Abstract
Breast cancer remains one of the leading causes of death in women diagnosed with cancer. In breast cancer, aberrant expression of the CYP19A1 gene, which encodes the aromatase enzyme, contributes to increased intratumoral levels of estradiol. Regardless of whether this estrogen is produced by peripheral tissues or within specific subpopulations of cells within the breast tumor, it is clear that the aromatase enzymatic activity is critical for the growth of estrogen-dependent tumors. Currently, aromatase inhibitors have proven to be highly effective in blocking the growth of estrogen-dependent forms of breast cancer. CYP19A1 transcription is tightly controlled by 10 tissue-specific promoters. In breast cancer, however, aromatase transcription is driven by multiple promoters that somehow override the tissue-specific regulation of normal tissue. Here, we explore the role that the deacetylase, sirtuin-1 (SIRT1), plays in positively regulating aromatase in breast cancer. We demonstrate that the use of cambinol and the SIRT1/2 inhibitor VII, 2 small molecule inhibitors of SIRT1 and SIRT2, as well as small molecule inhibitors and small interfering RNA specific to SIRT1, all reduce the levels of aromatase mRNA. We further demonstrate that pharmacologic inhibition causes a marked reduction in aromatase protein levels. Additionally, by chromatin immunoprecipitation, we demonstrate that SIRT1 occupies the promoter regions PI.3/PII and PI.4, and its inhibition leads to increased acetylation of estrogen-related receptorα, a transcription factor that positively regulates CYP19A1 transcription in epithelial cells. Finally, we demonstrate by immunohistochemistry that SIRT1 is significantly up-regulated in invasive ductal carcinoma relative to normal tissue adjacent to tumor, further suggesting a role of SIRT1 in breast cancer. This work uncovers a new mechanism for the regulation of aromatase and provides rationale for further investigation of how the inhibition of specific sirtuins may provide a unique strategy for inhibiting aromatase that may complement or synergize with existing therapies.
Breast tumors are characterized by a heterogeneous population of cells that generate cell type-specific and cell type-codependent survival signals, which collectively contribute to malignancy (1). Because of this heterogeneity, significant obstacles remain in developing better therapies for patients. Recent reports have shown that a number of genomic rearrangements (2) and epigenetic changes (3, 4) in breast cancer appear to contribute to this heterogeneity. One gene in particular, CYP19A1, which encodes the protein aromatase, contributes significantly to the malignancy of most breast cancers (5). The aromatase enzyme converts androgens into estrogens (6) and has been shown to be overexpressed in breast tumor tissue (7, 8). Because approximately 75% of diagnosed breast cancers are estrogen dependent (5), antiestrogen therapies will be used to treat most breast cancers.
Aromatase inhibitors (AIs) are effective in the treatment of postmenopausal breast cancer. Clinical trials show higher clinical benefit in patients treated with AIs compared with those treated with tamoxifen (9–11), and recommendations have been made to incorporate an AI as a primary, sequential, or extended therapy to reduce the risk of breast cancer recurrence compared with tamoxifen alone (12). Aromatase is expressed widely in ovarian (granulosa cells) and in extraovarian tissues, such as the hypothalamus, cerebral cortex, hippocampus, cerebellum, skin, bone, placenta, and adipose tissue (6). To improve current therapeutics and decrease side effects, it will be important to characterize the mechanisms by which aromatase expression is increased in specific populations of cells, which may contribute to higher systemic or intratumoral conversion of androgens to estrogens.
CYP19A1 (aromatase) transcription is normally tightly regulated and tissue specific. To date, 10 tissue-specific promoters have been identified within the 93-kilobase promoter region upstream of the aromatase coding sequence (6, 13). Despite differences in the transcriptional start site, the aromatase protein is identical in all tissues in which it is expressed. In normal breast tissue, aromatase expression is controlled through activation of promoter I.4 (6). In breast cancer, however, 3 additional promoters are used to initiate transcription (8). In breast cancer, promoters I.7, I.4, I.3, and II are responsible for driving the transcription of CYP19A1. The region between promoters I.3 and II is the most active transcriptional start site and the most well characterized of these promoters (14, 15). Additionally, the pII promoter has been shown to be activated by the liver receptor homologue-1 (LRH-1) in preadipocytes (16) and in breast cancer lines (17). This suggests that specific factors might drive the increase in intratumoral levels of estrogen by activating the pII CYP19A1 promoter in both preadipocytes and epithelial cell populations of the breast tumors. By characterizing transcriptional regulators and pharmacologic agents that regulate the activation from these distinct aromatase promoters, a more selective therapeutic target could be identified.
Sirtuin-1 (SIRT1) is an nicotinamide adenine dinucleotide+-dependent deacetylase and has been shown to be up-regulated in a variety of cancers (18) and influences multiple hallmarks of cancer by regulating the function of histone proteins (19, 20) as well as several nonhistone proteins, including Dishevelled (Dvl) (21), Cortactin (22), SUV39H1, and estrogen-related receptor (ERR)α (23). Moreover, inhibition of SIRT1 has been linked to a suppression of estrogen receptor signaling (24) and has been reported to be essential for oncogenic signaling by estrogen receptor (ER)α in breast cancer (25). Given the impact of the sirtuins on several physiological properties frequently altered in breast cancer, we sought to identify novel targets of small molecule inhibitors of the sirtuins in breast cancer cell lines. Interestingly, we found that multiple small molecule inhibitors of SIRT1/2 and SIRT1 decrease the mRNA and protein levels of aromatase. We found that SIRT1 is a positive regulator of aromatase mRNA levels. Moreover, we found that SIRT1 localizes to the PII/I.3 and PI.4 promoters in breast cancer cells and regulates ERRα acetylation and protein levels. Finally, we found that SIRT1 is significantly overexpressed in invasive ductal carcinoma (IDC) relative to normal breast tissue.
Results
Given the role of the sirtuins on several physiological properties frequently altered in breast cancer (19, 24) and its role in mammary gland development (26), we sought to identify novel targets of the sirtuins in breast cancer cell lines. Because SIRT1-null animals show defects in corpus luteum formation (27) and defects in spermatogenesis (27, 28), processes which both require estrogen biosynthesis, we reasoned that SIRT1 must be important in estrogen biosynthesis. Aromatase expression is significantly increased in breast cancer tissue, and the dependency of ERα-positive cells on the estradiol produced by the aromatase-expressing ERα-negative cells directly contributes to malignancy (29, 30). Whether the production of estrogen comes from mesenchymal stromal cells, adipocytes, ERα-negative epithelial cells, peripheral tissue, or a combination of mixed cell populations, it is clear that the paracrine/autocrine signaling is critical for tumor progression (30–33).
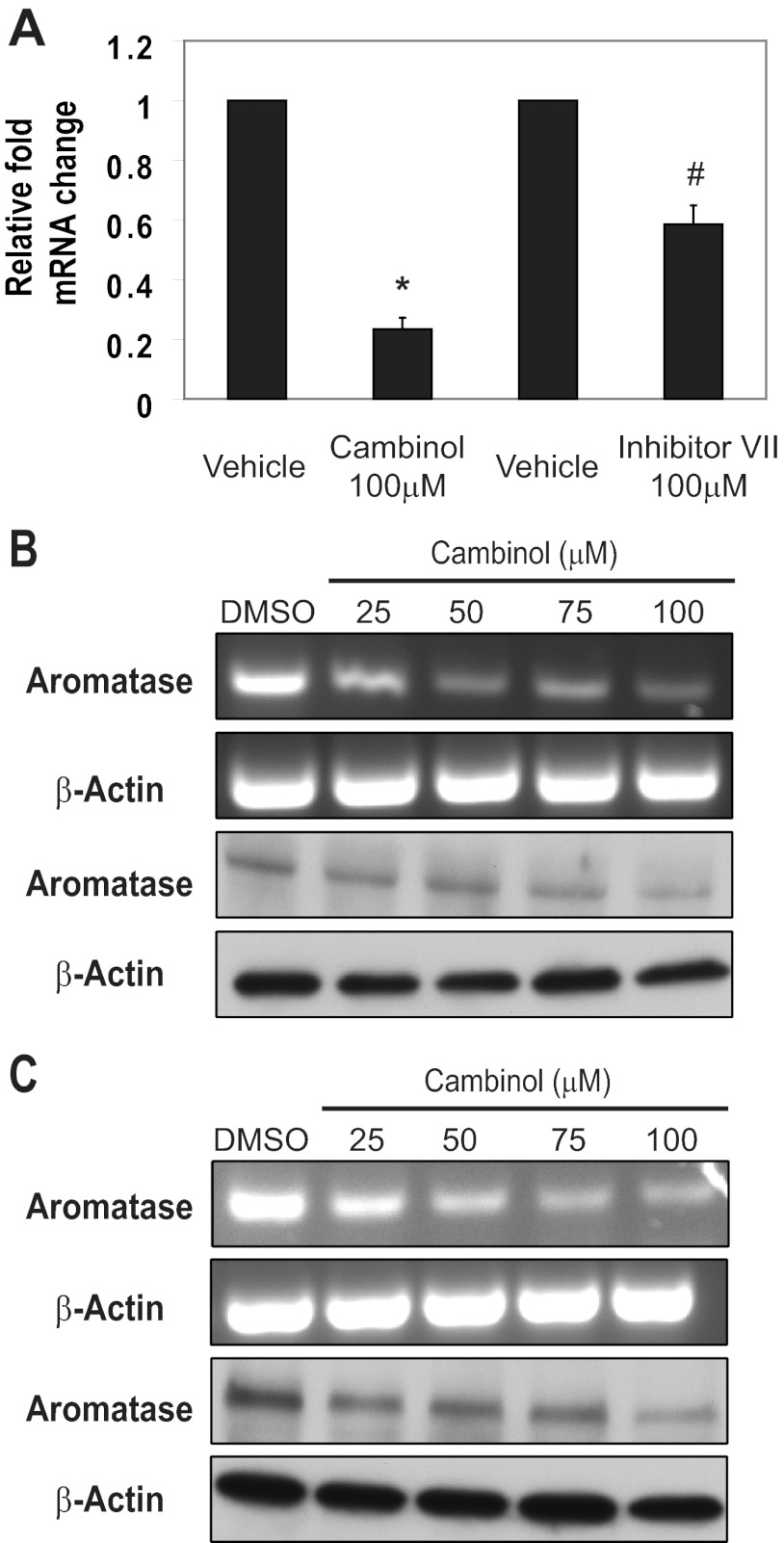
We used cambinol, a specific inhibitor of SIRT1 and SIRT2 (34), and inhibitor VII, another SIRT1/2 inhibitor, to treat MDA-MB-231 cells that express aromatase (35). Previous studies demonstrated that cambinol specifically inhibits SIRT1 and SIRT2 but none of the other sirtuins or class I/II histone deacetylases (HDACs) (34). The SIRT1/2 inhibitor VII is a thiobarbiturate compound, where computer-aided molecular structural analysis suggests that it docks into the acetyl lysine substrate channel (EMD Biosciences, San Diego, CA). We treated MDA-MB-231 cells with either cambinol or inhibitor VII for 32 hours and observed a significant decrease in the levels of aromatase transcript via real-time PCR analysis (Figure 1A). These results show that using 2 different pharmacologic inhibitors of SIRT1 and SIRT2 leads to a striking reduction in aromatase mRNA by 80% (cambinol) or 40% (inhibitor VII) relative to vehicle control in MDA-MB-231 cells. To determine the range of cambinol that would lead to decreases in aromatase mRNA, we treated MDA-MB-231 cells for 32 hours with cambinol ranging from 25μM to 100μM. We observed a reduction in mRNA with the lowest dose of cambinol (Figure 1B, top panels). Next, to determine whether this change in mRNA lead to a reduction in protein, we performed Western blottings on parallel samples. We found that cambinol treatment of MDA-MB-231 cells caused a dose-dependent reduction in aromatase protein (Figure 1B, bottom panels). To determine whether a shorter treatment of cambinol lead to similar changes in aromatase levels, we treated cells for 24 hours and again found that there was a dose-dependent reduction in aromatase mRNA (Figure 1C, top) and protein (Figure 1C, bottom). Although the AIs inhibit the enzymatic activity reversibly (letrozole, anastrozole) or irreversibly (exemestane), the SIRT1/2 inhibitors cambinol and inhibitor VII lead to a decrease in the mRNA and protein levels of aromatase.
Figure 1.
Inhibition of SIRT1 and SIRT2 in MDA-MB-231 cells causes a decrease in aromatase transcript and protein. (A) MDA-MB-231 cells were treated with 100μM cambinol or inhibitor VII for 32 hours, and RNA was isolated and reverse transcribed. Quantitative PCR was performed to determine relative changes in mRNA transcripts. (B and C) MDA-MB-231 cells were treated with varying amounts of cambinol over a (B) 32-hour or (C) 24-hour period with DMSO used as a vehicle control. RNA was isolated, and cDNA was generated. PCR was performed using intron-spanning primers. For Western blot analysis, equal protein was analyzed using antibodies against aromatase and β-actin. Error bars reflect SD from at least 3 independent experiments.*P < .0001, #P < .0001.
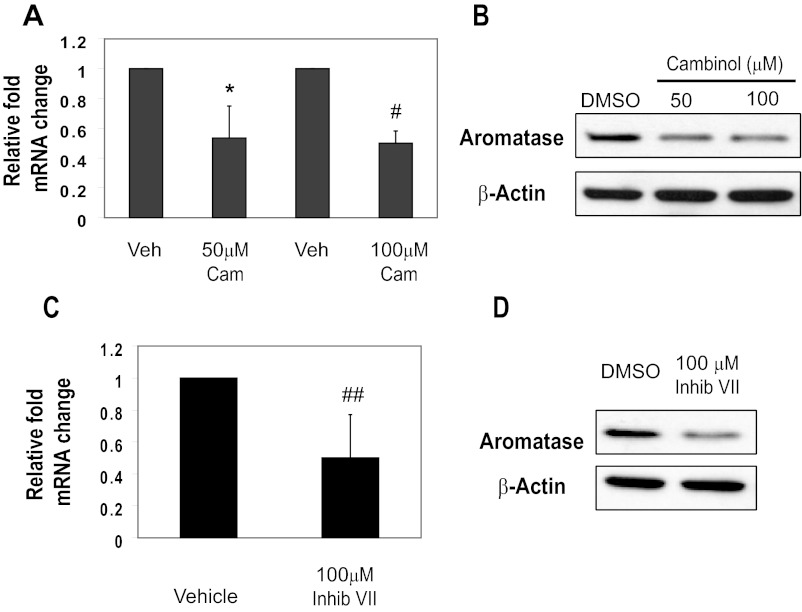
Hormone-dependent breast tumors are characterized by a heterogeneous population of ERα positive and negative cells that maintain autocrine and/or paracrine signaling necessary for tumor progression. Therefore, we extended our analysis to ERα-positive cell lines. We treated the ERα-positive MCF-7 cell line with 50μM or 100μM cambinol for 24 hours and observed a decrease in the levels of aromatase transcript via real-time PCR analysis (Figure 2A). Next, to determine whether decrease in aromatase mRNA resulting from cambinol treatment would lead to a reduction in protein, we performed Western blottings. We found that treatment of MCF-7 with cambinol at both 50μM and 100μM for 24 hours caused a reduction in aromatase protein (Figure 2B). Moreover, we found treatment of MCF-7 cells with 100μM SIRT1/2 inhibitor VII for 24 hours caused a decrease in the levels of aromatase transcript via real-time PCR analysis (Figure 2C) and a reduction in aromatase protein (Figure 2D). Together, these results show that using 2 different pharmacologic inhibitors of SIRT1 and SIRT2 leads to a significant reduction in aromatase mRNA and protein in both ERα positive and negative cells.
Figure 2.
Inhibition of SIRT1 and SIRT2 in MCF-7 cells causes a decrease in aromatase transcript and protein. (A) MCF-7 cells were treated with 50μM or 100μM cambinol for 24 hours. RNA was isolated, cDNA was generated, and quantitative PCR was performed to determine relative changes in mRNA transcripts. Error bars reflect SD from at least 3 independent experiments.*P = .012, #P = .0005. (B) MCF-7 cells were treated with DMSO, 50μM, or 100μM cambinol for 24 hours, and aromatase and β-actin levels were analyzed by Western blot analysis of equal protein. (C) MCF-7 cells were treated with 100μM inhibitor VII for 24 hours, and mRNA changes were assessed as described in A. Error bars reflect SD from at least 3 independent experiments. ##P = .033. (B) MCF-7 cells were treated with DMSO or 100μM cambinol for 24 hours, and aromatase and β-actin levels were analyzed by Western blot analysis of equal protein. Veh, vehicle; Cam, cambinol; inhib, inhibitor.
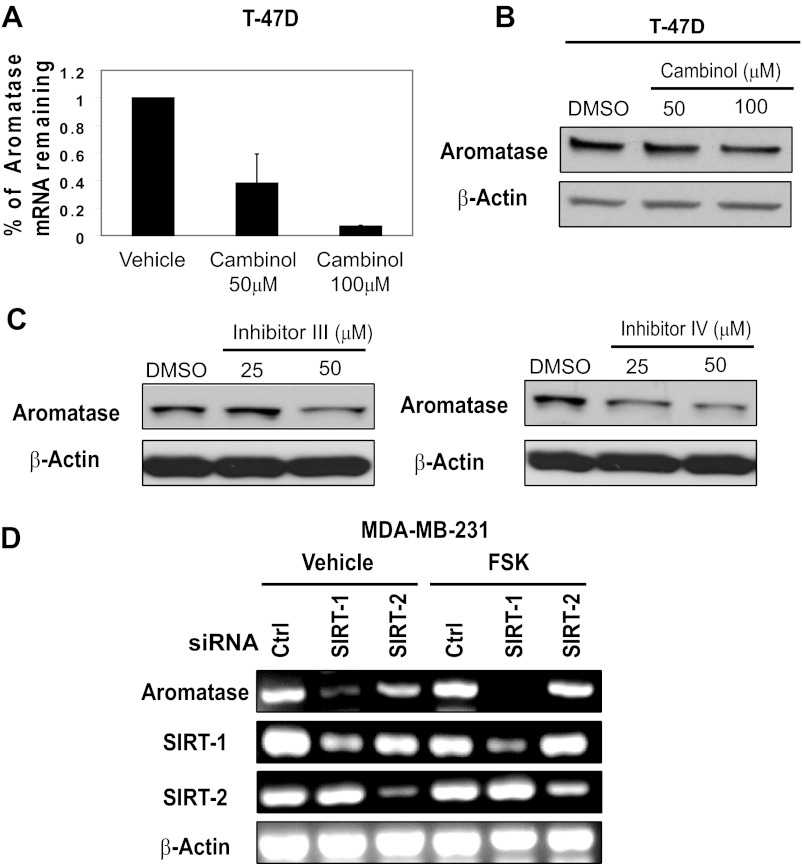
We further extended our analysis to a second ERα-positive cell line. We treated the ERα-positive cell line T-47D with either 50μM or 100μM cambinol for 24 hours and observed a 50% and 90% reduction in aromatase mRNA, respectively (Figure 3A). Next, to determine whether decrease in aromatase mRNA resulting from cambinol would lead to a reduction in protein, we performed Western blot analysis. We found that treatment of T-47D with cambinol at both 50μM and 100μM for 24 hours caused minimal reduction in aromatase protein (Figure 3B). Although aromatase mRNA reduction corresponds with a reduction in protein in MCF-7 cells, aromatase mRNA reduction does not show a proportional reduction in protein in T-47D cells. This suggests that although both cell lines are ERα-positive, their transcriptional or translational control of aromatase may be distinct. Because cambinol and inhibitor VII inhibit both SIRT1 and SIRT2, we wanted to determine the specific contribution of SIRT1 in the regulation of aromatase expression. We extended our analysis using SIRT1-specific pharmacologic inhibitors (inhibitor IV and III). We treated MDA-MB-231 cells with SIRT1 inhibitor III (Figure 3C, left) and SIRT1 inhibitor IV (Figure 3C, right) and observed a decrease in aromatase starting at 50μM and 25μM, respectively.
Figure 3.
Inhibition of SIRT1 and SIRT2 in T-47D cells and SIRT1 inhibition in MDA-MB231 cells causes a decrease in aromatase transcript. (A) T-47D cells were treated with 50μM or 100μM cambinol for 24 hours, RNA was isolated, cDNA was generated, and quantitative PCR was performed to determine relative changes in mRNA transcripts. Error bars reflect SD from 2 independent experiments. (B) T-47D cells were treated with DMSO, 50μM, or 100μM cambinol for 24 hours, and Western blot analysis was performed on equal protein using antibodies against aromatase and β-actin. (C) MDA-MB-231 cells were treated with vehicle (DMSO), 25μM, or 50μM inhibitor III (left) or inhibitor IV (right) for 24 hours, protein was isolated quantitated, and equal amounts were analyzed by Western blot analysis for aromatase and β-tubulin. Data shown are representative of at least 3 independent experiments. (D) MDA-MB-231 cells were transfected with either a control, SIRT1, or SIRT2 siRNA (100nM) for a total of 72 hours in the presence or absence of 10μM forskolin for 24 hours after knockdown. Semiquantitative RT-PCR was used to monitor changes in aromatase, SIRT1, and SIRT2 expression under each of the conditions shown. β-Actin is shown as a loading control. Data shown are representative of at least 3 independent experiments. Ctrl, control; FSK, forskolin.
Because we saw a robust decrease with 32-hour pharmacological inhibition, given the faster kinetics of drug inhibition, we performed a 72-hour knockdown of SIRT1 and SIRT2 in MDA-MB-231 cells and observed a striking decrease in the levels of aromatase mRNA (Figure 3D). Both SIRT1 and SIRT2 knockdown cause a decrease in aromatase, although the knockdown of SIRT1 caused a more marked reduction in aromatase. Forskolin increases intracellular cAMP levels, and cAMP is a well-known activator of aromatase within the ovary. Additionally, because forskolin had been reported to regulate aromatase expression (36), we treated cells for 24 hours with forskolin after SIRT1 or SIRT2 knockdown. As shown in Figure 3D, forskolin treatment did not overcome the effects of the SIRT1 knockdown, suggesting that SIRT1 may contribute more to maintaining aromatase mRNA levels than SIRT2 in the presence or absence of increased cAMP levels. Together, these data suggest that both SIRT1 and SIRT2 are involved in regulating aromatase mRNA levels and that dual inhibition of both enzymes leads to significant decreases in aromatase mRNA.
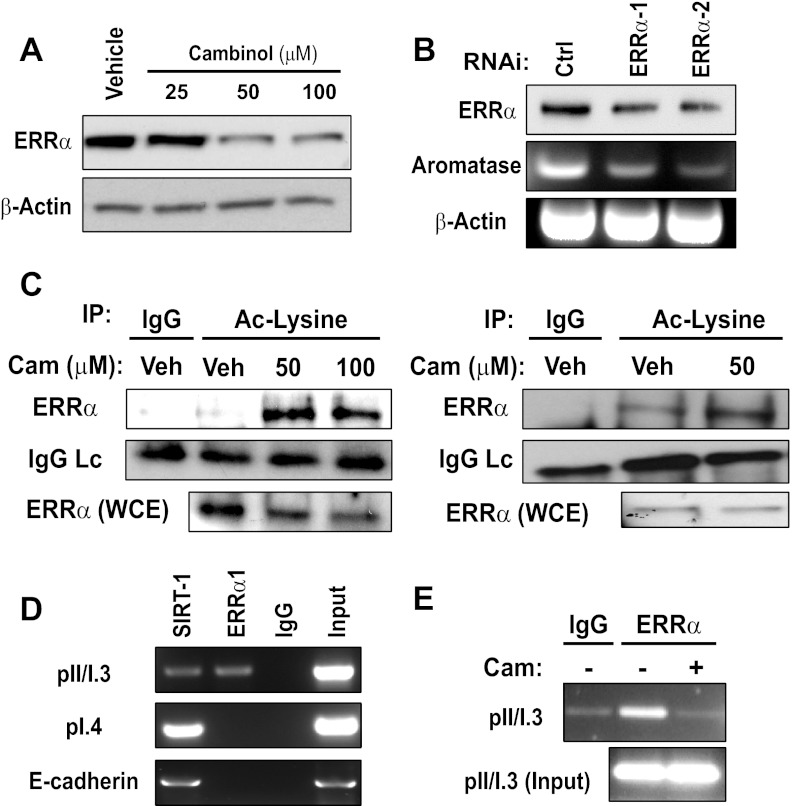
SIRT1 has been demonstrated to deacetylate ERRα, enabling it to directly bind DNA and regulate transcription (23). Moreover, ERRα has been shown to occupy the PI.3/PII promoter and activate aromatase transcription (37). Although these reports suggest a connection between SIRT1 and ERRα, their relationship with respect to aromatase expression in breast cancer has not been investigated. First, to determine whether SIRT1/2 is linked with ERRα, we treated MDA-MB-231 cells with increasing doses of cambinol for 24 hours. Strikingly, we found that SIRT1/2 inhibition caused a decrease in the levels of ERRα protein (Figure 4A). Next, to determine whether ERRα regulates aromatase expression in MDA-MB-231 cells, we knocked down ERRα with 2 distinct sequences targeting different regions of mRNA. We found that depletion of ERRα caused a decrease in aromatase mRNA levels (Figure 4B). Next, we wanted to determine whether cambinol-induced decrease in aromatase corresponded with changes in ERRα acetylation. To do this, we treated exponentially growing MDA-MB-231 cells with vehicle, 50μM, or 100μM cambinol for 30 minutes and assessed changes in acetylation of ERRα. As a control, we blotted for the antibody light chain to ensure that the increase in acetylation did not correspond with an increase in the amount of ERRα antibody captured by the beads. We also blotted for ERRα in whole cell extracts to ensure that increased ERRα acetylation was not due to an increase in total ERRα levels. We found that SIRT1/2 inhibition markedly increased ERRα acetylation (Figure 4C, left panel). We repeated this experiment in confluent cells and observed similar induction of ERRα acetylation (Figure 4C, right panels). These data demonstrate that the doses of cambinol that causes a reduction in aromatase also increase the acetylation of ERRα as early as 30 minutes after treatment.
Figure 4.
SIRT1 occupies aromatase promoters PII/PI.3 and PI.4 and inhibition causes a decrease in aromatase transcripts and ERRα protein. (A) MDA-MB-231 cells were treated with vehicle (DMSO) or increasing concentrations of cambinol (25μM, 50μM, and 100μM) for 24 hours, and Western blottings for ERRα and β-actin were performed on equal protein. (B) MDA-MB-231 cells were stably infected with either a control or virus targeting 2 distinct regions of ERRα. Western blottings for ERRα (top panel) and semiquantitative RT-PCR were used to monitor changes in aromatase (middle panel). β-Actin is shown as a loading control (bottom panel). (C) (left panel) Subconfluent MDA-MB-231 cells were treated with DMSO or increasing concentrations of cambinol (50μM and 100μM) for 30 minutes, and acetyl-lysine (#9681) was immunoprecipitated followed by Western blottings for ERRα. The antibody light chain (LC) was probed, and a Western blotting for ERRα in whole cell extracts (WCE) was performed. (C) (right panel) Confluent MDA-MB-231 were treated with DMSO or 50μM cambinol for 30 minutes, and acetyl-lysine was immunoprecipitated followed by Western blottings for ERRα. (D) ChIP of MDA-MB-231 cells for SIRT1, ERRα, and rabbit IgG were performed. Primers against the promoter II/I.3 and promoter I.4 were used to determine the presence of both SIRT1 and ERRα at these promoters. E-cadherin primers were also included as a control, because SIRT1 is known to bind to its promoter. The input lane represents sonicated material that was not exposed to antibody immunoprecipitation. (E) ChIP for rabbit IgG and ERRα in MDA-MB-231 cells treated with either vehicle (−) or 100μM cambinol (+) for 3 hours. Primers against the promoter II/I.3 were used to determine the presence of ERRα at this promoter region. The input lane represents sonicated material that was not exposed to antibody immunoprecipitation. Data shown are representative of at least 3 independent experiments. RNAi, RNA interference; Ctrl, control; Veh, vehicle; Cam, cambinol; IP, immunoprecipitation; AC, acetyl.
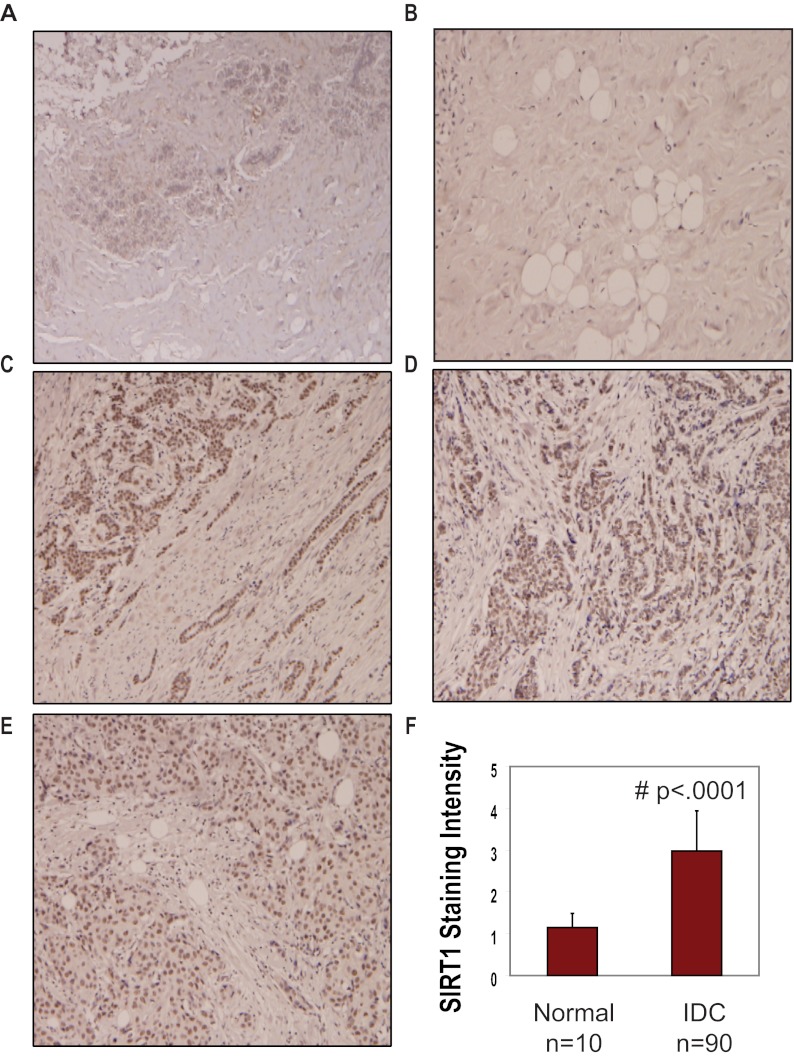
These data suggested that SIRT1 and ERRα collaborate to regulate CYP19A1 transcription. To determine whether SIRT1 and ERRα are localized to the key promoters driving CYP19A1 transcription, we performed chromatin immunoprecipitation (ChIP) followed by PCR to identify regions of the aromatase promoter occupied by SIRT1 and ERRα. Antibodies specific for SIRT1, ERRα, and rabbit IgG were used to determine localization of each protein at 2 CYP19A1 promoters. Primers were designed to detect occupancy of each protein at the region between aromatase promoter II and I.3, the region previously established to be occupied by ERRα. ERRα was found bound to the promoters II/I.3 as previously demonstrated (Figure 4A, top panel) (37). Interestingly, we also observed SIRT1 binding to the same region to which ERRα was bound (promoter II/I.3). Additionally, SIRT1 binding was observed at promoter I.4, a region not bound by ERRα (Figure 4A, middle panel). These data are the first to demonstrate that SIRT1 not only positively regulates the expression of aromatase but also localizes to multiple promoters that have been shown to drive expression in a tissue-specific manner and in breast cancer. Finally, we observed that SIRT1 also localizes to the E-cadherin promoter as demonstrated previously (Figure 4A, bottom panel) (19). Localization of SIRT1 to an actively transcribed gene, such as CYP19A1, and to a silenced tumor suppressor gene, such as E-cadherin, demonstrates its ability to activate and repress specific genes. If SIRT1 is deacetylating ERRα in its DNA binding domain as previously demonstrated (23), it may thereby enable ERRα to bind and transcribe CYP19A1. This has important implications for targeting SIRT1 pharmacologically in tumors where ERRα expression is associated with an unfavorable outcome (38, 39). This suggests that perhaps the inhibition of SIRT1 and SIRT2 leads to a reduction in aromatase mRNA by decreasing the levels of ERRα in MDA-MB-231 cells, which is preceded by increased acetylation. Next, we wanted to see whether SIRT1/2 inhibition caused ERRα to vacate the PII/I.3 promoter. Strikingly, we found that treatment of MDA-MB-231 cells with 100μM cambinol for only 3 hours caused a decrease in the levels of ERRα protein that localized to the pII/I.3 promoter (Figure 4D). We also observed a similar decrease at 2-hour cambinol treatment (data not shown). Treatment of MDA-MB-231 cells with cambinol for 10 hours or less shows no change in aromatase protein (data not shown). Together, these data demonstrate that inhibition of SIRT1/2 induces ERRα acetylation, which precedes a reduction in aromatase protein. The timing of this ERRα acetylation correlated with its leaving the CYP19A1 promoter. Previously, we demonstrated that SIRT1 is a positive regulator of Dvl proteins and constitutive Wnt signaling in breast cancer cells (21). In this study, we found that SIRT1 is a positive regulator of aromatase. Next, we wanted to determine whether SIRT1 is differentially expressed in human malignant breast tumors relative to normal tissue. To address this, we performed immunohistochemistry on 10 normal breast tissues adjacent to tumors and 90 IDCs. Representative images of SIRT1 staining in benign breast tissue (Figure 5A) and normal breast tissue adjacent to cancer (Figure 5B) are shown. As shown, SIRT1 expression was found to increase in IDC at different grades and stages, such as grade-1/stage-IIIb (Figure 5C), grade-2/stage-I (Figure 5D), and grade-2/stage-IIIb) (Figure 5E). SIRT1 staining intensity was scored as 1 (negative), 2 (minimal), 3 (moderate), or 4 (strong). A sample was deemed positive for SIRT1 if the staining intensity was scored between 2 and 4. We found that 20% (2/10) of the benign tissue and 90% (81/90) of the IDC were positive for SIRT1 expression. We also found a statistically significant difference in the level of expression of SIRT1 in the IDC vs benign or normal tissue. The mean staining intensity for SIRT1 was 2.98 ± 0.96 in IDC relative to 1.15 ± 0.34 in the benign tissue (P < .0001) (Figure 5C). This corresponds to a 2.6-fold increase in SIRT1 expression in IDC relative to benign tissue. Together, these data demonstrate that SIRT1 is not only a positive regulator of aromatase in human breast cancer cell lines but also is increased significantly in IDC of the breast relative to benign or normal breast tissue adjacent to tumor.
Figure 5.
SIRT1 is expressed at higher levels in IDC relative to normal tissue adjacent to tumor. Immunohistochemistry was performed on 90 IDCs and 10 normal tissues adjacent to tumor. (A–E) Shown are representative images of SIRT1 staining in benign breast tissue (A), normal breast tissue adjacent to cancer (B), grade-1/stage-IIIb IDC (C), grade-2/stage-I IDC (D), and grade-2/stage-IIIb IDC (E). (F) SIRT1 staining intensity was scored as 1 (negative), 2 (minimal), 3 (moderate), or 4 (strong). Shown is a statistically significant difference in the level of expression of SIRT1 in the IDC vs normal tissue adjacent to tumor. The mean staining intensity for SIRT1 was 2.98 ± 0.96 in IDC relative to 1.15 ± 0.34 in the normal adjacent to tumor tissue (P < .0001). This corresponds to a 2.6-fold increase in SIRT1 expression in IDC relative to benign tissue.
Discussion
Historically, HDACs have been associated with formation of heterochromatin and gene repression as is the case with growth control or tumor suppressor genes (19). However, more recent reports are demonstrating that deacetylation of transcription factors and DNA binding proteins can be just as important in the activation of gene expression (23, 40) and thus the term lysine deacetylase is gaining more popularity. This report provides the first evidence that SIRT1 directly regulates expression of aromatase by acting at specific promoter regions and thereby may contribute to its overexpression in breast cancer.
AIs have proven to be one of the most clinically effective breast cancer therapies. However, the broad function of the inhibitors causes a significant decrease in estrogen biosynthesis in all estrogen-producing tissues resulting in side effects such as bone degradation, cognitive dysfunction, and arthralgia (5). Finding a way to selectively suppress transcription from promoters driving the aberrant increase in aromatase without blocking transcription from other promoters not associated with aromatase overexpression in breast cancer may be valuable in further tailoring therapy and potentially decreasing side effects. One challenge with this goal is that only in primates has aromatase been shown to function in tissues other than the gonads or brain (41, 42). In female mice, aromatase transcription is driven by 2 promoters and is expressed only in the ovaries and brain, whereas aromatase is expressed in many peripheral tissues, such as the breast in women (6, 43). Because of this, generation of mouse models to study promoter switching in cancer has been a challenge. The results described in this article could help in understanding how SIRT1 might be involved in promoting switching that occurs in breast cancer. Moreover, the link between SIRT1 and aromatase, which has a drastically different pattern of expression in humans vs rodents, may also explain some of the conflicting results found concerning the role of SIRT1 in human vs mouse-derived tumors. Transcriptional control of CYP19A1 presents a novel target for developing new therapies in which individual promoters could be effectively targeted through unique regulatory factors. Only very recently have the class I/II (nonsirtuin) HDACs been implicated in the regulation of aromatase transcription of sensitivity to AIs (44, 45). Here, for the first time, we demonstrate that multiple small molecule inhibitors of SIRT1 and SIRT2 and SIRT1-specific small interfering RNA (siRNA) result in a significant decrease in aromatase expression.
We also show that SIRT1 physically associates with aromatase promoters II/I.3, suggesting that SIRT1 has an even more direct role in the transcriptional control of aromatase expression by targeting a nonhistone protein or transcription factor that activates CYP19A1 transcription. SIRT1 has previously been shown to regulate the activity of a number of nonhistone proteins, including Peroxisome proliferator-activated receptor γ coactivator 1α (46), ERRα (23), and β-catenin (21). Each factor has recently been implicated in the regulation of aromatase transcription on some level (47, 48). Moreover, Kuperwasser and coworkers (49) identified a poor prognosis gene signature, in which several components of the Wnt signaling pathway were overexpressed in early lung metastases. They demonstrated that the Wnt genes identified in this signature were strongly associated with human basal-like breast cancers. Interestingly, we previously reported that SIRT1 is a major regulator of all 3 Dvl proteins, which relay diverse signals from as many as 19 Wnt ligands and 10 receptors (Frizzleds) (21). This perhaps hints of an important link between SIRT1, Wnt/Dvl signaling, and aromatase in subsets of breast cancer cells. Finally, we demonstrate that SIRT1 also is significantly increased in IDC of the breast relative to benign or normal breast tissue by 2.6-fold. This increase in SIRT1 expression is consistent with other reports of SIRT1 up-regulation in malignant human breast carcinoma (50), hepatocellular carcinoma (51), diffuse B-cell lymphoma (52), gastric carcinoma (53), and colorectal cancer with microsatellite instability and CpG island methylator phenotype (54).
Finally, ERRα has been shown to be a potent prognostic factor in human breast carcinoma and was found to be significantly associated with an increased risk of recurrence and adverse clinical outcome (39). Moreover, genome-wide studies have implicated ERRα as a determinant of breast cancer heterogeneity (55). More recently, ERRα has been explored as a therapeutic target especially in cases of ERα-negative breast cancer (38, 39). This is important because data shown here suggest that sirtuin inhibitors regulate ERRα and aromatase expression at least in part through SIRT1-specific inhibition. Here, we demonstrated that cambinol caused a decrease in ERRα, which could provide a specific means of targeting tumors in which ERRα is a prognostic indicator. However, it is possible that the relative contribution of LRH-1 vs ERRα may depend on the ERα status (positive vs negative) or on the tissue type (stromal/mesenchymal/adipose vs epithelial) (17). Therefore, it is possible that part of the SIRT1-mediated regulation in MCF-7 cells may be LRH-1 dependent, although we did not test this possibility. However, regardless of whether it is LRH-1 or ERRα dependent, these findings provide a potentially unique therapeutic framework with some of the more potent next generation sirtuin inhibitors currently being developed. Perhaps these could be administered in combination with lower doses of AIs. Consistent with the cambinol-mediated inhibition, this report is the first to demonstrate that the SIRT1/2 inhibitor (inhibitor VII) and the SIRT1-specific inhibitors (inhibitors III and IV) also cause a reduction in aromatase levels. This study provides the first implication of the sirtuin family of deacetylases in the regulation of aromatase. It will be important to characterize all 10 CYP19A1 promoters in greater detail to potentially identify additional transcriptional regulators for more tailored treatments with fewer side effects.
Materials and Methods
Cell culture
Cell lines were obtained from the American Type Culture Collection (Manassas, Virginia). MDA-MB231 (HTB-26), MCF-7, and T-47D cells were cultured in L-15, MEM, or RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen, Carlsbad, California). Transfections were performed with Oligofectamine according to the manufacturer's protocol. Unless stated otherwise, cells were seeded at approximately 2 × 105 per well in a 6-well dish and transfected with 100nM siRNA for 72 hours with transfections repeated every 24 hours. ONTarget siRNAs were obtained from Dharmacon (Lafayette, Colorado) for the following targets: random sequence control (D-001810-01-20), SIRT1 (M-003540-01-0005), and SIRT2 (M-004826-02-0005). For inhibitor studies, cambinol (C0494; Sigma, St Louis, Missouri) and inhibitors III, IV, and VII (566322, 566323, and 566327; EMD-Millipore, Bedford, Massachusetts) were resuspended in dimethylsulfoxide (DMSO), and cells were treated with DMSO as a vehicle control and varying amounts as noted in the figure legends. Media were changed on cells immediately preceding treatment with cambinol. Forskolin (F6886; Sigma) was resuspended in DMSO and used at a final concentration of 10μM. Media were changed on cells immediately preceding treatment with forskolin.
Endpoint and real-time quantitative PCR
Intron-spanning primers specific for human aromatase, aromatase PI.3, PI.4, ERRα-1, SIRT1, and β-actin. Total RNA was isolated using TRIzol (Invitrogen), and 2 μg of RNA were reverse transcribed with Moloney murine leukemia virus Reverse Transcriptase (Promega, Madison, Wyoming). Reactions to detect aromatase products incorporated PCRx Enhancer System (Invitrogen) and JumpStart RedTaq (Sigma). The Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, California) and PerfeCTa SYBR Green FastMix, ROX (catalog number 95073-012; Quanta BioSciences, Inc, Gaithersburg, Maryland) were used for real-time analyses. Statistical analysis involving unpaired Student's t tests were performed for assessing whether aromatase mRNA levels between the samples treated either with vehicle control or SIRT1 inhibitor were significant (see Figure 5F).
Western blottings
Antibodies used in Western blottings were as follows: aromatase (A7981; Sigma), ERRα-1 (07-662; Millipore), β-actin (Santa Cruz Biotechnology, Inc, Santa Cruz, California), and β-tubulin (Sigma). Membranes were incubated in 5% milk/Tris-buffered saline with Tween 20 (TBST) with primary antibody overnight at 4°C. Membranes were washed briefly with TBST and probed with horseradish peroxidase-conjugated secondary antibodies in 5% milk/TBST for 1 hour at room temperature. Membranes were washed with TBST and deionized water before visualization by enhanced chemiluminescence.
Immunohistochemistry and statistical analysis
All tissue was collected under the highest ethical standards with the donor being informed completely and with their consent. All human tissues were collected under Health Insurance Portability and Accountability Act-approved protocols, and tissue microarrays consisted of 100 malignant IDCs and 10 adjacent normal tissues with 110 cores representing 110 cases. Of the cases, 20% were grade 1, 67% were grade 2, and 5% were grade 3 with the remainder being scored as grade 1–2 or 2–3. The tumor stage ranged from I to IIIb, and the average age of the patients was 48 (BC081120; US Biomax, Inc, Rockville, Maryland). Paraffin sections were labeled and dried in 60°C oven for at least 1 hour, cooled, deparaffinized, and incubated in antigen retrieval solution (#H-3300; Vector Laboratories, Burlingame, California). For antigen retrieval, slides were heated and cooled in antigen retrieval solution for 25 and 20 minutes, respectively. Slides were then rinsed 4–5 times in dH2O; once in 0.3% peroxide in 50% methanol for 30 minutes, and 3 times for 5 minutes in wash buffer. Slides were then are put on the BioGenex i6000 Automated Staining System, and wash buffer was added onto each slide to keep tissue from drying out while machine is putting antibody on all the slides. The block step consisted of 10% goat serum in PBS, for 15 minutes, 5% casien block in PBS, for 10 minutes, and 10% goat serum in PBS, for 1 minute. The primary antibody (SIRT1, 1104-1; Epitomics, Burlingame, California) was added to Dako antibody diluent (Dako, Glostrup, Denmark) for 1 hour, rinsed 5 times in wash buffer. The secondary (MultiLink-BioGenex Super Sensitive Link-Label IHC Detection System #QP900-9L; BioGenex, Fremont, California) was applied for 20 minutes, rinsed 3 times in wash buffer, and horseradish peroxidase labeled (BioGenex Super Sensitive Link-Label IHC Detection System #QP900-9L) for 15 minutes, and rinsed 3 times in wash buffer. 3,3-Diaminobenzidine (#K3466; DakoCytomation Liquid DAB Substrate Chromogen System) was applied for 5 minutes, followed by 3 rinses in wash buffer. Hematoxylin (#S3301; DakoCytomation Automation Hematoxylin) was applied for 2 minutes, followed by 3 rinses in wash buffer. Slides were then rinsed with dH2O for 4 minutes, dehydrated sequentially with ethanol and xylene.
Table 1.
Primer Sequences
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Expression | ||
| Aromatase For/Rev | ACCCTTCTGCGTCGTGTCA | TCTGTGGAAATCCTGCGTCTT |
| Aromatase F1/R1 | TGCGGTACCAGCCTGTCGTG | CCACGGGGCCCAAAGCCAAA |
| Aromatase F2/R2 | GGGATCGGCAGTGCCTGCAA | AACAAGGCCGGGGCCTGACA |
| SIRT1 | GCAGATTAGTAGGCGGCTTG | TCTGGCATGTCCCACTATCA |
| SIRT2 | TCGCAGAGTCATCTGTTTGG | GCTTGAACTGCCCAGGATAG |
| β-Actin | GGACTTCGAGCAAGAGATGG | AGCACTGTGTTGGCGTACAG |
| ChIP | ||
| PII/I.3 | AACCTGCTGATGAAGTCACAA | TCAGACATTTAGGCAAGACT |
| PI.4 | ATGCCAACCAAGACTAAGAG | CAGTTGGTCACGTTCTACTTGG |
| E-cadherin | TAGAGGGTCACCGCGTCGATG | GGGTGCGTGGCTGCAGCCAGG |
| Promoter specific | ||
| PI.3 | GTCTTGCCTAAATGTCTGATC | CAGAGATCCAGACTCGCATG |
| PI.4 | GTAGAACGTGACCAACTGGAG | AGGCACGATGCTGGTGATG |
In evaluation of SIRT1 stained sections, interior tissue sections were examined. SIRT1 staining intensity was scored as 1 (negative, no nuclear staining), 2 (minimal, recognizable but weak staining in the nuclei), 3 (moderate, unequivocal staining in the nuclei), or 4 (strong staining in the nuclei) by a board certified pathologist. A sample was deemed positive for SIRT1 if the staining intensity was scored between 2 and 4. Statistical analysis involving unpaired Student's t tests was performed for assessing whether the difference in SIRT1 expression between the IDC and normal adjacent to tumor was significant. The mean and SD was calculated at a 95% confidence interval for the mean.
Chromatin immunoprecipitation
MDA-MB-231 cells were grown to approximately 70% confluence. Proteins were cross-linked to DNA using formaldehyde added directly to the culture medium at a final concentration of 1% for 10 minutes at room temperature. The cross-linking reaction was quenched by adding glycine to a final concentration of 0.125M for 5 minutes at room temperature. The medium was then removed, and cells were washed with 1× PBS containing a protease inhibitor cocktail. The PBS was removed, and 0.2× trypsin was added to the cells. After a 5-minute incubation at 37°C, ice-cold 1× PBS containing 10% fetal bovine serum was added to stop trypsinization. The cells were scraped, pelleted, and washed twice with PBS plus protease inhibitor cocktail as described above. Cells were resuspended in sodium dodecyl sulfate lysis buffer (20-163; Millipore) plus protease inhibitor cocktail. Cells were sonicated in a Diagenode Bioruptor sonicator for 18 rounds of sonication (30-second pulses and 30-second rest). The soluble chromatin fraction was collected and incubated overnight at 4°C with either SIRT1 (DB083; Delta Biolabs, Gilroy, California), ERRα (ab16363; Abcam, Cambridge, MA), or rabbit IgG (I5006; Sigma). Magnetic agarose beads (Invitrogen) were added to the chromatin-antibody mixture and incubated with rotation for 1 hour at 4°C. ChIPs were washed with a low salt wash buffer (20-154; Millipore), high-salt wash buffer (20-155; Millipore), and Tris-EDTA (20-157; Millipore). Cross-links were reserved o/n at 65°C followed by treatment with RNAse A and Proteinase K (Promega) for 2 hours at 37°C and 55°C, respectively. DNA was eluted using Qiaquick PCR purification kit (QIAGEN, Valencia, California) and amplified as described above.
Acknowledgments
We thank Tara Calhoun for excellent technical assistance during the early stages of establishing the link between SIRT1 and aromatase.
This study was funded by a grant (CA155223) from the National Cancer Institute of the National Instututes of Health and an intramural grant from the Feist-Weiller Cancer Center.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AI
- aromatase inhibitor
- ChIP
- chromatin immunoprecipitation
- DMSO
- dimethylsulfoxide
- Dvl
- Dishevelled
- ER
- estragen receptor
- ERR
- estragen-related receptor
- HDAC
- histone deacetylase
- IDC
- invasive ductal carcinoma
- LRH-1
- liver receptor homologue-1
- siRNA
- small interfering RNA
- SIRT1
- sirtuin-1
- TBST
- Tris-buffered saline with Tween 20.
References
- 1. Kuperwasser C. The tumor stromal microenvironment as modulator of malignant behavior. J Mammary Gland Biol Neoplasia. 2010;15:377–379 [DOI] [PubMed] [Google Scholar]
- 2. Stephens PJ, McBride DJ, Lin ML, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068 [DOI] [PubMed] [Google Scholar]
- 4. Jovanovic J, Ronneberg JA, Tost J, Kristensen V. The epigenetics of breast cancer. Mol Oncol. 2010;4:242–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnston SR, Dowsett M. Aromatase inhibitors for breast cancer: lessons from the laboratory. Nat Rev Cancer. 2003;3:821–831 [DOI] [PubMed] [Google Scholar]
- 6. Bulun SE, Lin Z, Imir G, et al. Regulation of aromatase expression in estrogen-responsive breast and uterine disease: from bench to treatment. Pharmacol Rev. 2005;57:359–383 [DOI] [PubMed] [Google Scholar]
- 7. Yue W, Wang JP, Li Y, et al. Effects of estrogen on breast cancer development: role of estrogen receptor independent mechanisms. Int J Cancer. 2010;127:1748–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bulun SE, Lin Z, Zhao H, et al. Regulation of aromatase expression in breast cancer tissue. Ann NY Acad Sci. 2009;1155:121–131 [DOI] [PubMed] [Google Scholar]
- 9. Bonneterre J, Buzdar A, Nabholtz JM, et al. Anastrozole is superior to tamoxifen as first-line therapy in hormone receptor positive advanced breast carcinoma. Cancer. 2001;92:2247–2258 [DOI] [PubMed] [Google Scholar]
- 10. Nabholtz JM, Buzdar A, Pollak M, et al. Anastrozole is superior to tamoxifen as first-line therapy for advanced breast cancer in postmenopausal women: results of a North American multicenter randomized trial. Arimidex Study Group. J Clin Oncol. 2000;18:3758–3767 [DOI] [PubMed] [Google Scholar]
- 11. Mouridsen H, Gershanovich M, Sun Y, et al. Superior efficacy of letrozole versus tamoxifen as first-line therapy for postmenopausal women with advanced breast cancer: results of a phase III study of the International Letrozole Breast Cancer Group. J Clin Oncol. 2001;19:2596–2606 [DOI] [PubMed] [Google Scholar]
- 12. Burstein HJ, Prestrud AA, Seidenfeld J, et al. American Society of Clinical Oncology clinical practice guideline: update on adjuvant endocrine therapy for women with hormone receptor-positive breast cancer. J Clin Oncol. 2010;28:3784–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Santen RJ, Brodie H, Simpson ER, Siiteri PK, Brodie A. History of aromatase: saga of an important biological mediator and therapeutic target. Endocr Rev. 2009;30:343–375 [DOI] [PubMed] [Google Scholar]
- 14. Harada N. [Estrogen synthetase (P-450. aromatase) as a regulatory factor concerning sexual differentiation of brain and sexual behavior—physiological functions and regulation of gene expression of aromatase]. Seikagaku. 1993;65:67–85 [PubMed] [Google Scholar]
- 15. Agarwal VR, Bulun SE, Leitch M, Rohrich R, Simpson ER. Use of alternative promoters to express the aromatase cytochrome P450 (CYP19) gene in breast adipose tissues of cancer-free and breast cancer patients. J Clin Endocrinol Metab. 1996;81:3843–3849 [DOI] [PubMed] [Google Scholar]
- 16. Clyne CD, Speed CJ, Zhou J, Simpson ER. Liver receptor homologue-1 (LRH-1) regulates expression of aromatase in preadipocytes. J Biol Chem. 2002;277:20591–20597 [DOI] [PubMed] [Google Scholar]
- 17. Chand AL, Wijayakumara DD, Knower KC, Herridge KA, Howard TL, Lazarus KA, Clyne CD. The orphan nuclear receptor LRH-1 and ERα activate GREB1 expression to induce breast cancer cell proliferation. PLoS One. 2012;7:e31593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009;69:1702–1705 [DOI] [PubMed] [Google Scholar]
- 19. Pruitt K, Zinn RL, Ohm JE, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hussain M, Rao M, Humphries AE, et al. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 2009;69:3570–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holloway KR, Calhoun TN, Saxena M, et al. SIRT1 regulates Dishevelled proteins and promotes transient and constitutive Wnt signaling. Proc Natl Acad Sci USA. 2010;107:9216–9221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y, Zhang M, Dong H, et al. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene. 2008 [DOI] [PubMed] [Google Scholar]
- 23. Wilson BJ, Tremblay AM, Deblois G, Sylvain-Drolet G, Giguere V. An acetylation switch modulates the transcriptional activity of estrogen-related receptor α. Mol Endocrinol. 2010;24:1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yao Y, Li H, Gu Y, Davidson NE, Zhou Q. Inhibition of SIRT1 deacetylase suppresses estrogen receptor signaling. Carcinogenesis. 2010;31:382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Elangovan S, Ramachandran S, Venkatesan N, et al. SIRT1 is essential for oncogenic signaling by estrogen/estrogen receptor α in breast cancer. Cancer Res. 2011;71:6654–6664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Rajendran GK, Liu N, Ware C, Rubin BP, Gu Y. SirT1 modulates the estrogen-insulin-like growth factor-1 signaling for postnatal development of mammary gland in mice. Breast Cancer Res. 2007;9:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McBurney MW, Yang X, Jardine K, et al. The mammalian SIR2α protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coussens M, Maresh JG, Yanagimachi R, Maeda G, Allsopp R. Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One. 2008;3:e1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen D, Reierstad S, Lu M, Lin Z, Ishikawa H, Bulun SE. Regulation of breast cancer-associated aromatase promoters. Cancer Lett. 2009;273:15–27 [DOI] [PubMed] [Google Scholar]
- 30. Brisken C, O'Malley B. Hormone action in the mammary gland. Cold Spring Harb Perspect Biol. 2010;2:a003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miki Y, Suzuki T, Tazawa C, et al. Aromatase localization in human breast cancer tissues: possible interactions between intratumoral stromal and parenchymal cells. Cancer Res. 2007;67:3945–3954 [DOI] [PubMed] [Google Scholar]
- 32. Wiseman BS, Werb Z. Stromal effects on mammary gland development and breast cancer. Science. 2002;296:1046–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dunbier AK, Martin LA, Dowsett M. New and translational perspectives of oestrogen deprivation in breast cancer. Mol Cell Endocrinol. 2011;340:137–141 [DOI] [PubMed] [Google Scholar]
- 34. Heltweg B, Gatbonton T, Schuler AD, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377 [DOI] [PubMed] [Google Scholar]
- 35. Okubo T, Truong TK, Yu B, et al. Down-regulation of promoter 1.3 activity of the human aromatase gene in breast tissue by zinc-finger protein, snail (SnaH). Cancer Res. 2001;61:1338–1346 [PubMed] [Google Scholar]
- 36. Tseng L, Malbon CC, Lane B, et al. Progestin-dependent effect of forskolin on human endometrial aromatase activity. Hum Reprod. 1987;2:371–377 [DOI] [PubMed] [Google Scholar]
- 37. Yang C, Zhou D, Chen S. Modulation of aromatase expression in the breast tissue by ERRα-1 orphan receptor. Cancer Res. 1998;58:5695–5700 [PubMed] [Google Scholar]
- 38. Ariazi EA, Clark GM, Mertz JE. Estrogen-related receptor α and estrogen-related receptor γ associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res. 2002;62:6510–6518 [PubMed] [Google Scholar]
- 39. Suzuki T, Miki Y, Moriya T, et al. Estrogen-related receptor α in human breast carcinoma as a potent prognostic factor. Cancer Res. 2004;64:4670–4676 [DOI] [PubMed] [Google Scholar]
- 40. Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simpson ER, Mahendroo MS, Means GD, et al. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr Rev. 1994;15:342–355 [DOI] [PubMed] [Google Scholar]
- 42. Sebastian S, Bulun SE. A highly complex organization of the regulatory region of the human CYP19 (aromatase) gene revealed by the Human Genome Project. J Clin Endocrinol Metab. 2001;86:4600–4602 [DOI] [PubMed] [Google Scholar]
- 43. Zhao H, Innes J, Brooks DC, et al. A novel promoter controls Cyp19a1 gene expression in mouse adipose tissue. Reprod Biol Endocrinol. 2009;7:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen S, Ye J, Kijima I, Evans D. The HDAC inhibitor LBH589 (panobinostat) is an inhibitory modulator of aromatase gene expression. Proc Natl Acad Sci USA. 2010;107:11032–11037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sabnis GJ, Goloubeva OG, Chumsri S, Nguyen N, Sukumar S, Brodie AH. Functional activation of ER-α and aromatase by the HDAC inhibitor entinostat increases the sensitivity of ER-negative tumors to letrozole. Cancer Res. 2011;71(5):1893–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118 [DOI] [PubMed] [Google Scholar]
- 47. Parakh TN, Hernandez JA, Grammer JC, et al. Follicle-stimulating hormone/cAMP regulation of aromatase gene expression requires β-catenin. Proc Natl Acad Sci USA. 2006;103:12435–12440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Safi R, Kovacic A, Gaillard S, et al. Coactivation of liver receptor homologue-1 by peroxisome proliferator-activated receptor γ coactivator-1α on aromatase promoter II and its inhibition by activated retinoid X receptor suggest a novel target for breast-specific antiestrogen therapy. Cancer Res. 2005;65:11762–11770 [DOI] [PubMed] [Google Scholar]
- 49. DiMeo TA, Anderson K, Phadke P, et al. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009;69:5364–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee H, Kim KR, Noh SJ, et al. Expression of DBC1 and SIRT1 is associated with poor prognosis for breast carcinoma. Hum Pathol. 2011;42:204–213 [DOI] [PubMed] [Google Scholar]
- 51. Chen J, Zhang B, Wong N, et al. Sirtuin 1 is upregulated in a subset of hepatocellular carcinomas where it is essential for telomere maintenance and tumor cell growth. Cancer Res. 2011;71:4138–4149 [DOI] [PubMed] [Google Scholar]
- 52. Jang KY, Hwang SH, Kwon KS, et al. SIRT1 expression is associated with poor prognosis of diffuse large B-cell lymphoma. Am J Surg Pathol. 2008;32:1523–1531 [DOI] [PubMed] [Google Scholar]
- 53. Cha EJ, Noh SJ, Kwon KS, et al. Expression of DBC1 and SIRT1 is associated with poor prognosis of gastric carcinoma. Clin Cancer Res. 2009;15:4453–4459 [DOI] [PubMed] [Google Scholar]
- 54. Nosho K, Shima K, Irahara N, et al. SIRT1 histone deacetylase expression is associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Mod Pathol. 2009;22:922–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Deblois G, Hall JA, Perry MC, et al. Genome-wide identification of direct target genes implicates estrogen-related receptor α as a determinant of breast cancer heterogeneity. Cancer Res. 2009;69:6149–6157 [DOI] [PubMed] [Google Scholar]