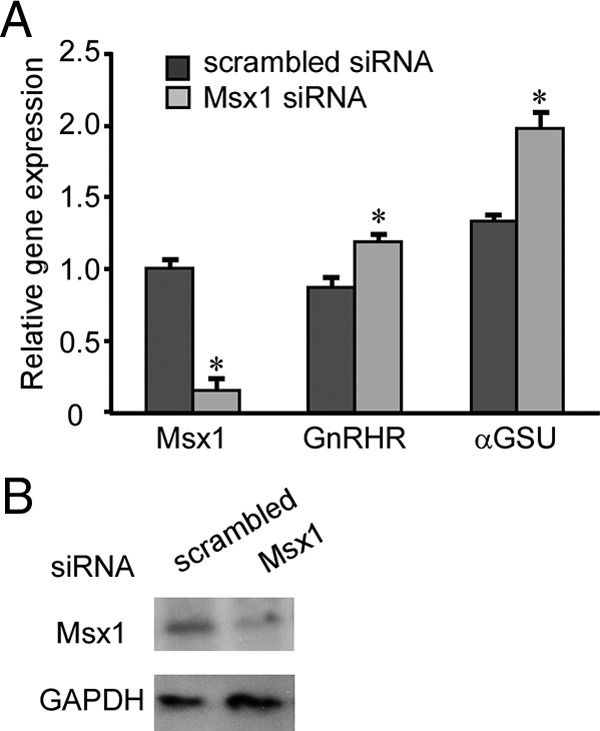
Knockdown of endogenous Msx1 in αT3-1 cells increases endogenous mouse αGSU and GnRHR mRNAs. (A) αT3-1 cells were transfected with SMARTpools of siRNA for Msx1 or a scrambled control. Cells were transfected for 72 hours, at which point total RNA were harvested and reverse transcribed. The resulting cDNA was used in qPCR reactions with primers specific for Msx1, αGSU, GnRHR, and GAPDH. All values represent the SQ means from 4 independent experiments with all means adjusted to corresponding GAPDH values within experiment. All values are normalized to scrambled control. (B) Whole-cell lysates were used for protein expression by Western blotting. An asterisk indicates significant differences (P < .05) from empty vector control.
Msx1 is expressed in the ventral aspect of the developing mouse anterior pituitary gland
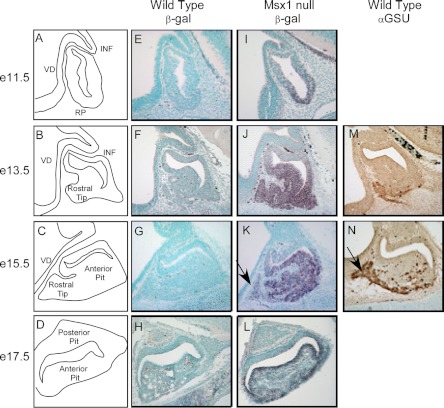
Msx1 mRNA is expressed during pituitary development in Rathke's pouch (Allen Brain Atlas). However, protein expression is not well characterized in either a spatial or temporal fashion in regards to gonadotrope development (22). We wanted to confirm and expand on the initial in situ hybridization studies of Msx1 expression by examining the spatial and temporal pattern of Msx1 gene expression during anterior pituitary development. To do this, we used Msx1/β-gal mice, in which the Msx1 coding sequence is deleted and replaced with that of β-gal (49). Consequently, β-gal is expressed from the Msx1 gene and is detectable using standard immunohistochemical techniques with a primary antibody against β-gal. Because Msx1-null mice are perinatal lethal, heterozygote mice were used for timed matings to generate e11.5, e13.5, e15.5, and e17.5 embryos. Figure 7, A–D, presents diagrams of pituitary development at these stages. No β-gal expression can be detected in wild-type mice (Figure 7, E–H). Expression of Msx1/β-gal becomes detectable on e11.5 (Figure 7I), reaches its strongest expression at e13.5 (Figure 7J), and, in both cases, is highest in the ventral aspect of the pituitary, the area destined to become the anterior pituitary gland. By e15.5, expression of β-gal, representing Msx1, begins to decline especially in the rostral tip of anterior pituitary (marked with arrow in Figure 7K) showing sparse expression. By e17.5, β-gal has significantly reduced expression (Figure 7L). Thus, Msx1 is highly expressed in the ventral region of the developing anterior pituitary, which is also the primary location of developing gonadotropes. Furthermore, Msx1 expression levels begin to decline at the approximate time of onset of gonadotrope-specific gene expression. For example, at e13.5, αGSU expression is emerging but is confined to the rostral tip of the developing anterior pituitary (Figure 7M). By e15.5, the decrease in Msx1/β-gal expression correlates with the increase in αGSU expression in the ventral aspect of the developing anterior pituitary (marked with arrow in Figure 7N). Thus, the decline in Msx1 expression levels in the developing mouse anterior pituitary may alleviate Msx1-induced repression of αGSU and GnRHR expression levels and thereby act to time expression of the gonadotrope gene program.
Figure 7.
Msx1/β-gal expression peaks during mouse anterior pituitary development on e13.5 and begins to decline by e15.5 concurrent with increased αGSU expression. Embryos produced by mating heterozygote Msx1/βgal males and females were fixed, embedded, sectioned, and subjected to immunohistochemical. (A–D) Schematic diagrams of pituitary development at different stages. Rathke's pouch (RP) is formed and makes direct contact with the portion of the ventral diencephalon (VD) comprising the infundibulum (INF). Five different pituitary cell types are hypothesized to proliferate ventrally from the epithelial cells of RP to populate the anterior lobe of the pituitary gland. Wild-type embryos (E–H) and Msx1-null/β-gal insertion (I–L) illustrate sections of e11.5, e13.5, e15.5, and e17.5 developing mouse anterior pituitaries with methyl green as a counter stain, whereas the purple/red peroxidase staining represents β-gal in the place of normal Msx1 expression. (M and N) Brown DAB (3,3'-diaminobenzidine) staining representing αGSU expression in wild-type embryos at e13.5 and e15.5, respectively. The arrow indicates the rostral tip of the developing anterior pituitary.
GnRHR and αGSU mRNA expression in developing pituitary are induced in Msx1-null mice
To determine the repressive roles of Msx1 on GnRHR and αGSU in vivo, total pituitary RNA was harvested from e18.5 wild-type or Msx1-null embryos (Msx1/β-gal). Four gonadotrope-specific mRNAs, GnRHR, αGSU, LHβ, and FSHβ, were analyzed by qPCR (Figure 8, A–D). To collect enough RNA, each group contained 5 individual embryonic pituitaries, and 3 (GnRHR and αGSU) or 5 groups were analyzed (LHβ and FSHβ). mRNA expression levels of both GnRHR (Figure 8A) and αGSU (Figure 8B) in Msx1-null embryos (Msx1/β-gal) at e18.5 were significantly induced compared with wild type, further confirming the repression by Msx1 of both genes in vivo. Consistent with our in vitro studies (Figure 1C), Msx1 knockdown did not have a statistically significant effect on either LHβ (Figure 8C) or FSHβ (Figure 8D) mRNA expression.
Figure 8.
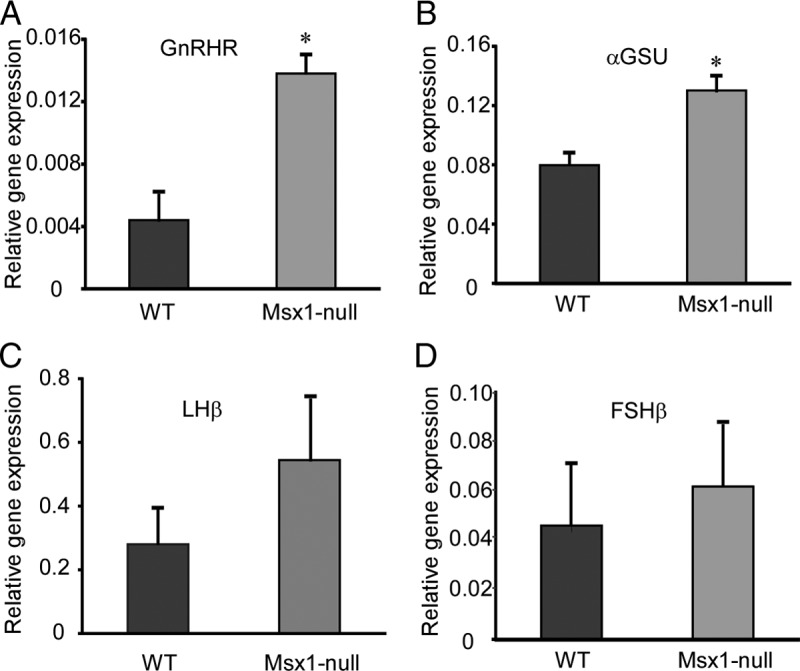
GnRHR and αGSU mRNA expressions in developing pituitary are highly induced in Msx1-null mice. Embryo of both wild-type (WT) and null mice at e18.5 were harvested by timed mating of heterozygote Msx1/β-gal males and females. Total RNAs were isolated from pituitaries and then reverse transcribed to generate cDNA. qPCRs were performed with specific primers for (A) GnRHR, (B) αGSU, (C) LHβ, and (D) FSHβ. Values represent the SQ means from 3 (A and B) or 5 (C and D) groups of embryonic pituitaries, and each group contains 5 pituitaries, with all means adjusted to corresponding GAPDH values within experiment. An asterisk represents values (P < .05) different from wild type (+/+).
Discussion
The mature gonadotropes of the anterior pituitary relay hypothalamic GnRH input via GnRHR to modulate expression and release of LH and FSH. Thus, proper expression of αGSU, GnRHR, LHβ, and FSHβ subunits is critical for mammalian reproductive function. A particularly interesting set of transcription factors critical for gonadotrope development are the homeobox proteins, which can be either stimulatory or inhibitory, such as Lhx, Pit-1, Pitx1, and now Msx1 (10, 55, 56). In our studies, overexpression of Msx1 reduced transcriptional activity of the GnRHR and αGSU promoters, and siRNA knockdown rescued the mRNA expression of both genes in αT3-1 cells, indicating that Msx1 may play a role in repressing expression of these genes during gonadotrope differentiation. Studies from Msx1-null mice in vivo confirmed that replacement of Msx1 by β-gal induced the transcription of both GnRHR and αGSU. Meanwhile, expression of β-gal, representing Msx1, begins to decline, especially in the rostral tip of anterior pituitary, at the approximate onset of αGSU gene expression, indicating that Msx1 may act to time expression of the gonadotrope gene program.
Msx1 repression of pituitary-specific genes during development is not without precedent, because the onset of melatonin receptor expression in thyrotropes coincides with a decline in Msx1 expression at e15.5 in the rat pituitary (57). Msx1 potently inhibits Pitx1-induced melatonin receptor promoter activity and therefore represents a key inhibitor of melatonin receptor, in effect, timing its expression (57). Additionally, Msx1 is believed to inhibit differentiation of the pituitary cell lineages; for example, the decline of Msx1 expression in the developing mouse anterior pituitary coincides with the onset of gonadotrope-lineage-specific transcription factors, including Pit-1 (58).
Msx1 repression of the GnRHR promoter maps to a consensus Msx-binding motif in DARE, which binds Msx1 in vitro (Figure 2). DARE is a composite regulatory element that is known to bind multiple transcription factors, such as Lhx3, NF-Y, and Oct-1, all of which are expressed in αT3-1 cells (45). In other systems, the consensus Msx site can bind the homeodomain transcription factors Dlx3 and Dlx5, which share the same binding motif as Msx1 (23). As such, the binding of multiple transcription factors to the DARE region indicates that competition likely exists for this binding site and that the relative levels and affinities of the factors are critical. Another possibility is that various transcription factors in this small area form higher order complexes with one another, circumventing the need for direct DNA binding.
As the common subunit of all glycoprotein hormones, αGSU expression is restricted to pituitary (gonadotropes and thyrotropes) and placenta (trophoblasts). Msx1 expression seems specific to αGSU-producing cells, because other anterior pituitary cells, such as the somatolactotrope GH3 cells, do not express the αGSU and likewise do not express Msx1 (52). In thyrotropes, the overexpression of Msx1 reduces activity of the αGSU promoter in thyrotrope by binding to the αGSU promoter (−435/−413) that contains 2 tandem repeats of GXAATTG, a 6- out of 7-bp match with the defined Msx1-consensus binding sequence. Our studies indicate that Msx1 also strongly represses αGSU in gonadotropes and that this repression maps to a known Dlx3-binding element located at JRE (Figure 3). Thus, a common developmental mechanism may have emerged, in which Msx1 is expressed in undifferentiated gonadotropes and thyrotropes, binds to the αGSU promoter, and represses the expression of αGSU. Msx1 repression is eventually alleviated by declining levels of Msx1 as the pituitary develops and the thyrotrope/gonadotrope lineage differentiates from other cell types. Overexpression of Msx1 also represses transcription of both GnRHR and αGSU promoters in the more mature LβT2 gonadotrope cell line, although Msx1 repression of the αGSU promoter is not as robust as in αT3-1 cells (Figure 1, B and C). Msx1 binds to the same element on αGSU promoter in both immature αT3-1 and mature cell lines, although the supershift in αT3-1 nuclear extracts is weak due to the low expression of Msx1.
Distinct sets of regulatory regions of the αGSU gene confer cell-specific expression via combinations of cis-acting elements and their cognate binding transcription factors (59, 60). The αGSU promoter has several fundamental elements that are necessary for gonadotrope-specific activity (11, 12, 61). In placental cells, mutations within the JRE reduced basal αGSU expression despite the presence of intact dual cyclic AMP response elements (CREs) (62). Dlx3 is a placental-specific transcriptional regulator that binds to the JRE and contributes to expression of the αGSU gene in cells of trophoblast origin (54). In contrast to transcriptional repression in placental cells, overexpression of Dlx3 in LβT2 cells inhibits transcriptional activation by glucocorticoids of the human αGSU gene by binding at the JRE located from −114 to −106 (46). Our results show that this −114 GTAATTA site binds Msx1 for transcriptional repression of αGSU during early gonadotrope development. Therefore, the same site recruits different transcription factors at different developmental stages and in different tissues.
We hypothesized that removal of Msx1 to alleviate Msx1-induced repression would result in an increase in αGSU and GnRHR mRNA expression. Our results proved this point both in vitro and in vivo: Msx1 knockdown in αT3-1 cells increased both endogenous mouse αGSU and GnRHR mRNAs, and Msx1 deletion in mice induced higher expression of both mRNAs as well (Figures 6 and 8). Thus, we conclude that Msx1 plays a functional role in repressing αGSU and GnRHR expression. Regulation of the gonadotrope-specific gene program appears specific, because Msx2 is not expressed during anterior pituitary development in vivo despite weak detection of its mRNA in the αT3-1 cell line (25). In addition, Mxs3 is not detected in αT3-1 cells or Rathke's pouch and thus was not included in our analysis of initiation of the gonadotrope-specific gene program (50). Analysis of the expression pattern of both Msx1 and αGSU showed that αGSU emerges at the rostral tip of the anterior pituitary after the decline of Msx1 expression at e13.5 (Figure 7), further suggesting that Msx1 times the onset of gonadotrope development. Overall, Msx1 plays a critical role in inhibiting differentiation of the anterior pituitary cells by suppressing transcription factors and proteins that are ultimately responsible for initiating, differentiating, and defining the 5 endocrine cell types of the anterior pituitary potentially timing their expression in the developing gonadotrope.
With respect to their biochemical properties and expression patterns, individual Dlx family members are required for relieving Msx1-mediated transcriptional repression of specific genes and function as transcriptional activators by replacing association of repressor Msx1 on target genes during development. In our studies, Dlx3 and Dlx2 act as transcription activators that antagonize the repressive effect of Msx1 on GnRHR and αGSU, respectively, and both bind the same response element as Msx1 (Figure 4). Interestingly, introduction of Dlx3 into LβT2 cells interferes with glucocorticoid receptor induction of human αGSU expression by binding to the same JRE element and dramatically reducing dexamethasone induction (46). Therefore, Msx1 and Dlx proteins may play a role in the cell type- and stimuli-specific effect of αGSU transcription during gonadotrope development.
Cotransfections of Msx1 and TLE4 indicate that TLE4 functions as a corepressor of Msx1 regulation of αGSU and GnRHR promoter activity in the αT3-1 cell line. The TLE family of corepressors repress transcription by inhibiting the basal transcriptional machinery (63) and recruiting histone deacetylases. Because TLE proteins oligomerize, associate with histones (64), and bind histone deacetylases, we hypothesize that Msx1 and the corepressor TLEs repress αGSU and GnRHR expression by establishing regional repressive chromatin structures. TLEs have also been shown to act as corepressors of other critical transcription factors during gonadotrope differentiation (42, 43). The TLE proteins act as corepressors by protein-protein binding to transcription factors such as HESX1, TCF/LEF, and SIX family members. Wnt signaling (TCF/β-catenin) pathways play important roles in targeting important biological processes in gonadotrope biology (65, 66). HESX1 and PROP1 are transcription factors involved in the commitment and differentiation of cell types in the anterior lobe of the pituitary. TCF4 and TLE5 can influence pituitary growth and development, and TCF4 is required for down-regulation of PROP1 expression (42). TLE1 and TLE3 not only function as corepressors as HESX1 by enhancing its repression of PROP1 activation but also may interact with other transcription factors to affect gonadotrope and thyrotrope differentiation (43).
The studies presented here further support the role of Msx1 as a critical transcriptional repressor necessary for anterior pituitary development, specifically for the gonadotrope-specific gene program. Not only does Msx1 strongly repress activity of the GnRHR and αGSU promoters in αT3-1 and LβT2 cells, but Msx1 specifically binds these 2 promoters in vitro. In vivo, Msx1 expression in the developing anterior pituitary declines at the approximate time that the gonadotrope begins to differentiate and starts expressing gonadotrope-specific gene products. Thus, Msx1 plays a critical role in timing the onset of the gonadotrope-specific gene program both in vitro, in the αT3-1 immature gonadotrope cell line, and in vivo, in the developing mouse anterior pituitary.
Acknowledgments
We thank Sunamita Leming, Cindy Ngo and Susan Mayo for technical assistance, Kellie Breen and Varykina Thackray for reading the manuscript, Melissa Brayman for RNA-seq analysis, and Mark Lawson for software and statistical assistance.
Present address for B.D.C.: Department of Zoology and Physiology, University of Wyoming, Laramie, Wyoming 82071.
This work was supported by National Institutes of Health (NIH) Grants R01 DK044838, R01 HD072754, and R01 HD020377 (to P.L.M.) and by National Institute of Child Health and Human Development/NIH through a cooperative agreement (U54 HD012303) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research (to P.L.M.). P.L.M. was partially supported by P30 DK063491, P42 ES101337, and P30 CA023100. H.X. was partially supported by T32 HD007203 and F32 HD070579. B.D.C. was partially supported by T32 HD007203. E.A.W. was partially supported by T32 GM008666, P42 ES010337, and T32 DA007315.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- bp
- base pair
- CRE
- cyclic AMP response element
- DARE
- downstream activin regulatory element
- e
- embryonic day
- β-gal
- β-galactosidase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GnRHR
- GnRH receptor
- αGSU
- α-glycoprotein subunit
- JRE
- junctional regulatory element
- qPCR
- quantitative PCR
- RNA-seq
- Whole Transcriptome RNA Sequencing
- SF
- steroidogenic factor
- siRNA
- small interfering RNA
- TLE
- transducin-like enhancer of split.
References
- 1. Simmons DM, Voss JW, Ingraham HA, et al. Pituitary cell phenotypes involve cell-specific Pit-1 mRNA translation and synergistic interactions with other classes of transcription factors. Genes Dev. 1990;4:695–711 [DOI] [PubMed] [Google Scholar]
- 2. Scully KM, Rosenfeld MG. Pituitary development: regulatory codes in mammalian organogenesis. Science. 2002;295:2231–2235 [DOI] [PubMed] [Google Scholar]
- 3. Windle JJ, Weiner RI, Mellon PL. Cell lines of the pituitary gonadotrope lineage derived by targeted oncogenesis in transgenic mice. Mol Endocrinol. 1990;4:597–603 [DOI] [PubMed] [Google Scholar]
- 4. Alarid ET, Windle JJ, Whyte DB, Mellon PL. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development. 1996;122:3319–3329 [DOI] [PubMed] [Google Scholar]
- 5. Lew D, Brady H, Klausing K, et al. GHF-1 promoter-targeted immortalization of a somatotropic progenitor cell results in dwarfism in transgenic mice. Genes Dev. 1993;7:683–693 [DOI] [PubMed] [Google Scholar]
- 6. Pernasetti F, Spady TJ, Hall SB, et al. Pituitary tumorigenesis targeted by the ovine follicle-stimulating hormone β-subunit gene regulatory region in transgenic mice. Mol Cell Endocrinol. 2003;203:169–183 [DOI] [PubMed] [Google Scholar]
- 7. Yusta B, Alarid ET, Gordon DF, Ridgway EC, Mellon PL. The thyrotropin β-subunit gene is repressed by thyroid hormone in a novel thryrotrope cell line, mouse TαT1 cells. Endocrinology. 1998;139:4476–4482 [DOI] [PubMed] [Google Scholar]
- 8. Pernasetti F, Vasilyev VV, Rosenberg SB, et al. Cell-specific transcriptional regulation of FSHβ by activin and GnRH in the LβT2 pituitary gonadotrope cell model. Endocrinology. 2001;142:2284–2295 [DOI] [PubMed] [Google Scholar]
- 9. Graham KE, Nusser KD, Low MJ. LβT2 gonadotroph cells secrete follicle stimulating hormone (FSH) in response to activin A. J Endocrinol. 1999;162:R1–R5 [DOI] [PubMed] [Google Scholar]
- 10. McGillivray SM, Bailey JS, Ramezani R, Kirkwood BJ, Mellon PL. Mouse GnRH receptor gene expression is mediated by the LHX3 homeodomain protein. Endocrinology. 2005;146:2180–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barnhart KM, Mellon PL. The orphan nuclear receptor, steroidogenic factor-1, regulates the glycoprotein hormone α-subunit gene in pituitary gonadotropes. Mol Endocrinol. 1994;8:878–885 [DOI] [PubMed] [Google Scholar]
- 12. Steger DJ, Hecht JH, Mellon PL. GATA-binding proteins regulate the human gonadotropin α-subunit gene in placenta and pituitary gland. Mol Cell Biol. 1994;14:5592–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alarid ET, Holley S, Hayakawa M, Mellon PL. Discrete stages of anterior pituitary differentiation recapitulated in immortalized cell lines. Mol Cell Endocrinol. 1998;140:25–30 [DOI] [PubMed] [Google Scholar]
- 14. Rosenberg SB, Mellon PL. An Otx-related homeodomain protein binds an LHβ promoter element important for activation during gonadotrope maturation. Mol Endocrinol. 2002;16:1280–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harris AN, Mellon PL. The basic helix-loop-helix leucine zipper transcription factor USF is a key regulator of SF-1 gene expression in pituitary gonadotrope and steroidogenic cells. Mol Endocrinol. 1998;12:714–727 [DOI] [PubMed] [Google Scholar]
- 16. Corpuz PS, Lindaman LL, Mellon PL, Coss D. FoxL2 is required for activin induction of the mouse and human follicle-stimulating hormone β-subunit genes. Mol Endocrinol. 2010;24:1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Breen KM, Thackray VG, Coss D, Mellon PL. Runt-related transcription factors impair activin induction of the follicle-stimulating hormone β-subunit gene. Endocrinology. 2010;151:2669–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghochani Y, Saini JK, Mellon PL, Thackray VG. FOXL2 is involved in the synergy between activin and progestins on the follicle-stimulating hormone β-subunit promoter. Endocrinology. 2012;153:2023–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lamba P, Fortin J, Tran S, Wang Y, Bernard DJ. A novel role for the forkhead transcription factor FOXL2 in activin A-regulated follicle-stimulating hormone β subunit transcription. Mol Endocrinol. 2009;23:1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Halvorson LM, Kaiser UB, Chin WW. Stimulation of luteinizing hormone β gene promoter activity by the orphan nuclear receptor, steroidogenic factor-1. J Biol Chem. 1996;271:6645–6650 [DOI] [PubMed] [Google Scholar]
- 21. Keri RA, Nilson JH. A steroidogenic factor-1 binding site is required for activity of the luteinizing hormone β subunit promoter in gonadotropes of transgenic mice. J Biol Chem. 1996;271:10782–10785 [DOI] [PubMed] [Google Scholar]
- 22. Mackenzie A, Fergusson MWJ, Sharpie PT. Hox-7 expression during murine craniofacial development. Development. 1991;113:601–611 [DOI] [PubMed] [Google Scholar]
- 23. Bendall AJ, Abate-Shen C. Roles for Msx and Dlx homeoproteins in vertebrate development. Gene. 2000;247:17–31 [DOI] [PubMed] [Google Scholar]
- 24. Zhang H, Catron KM, Abate-Shen C. A role for the Msx-1 homeodomain in transcriptional regulation: residues in the N-terminal arm mediate TATA binding protein interaction and transcriptional repression. Proc Natl Acad Sci USA. 1996;93:1764–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. MacKenzie A, Ferguson MW, Sharpe PT. Expression patterns of the homeobox gene, Hox-8, in the mouse embryo suggest a role in specifying tooth initiation and shape. Development. 1992;115:403–420 [DOI] [PubMed] [Google Scholar]
- 26. Alappat S, Zhang ZY, Chen YP. Msx homeobox gene family and craniofacial development. Cell Res. 2003;13:429–442 [DOI] [PubMed] [Google Scholar]
- 27. Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356 [DOI] [PubMed] [Google Scholar]
- 28. Catron KM, Wang H, Hu G, Shen MM, Abate-Shen C. Comparison of MSX-1 and MSX-2 suggests a molecular basis for functional redundancy. Mech Dev. 1996;55:185–199 [DOI] [PubMed] [Google Scholar]
- 29. Catron KM, Zhang H, Marshall SC, Inostroza JA, Wilson JM, Abate C. Transcriptional repression by Msx-1 does not require homeodomain DNA-binding sites. Mol Cell Biol. 1995;15:861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang H, Hu G, Wang H, et al. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol. 1997;17:2920–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zerucha T, Stuhmer T, Hatch G, et al. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20:709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu G, Zerucha T, Ekker M, Rubenstein JL. Evidence that GRIP, a PDZ-domain protein which is expressed in the embryonic forebrain, co-activates transcription with DLX homeodomain proteins. Brain Res Dev Brain Res. 2001;130:217–230 [DOI] [PubMed] [Google Scholar]
- 33. Givens ML, Rave-Harel N, Goonewardena VD, et al. Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J Biol Chem. 2005;280:19156–19165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koop KE, MacDonald LM, Lobe CG. Transcripts of Grg4, a murine groucho-related gene, are detected in adjacent tissues to other murine neurogenic gene homologues during embryonic development. Mech Dev. 1996;59:73–87 [DOI] [PubMed] [Google Scholar]
- 35. Leon C, Lobe CG. Grg3, a murine Groucho-related gene, is expressed in the developing nervous system and in mesenchyme-induced epithelial structures. Dev Dyn. 1997;208:11–24 [DOI] [PubMed] [Google Scholar]
- 36. Dehni G, Liu Y, Husain J, Stifani S. TLE expression correlates with mouse embryonic segmentation, neurogenesis, and epithelial determination. Mech Dev. 1995;53:369–381 [DOI] [PubMed] [Google Scholar]
- 37. Mallo M, Franco del Amo F, Gridley T. Cloning and developmental expression of Grg, a mouse gene related to the groucho transcript of the Drosophila enhancer of split complex. Mech Dev. 1993;42:67–76 [DOI] [PubMed] [Google Scholar]
- 38. Paroush Z, Finley RL, Kidd T, Weinwright SM, Ingham PI, Brent RI, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with Hairy-related bHLH proteins. Cell. 1994;79:805–815 [DOI] [PubMed] [Google Scholar]
- 39. Larder R, Mellon PL. Otx2 induction of the gonadotropin-releasing hormone promoter is modulated by direct interactions with Grg co-repressors. J Biol Chem. 2009;284:16966–16978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rave-Harel N, Miller NL, Givens ML, Mellon PL. The Groucho-related gene family regulates the gonadotropin-releasing hormone gene through interaction with the homeodomain proteins MSX1 and OCT1. J Biol Chem. 2005;280:30975–30983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mallo M, Gendron-Maguire M, Harbison ML, Gridley T. Protein characterization and targeted disruption of Grg, a mouse gene related to the groucho transcript of the Drosophila enhancer of split complex. Dev Dyn. 1995;204:338–347 [DOI] [PubMed] [Google Scholar]
- 42. Brinkmeier ML, Potok MA, Cha KB, et al. TCF and Groucho-related genes influence pituitary growth and development. Mol Endocrinol. 2003;17:2152–2161 [DOI] [PubMed] [Google Scholar]
- 43. Carvalho LR, Brinkmeier ML, Castinetti F, Ellsworth BS, Camper SA. Corepressors TLE1 and TLE3 interact with HESX1 and PROP1. Mol Endocrinol. 2010;24:754–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Duval DL, Nelson SE, Clay CM. The tripartite basal enhancer of the gonadotropin-releasing hormone (GnRH) receptor gene promoter regulates cell-specific expression through a novel GnRH receptor activating sequence. Mol Endocrinol. 1997;11:1814–1821 [DOI] [PubMed] [Google Scholar]
- 45. Cherrington BD, Farmerie TA, Lents CA, Cantlon JD, Roberson MS, Clay CM. Activin responsiveness of the murine gonadotropin-releasing hormone receptor gene is mediated by a composite enhancer containing spatially distinct regulatory elements. Mol Endocrinol. 2005;19:898–912 [DOI] [PubMed] [Google Scholar]
- 46. Sasson R, Luu SH, Thackray VG, Mellon PL. Glucocorticoids induce human glycoprotein hormone α-subunit gene expression in the gonadotrope. Endocrinology. 2008;149:3643–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cherrington BD, Bailey JS, Diaz AL, Mellon PL. NeuroD1 and Mash1 temporally regulate GnRH receptor gene expression in immortalized mouse gonadotrope cells. Mol Cell Endocrinol. 2008;295:106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miller NL, Wevrick R, Mellon PL. Necdin, a Prader-Willi syndrome candidate gene, regulates gonadotropin-releasing hormone neurons during development. Hum Mol Genet. 2009;18:248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Houzelstein D, Cohen A, Buckingham ME, Robert B. Insertional mutation of the mouse Msx1 homeobox gene by an nlacZ reporter gene. Mech Dev. 1997;65:123–133 [DOI] [PubMed] [Google Scholar]
- 50. Driegen S, Ferreira R, van Zon A, et al. A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res. 2005;14:477–482 [DOI] [PubMed] [Google Scholar]
- 51. Wang J, Kumar RM, Biggs VJ, et al. The Msx1 homeoprotein recruits Polycomb to the nuclear periphery during development. Dev Cell. 2011;21:575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sarapura VD, Strouth HL, Gordon DF, Wood WM, Ridgway EC. MsxI is present in thyrotropic cells and binds to a consensus site on the glycoprotein hormone α-subunit promoter. Mol Endocrinol. 1997;11:1782–1794 [DOI] [PubMed] [Google Scholar]
- 53. Kam KY, Jeong KH, Norwitz ER, Jorgensen EM, Kaiser UB. Oct-1 and nuclear factor Y bind to the SURG-1 element to direct basal and gonadotropin-releasing hormone (GnRH)-stimulated mouse GnRH receptor gene transcription. Mol Endocrinol. 2005;19:148–162 [DOI] [PubMed] [Google Scholar]
- 54. Roberson MS, Meermann S, Morasso MI, Mulvaney-Musa JM, Zhang T. A role for the homeobox protein Distal-less 3 in the activation of the glycoprotein hormone α subunit gene in choriocarcinoma cells. J Biol Chem. 2001;276:10016–10024 [DOI] [PubMed] [Google Scholar]
- 55. Jiang Q, Jeong KH, Horton CD, Halvorson LM. Pituitary homeobox 1 (Pitx1) stimulates rat LHβ gene expression via two functional DNA-regulatory regions. J Mol Endocrinol. 2005;35:145–158 [DOI] [PubMed] [Google Scholar]
- 56. DiMattia GE, Rhodes SJ, Krones A, et al. The Pit-1 gene is regulated by distinct early and late pituitary-specific enhancers. Dev Biol. 1997;182:180–190 [DOI] [PubMed] [Google Scholar]
- 57. Johnston JD, Klosen P, Barrett P, Hazlerigg DG. Regulation of MT melatonin receptor expression in the foetal rat pituitary. J Neuroendocrinol. 2006;18:50–56 [DOI] [PubMed] [Google Scholar]
- 58. Li S, Crenshaw EB, Rawson EJ, Simmons DM, Swanson LW, Rosenfeld MG. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature. 1990;347:528–533 [DOI] [PubMed] [Google Scholar]
- 59. Nilson JH, Bokar JA, Clay CM, et al. Different combinations of regulatory elements may explain why placenta-specific expression of the glycoprotein hormone α-subunit gene occurs only in primates and horses. Biol Reprod. 1991;44:231–237 [DOI] [PubMed] [Google Scholar]
- 60. Maurer RA, Kim KE, Schoderbek WE, Roberson MS, Glenn DJ. Regulation of glycoprotein hormone α-subunit gene expression. Recent Prog Horm Res. 1999;54:455–484 [PubMed] [Google Scholar]
- 61. Heckert LL, Schultz K, Nilson JH. Different composite regulatory elements direct expression of the human α subunit gene to pituitary and placenta. J Biol Chem. 1995;270:26497–26504 [DOI] [PubMed] [Google Scholar]
- 62. Budworth PR, Quinn PG, Nilson JH. Multiple characteristics of a pentameric regulatory array endow the human α-subunit glycoprotein hormone promoter with trophoblast specificity and maximal activity. Mol Endocrinol. 1997;11:1669–1680 [DOI] [PubMed] [Google Scholar]
- 63. Yu X, Li P, Roeder RG, Wang Z. Inhibition of androgen receptor-mediated transcription by amino-terminal enhancer of split. Mol Cell Biol. 2001;21:4614–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610 [DOI] [PubMed] [Google Scholar]
- 65. Salisbury TB, Binder AK, Nilson JH. Welcoming β-catenin to the GnRH transcriptional network in gonadotropes. Mol Endocrinol. 2008;22:1295–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Doyle JP, Dougherty JD, Heiman M, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]