Abstract
Substantial advances have been made in understanding critical molecular and cellular mechanisms driving tumor initiation, maintenance, and progression in non–small-cell lung cancer (NSCLC). Over the last decade, these findings have led to the discovery of a variety of novel drug targets and the development of new treatment strategies. Already, the standard of care for patients with advanced-stage NSCLC is shifting from selecting therapy empirically based on a patient's clinicopathologic features to using biomarker-driven treatment algorithms based on the molecular profile of a patient's tumor. This approach is currently best exemplified by treating patients with NSCLC with first-line tyrosine kinase inhibitors when their cancers harbor gain-of-function hotspot mutations in the epidermal growth factor receptor (EGFR) gene or anaplastic lymphoma kinase (ALK) gene rearrangements. These genotype-based targeted therapies represent the first step toward personalizing NSCLC therapy. Recent technology advances in multiplex genotyping and high-throughput genomic profiling by next-generation sequencing technologies now offer the possibility of rapidly and comprehensively interrogating the cancer genome of individual patients from small tumor biopsies. This advance provides the basis for categorizing molecular-defined subsets of patients with NSCLC in whom a growing list of novel molecularly targeted therapeutics are clinically evaluable and additional novel drug targets can be discovered. Increasingly, practicing oncologists are facing the challenge of determining how to select, interpret, and apply these new genetic and genomic assays. This review summarizes the evolution, early success, current status, challenges, and opportunities for clinical application of genotyping and genomic tests in therapeutic decision making for NSCLC.
INTRODUCTION
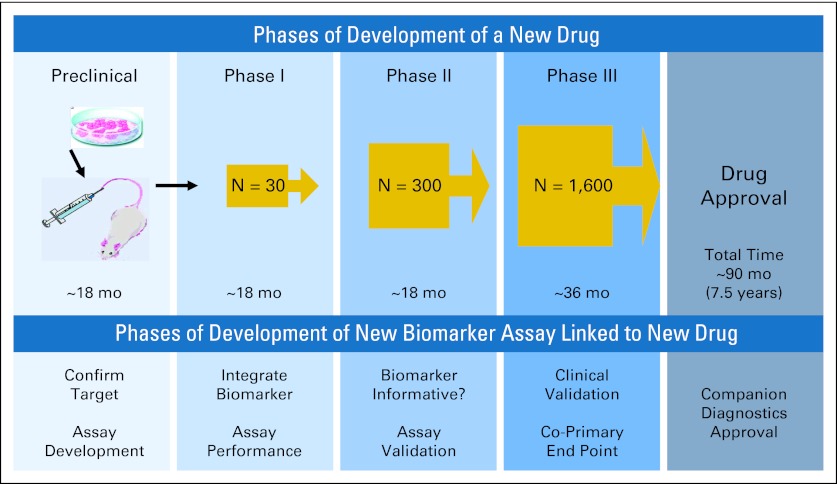
Non–small-cell lung cancer (NSCLC), regardless of histologic subtype, is one of the most genomically diverse and deranged of all cancers, creating tremendous challenges for both prevention and treatment strategies.1,2 Nevertheless, this same biologic diversity provides a number of opportunities for exploitation of interpatient tumor heterogeneity by ungrouping NSCLC into a variety of molecularly defined subsets for which mutations and/or abnormal gene expressions drive cancer cell growth and survival and can serve as druggable targets.3–5 Although the resulting transition from empiric to mechanism-based, molecular biomarker–driven therapeutic decision making remains in its early phases, new drug classes have already changed the paradigm for the management of advanced-stage NSCLC.6,7 A proof-of-principle example is the identification of gain-of-function tyrosine kinase–activating epidermal growth factor receptor (EGFR) mutations as the best predictive biomarker over clinicopathologic features in predicting tumor response and progression-free survival to EGFR tyrosine kinase inhibitors (TKIs).8–12 Similarly, gain-of-function tyrosine kinase–activating ALK gene rearrangements are valid predictive biomarkers in predicting tumor response and progression-free survival to the first-in-class ALK TKI crizotinib.13 Most importantly, crizotinib was one of the first two drugs granted US Food and Drug Administration (FDA) approval concurrently with an FDA-approved companion diagnostic test for selecting an uncommon (2% to 7%) subset of patients with NSCLC whose tumors harbor ALK gene rearrangements.14 Furthermore, the 4-year period from identification of the oncogenic ALK gene rearrangement in NSCLC to drug approval was remarkably short compared with the usual drug development process of approximately 10 years.7,15 This milestone highlights the importance of establishing a predictive biomarker assay early on during the development of a new mechanism-based drug for an uncommon subset of patients with NSCLC, with the goal of increasing the success rate in the phase III setting. Ideally, as recently reviewed and shown in Figure 1 , development of a companion new drug-associated predictive biomarker assay may parallel that of the new drug development process itself, so that the phase III trial for a new drug is used to validate the biomarker assay.7 Here, we provide a concise summary and perspective on clinical application of genotyping and genomic tests in NSCLC for therapeutic decision making.
Fig 1.
Improved drug-biomarker development paradigms: marriage of drug-biomarker development. Data adapted.7
EVOLUTION OF PERSONALIZED MEDICINE AND TECHNOLOGY ADVANCES
Personalized medicine is defined by the National Cancer Institute as “a form of medicine that uses information about a person's genes, proteins and environment to prevent, diagnose, and treat disease.”16 Compared with protein biomarkers, cancer genetic biomarkers are typically more reproducible and less subject to the influence of intrinsic and external stimuli. Decades of cancer research revealed that cancer results from accumulation of many genomic aberrations that ultimately govern tumor initiation, maintenance, and progression.17–19 Although genetics typically refers to the study of single genes, genomics refers to the study of the complete genes and their function in an individual.16 The central hypothesis of molecular-based personalized cancer therapy is that treatment decisions based on tumor genotype and genomic profile will improve clinical outcomes, as measured by response rate, survival, and safety.
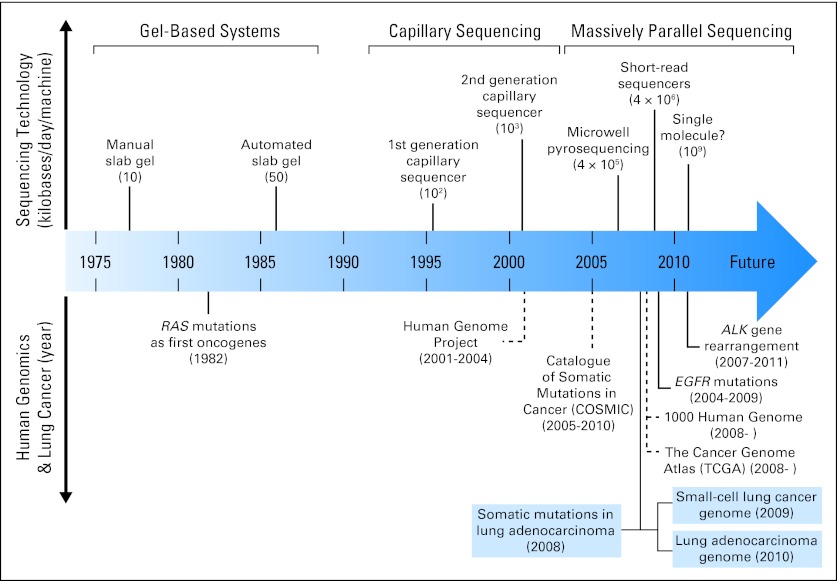
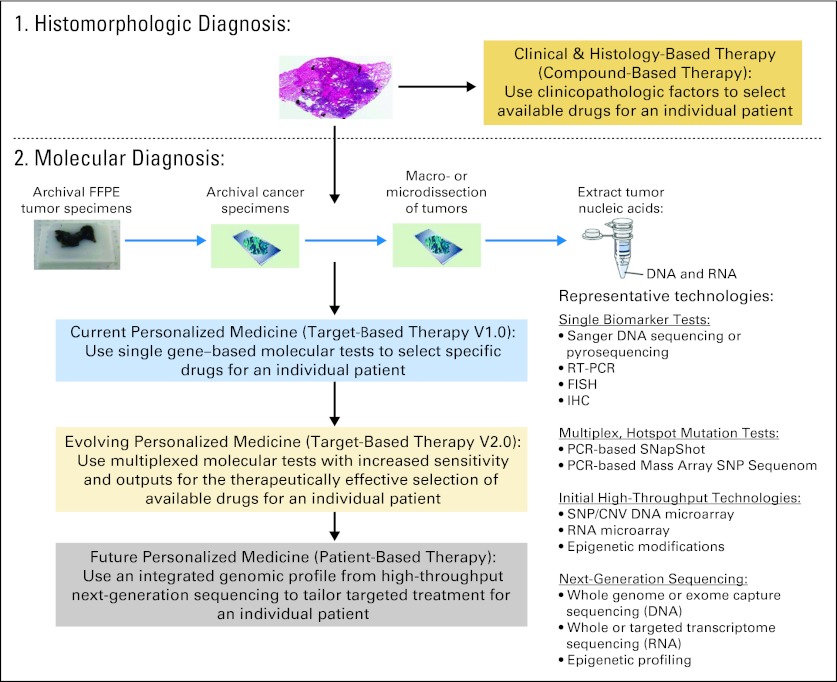
The evolution of personalized cancer medicine has been greatly accelerated by advances in DNA-based high-throughput genomic technologies.18,20 Figure 2 summarizes milestones in these technology advances over the last three decades and their implication in human genomics.4,21–32 The fundamental difference between first-generation Sanger sequencing technology and second-generation, or next-generation sequencing (NGS), technology is elimination of the need for gels or polymers as a sieving separation matrix and the need of prior knowledge of the genome sequence.20,22 These high-throughput technologies enable nucleic acid (DNA and RNA) sequencing at a faster speed with a reduced error and cost per base. The data output of NGS has been continuously increasing, more than doubling each year since it was invented. For example, a single sequencing run could produce a maximum of approximately 1 gigabase of data in 2007 and approximately 1 terabase of data in 2011, which is nearly a 1,000-fold increase in 4 years. However, the cost remains high given the large amount of nucleotides in the cancer genome. Early clinical application of these technologies has enabled rapid and comprehensive molecular annotation of an individual patient's cancer, facilitating identification of actionable and/or novel drug targets and treatment options, as well as characterization of underlying pathogenesis mechanisms. Matching targeted therapies against specific genetic aberrations is an important step for personalized cancer therapy that holds promise in ultimately improving patient outcomes.7,28 We propose to define the role of genotyping and genomic profiling in personalized medicine in lung cancer into three stages based on the therapeutic strategies used (Fig 3), which parallel the technology advances in genetic and genomic testing discussed in the following sections.
Fig 2.
Advances in sequencing technologies and human genomics.
Fig 3.
Genotyping and genomic profiling in personalized medicine: a scientific revolution of cancer molecular diagnosis and treatment. CNV, copy number variation; FFPE, formalin-fixed paraffin-embedded; FISH, fluorescent in situ hybridization; IHC, immunohistochemistry; RT-PCR, reverse transcriptase polymerase chain reaction; SNP, single nucleotide polymorphism.
GENOTYPE-BASED MOLECULAR BIOMARKERS
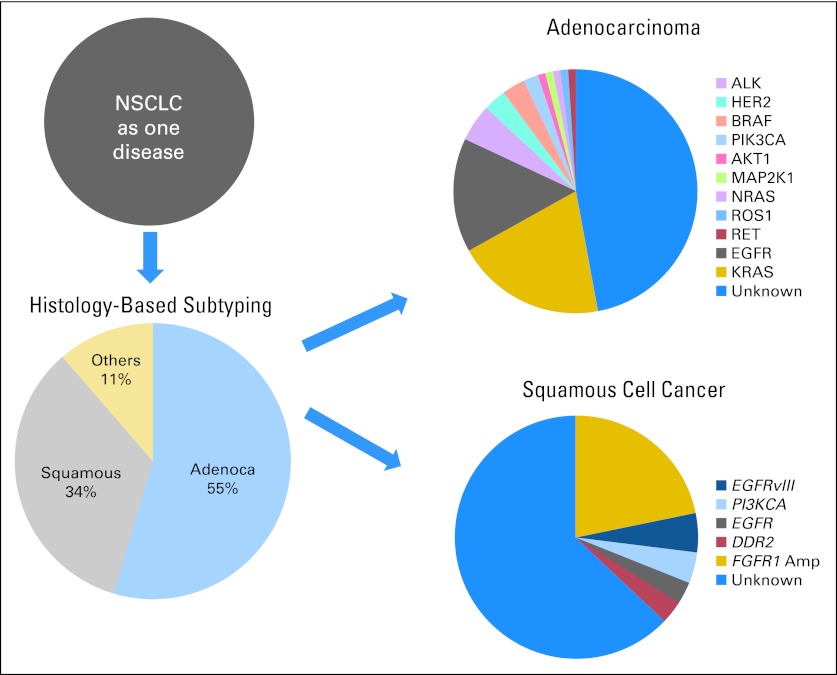
Clinical application of single gene–based biomarkers has already proven successful in guiding selection of molecularly targeted agents in NSCLC.6,33,34 The EGFR TKIs erlotinib and gefitinib were the first class of molecularly targeted agents approved for the treatment of advanced NSCLC in 2004.35 Although these agents were initially approved for use in unselected patients with NSCLC, subsequently the presence of gain-of-function tyrosine kinase–activating EGFR mutations was shown to be most predictive of response to EGFR TKIs.8–12 In August 2011, the FDA granted accelerated approval of the first-in-class ALK inhibitor crizotinib for treatment of ALK-positive advanced NSCLC.14 Subsequently, both the National Comprehensive Cancer Network and American Society of Clinical Oncology guidelines recommended EGFR mutation and ALK gene rearrangements testing on all NSCLCs that contain an adenocarcinoma component, regardless of histologic grade or dominant histologic subtype.36,37 EGFR mutation and ALK testing is not recommended for pure squamous cell carcinomas, pure small-cell carcinomas, or pure neuroendocrine carcinomas.37 Recent genotyping studies have revealed that distinct genetic abnormalities are present in adenocarcinomas and squamous cell carcinomas,5,38,39 providing opportunities for developing novel molecularly targeted and biomarker-driven therapeutic strategies for specific molecular subsets of patients (Fig 4; Table 1).
Fig 4.
Evolution of non–small-cell lung cancer (NSCLC) subtyping from histologic to molecular based. Data adapted.5 EGFR, epidermal growth factor receptor; HER2, human epidermal growth factor receptor 2; MAP2K1, mitogen-activated protein kinase kinase 1.
Table 1.
Oncogene Mutations Predict Likelihood of Response or Resistance to Current Targeted Therapies in Patients With NSCLC
| Oncogene | Mutation Prevalence | Mutation-Predicted Therapeutic Response | Predicted Response Rate |
|---|---|---|---|
| EGFR | Asians: 30%-40%; whites: 10%-20% | Sensitive to EGFR TKIs (most mutations)* | Erlotinib: 60%-83%11,12; gefitinib: ∼71%8–10 |
| KRAS | Asians: 10%; whites: 30% | Resistant to EGFR TKIs†; sensitive to MEK inhibitors? | Data are limited40,41 |
| EML4-ALK | 1%-7%; no clear racial difference | Sensitive to ALK inhibitors†; resistant to EGFR TKIs | Crizotinib: 50%-60%13; data are limited regarding resistance to EGFR TKIs42 |
| ROS1 | 1.7%; more in Asians? | Sensitive to ALK inhibitors† | Crizotinib: unknown43 |
| HER2 | More in Asians? | Sensitive to HER2 inhibitors | Trastuzumab: unknown; lapatinib, afatinib, and dacomitinib: unknown |
Abbreviations: EGFR, epidermal growth factor receptor; NSCLC, non–small-cell lung cancer; TKI, tyrosine kinase inhibitor.
Currently, EGFR mutation testing can be performed from nanograms of genomic DNA extracted from tumors cells on a few slides of archival formalin-fixed paraffin-embedded (FFPE) specimens, with a turnaround time of 5 to 10 days from some laboratories. Over the last few years, there have been significant improvements in the regulation of assay development and analytic validation and clinical validation of these genetic tests.47 Today, clinical molecular pathology laboratories can perform these tests using FDA-approved kits and equipment in Clinical Laboratory Improvement Act–certified academic or commercial laboratories.48 Recently, representatives of three professional organizations with interests in the diagnosis and management of lung cancer—the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology—convened to review the published data and to develop evidence-based guideline recommendations for the molecular testing of lung cancers for EGFR mutation and ALK gene arrangement testing.49 The draft report is currently available online for public comment. These molecular tests are being increasingly used worldwide. Notable is the government-sponsored program in France, initiated in 2009, which seeks to provide nationwide molecular testing for patients with a number of cancers, including NSCLC. For NSCLC, efforts began with testing for EGFR mutations. In 2010, more than 17,000 French patients were tested for EGFR mutations at a total of 28 laboratories in public hospitals, with a turnaround time of 13 days.50
MULTIPLEX GENOTYPING OF KNOWN HOTSPOT ONCOGENE MUTATIONS
Although landmark studies of molecularly targeted agents have largely focused on a single or small number of genetic mutations, there is an increasing need to develop clinically applicable methodologies that can simultaneously determine the mutational or expression status of many genes of interest and do so using small tumor samples. Multiplex polymerase chain reaction (PCR) is defined as the simultaneous amplification of at least two DNA or cDNA targets in a single reaction vessel.51 Both Sequenom (Sequenom, San Diego, CA) and SNaPShot (Applied Biosystems, Foster City, CA) platforms use multiplex PCR to identify potentially actionable molecular targets in lung cancer from genomic DNA derived from FFPE tumor specimens. These assays are being widely used in the cancer research community and show promise for clinical use as well.52 Table 2 compares the list of hotspot mutations and oncogenes included in the Sequenom and SNaPShot panels and corresponding approved and/or experimental drugs. It is noteworthy that these multiplex genomic tests only detect the expression of selected known hotspot mutations and oncogenes and do not have the ability to discover new or additional drug targets. Easily identifiable oncogenes compose most of the components, reflecting that precise changes in particular amino acids (ie, hotspots) are sufficient for oncogenic activation and oncogenic addition. In contrast, inactivation of a tumor suppressor can involve deletions or point mutations in wide regions of the loci, rendering analysis more challenging. Furthermore, there is presently no accepted treatment strategy to restore or repair the functions of tumor suppressors. Thus, hyperactive oncogenic mutations have been the primary focus of targeted therapy and associated predictive biomarker assays for NSCLC.
Table 2.
Comparison of Hotspot Mutations and Oncogenes in the Sequenom and SNaPShot Panels and Their Corresponding Approved and/or Experimental Drugs
| Sequenom Panel |
SNaPShot Panel |
Actionable Drugs |
||||||
|---|---|---|---|---|---|---|---|---|
| Genes in Sequenom (n = 19) | No. of Mutations in Sequenom (n = 238) | Sequenom OncoCarta V1.0 Oncomutations (n = 238) | Genes in SNaPShot (n = 8) | No. of Mutations in SNaPShot (n = 38) | Mutations | Drug Class | Approved Drugs | Representative Investigational Drugs |
| ABL-1 | 14 | G250E, Q252H, Y253H, Y253F, E255K, E255V, D276G, F311L, T315I, F317L, M351T, E355G, F359V, H396R | ||||||
| AKT-1 | 7 | V461L, P388T, L357T, E319G, V167A, Q43X, E17del | AKT-1 | 1 | E17K | Small-molecule TKI | MK-2206 | |
| AKT-2 | 2 | S302G, R371H | Small-molecule TKI | MK-2206 | ||||
| BRAF | 24 | G464R, G464V/E, G466R, F468C, G469S, G469E, G469A, G469V, G469R, D594V/G, F595L, G596R, L597S, L597R, L597Q, L597V, T599I, V600E, V600K, V600R, V600L, K601N, K601E | BRAF | 4 | G466V, G469A, L597V, V600E | Small-molecule TKI | Vemurafenib* | PLX4032, GSK1120212, GSK2118436, XL281 |
| CDK-4 | 2 | R24C, R24H | ||||||
| EGFR | 43 | R108K, T263P, A289V, G598V, E709K/H, E709A/G/V, G719S/C, G719A, M766_A767insAI, S768I, V769_D770insASV, L747_T750del, L747_T751del, L747_S752del, P753S, A750P, T751A, T751P, T751I, S752I/F, S752_I759del, L747_Q ins, E746_T751del, I ins (combined), E746_A750del, T751A (combined), L747_E749del, A750P (combined), L747_T750del, P ins (combined), L747_S752del, Q ins (combined) | EGFR | 6 (plus exon 20 insertion and 19 deletions by sizing assays in common)‡ | G719S/C, G719A, L747_T750del, L747_T751del, L747_S752del, T790M, L858R, L861Q | Small-molecule TKI, monoclonal antibody | Erlotinib, gefitinib, cetuximab* | Afatinib (BIBW2992), dacomitinib (PF-00299804), lapatinib,* cetuximab,* panitumumab* |
| ERBB2 | 7 | L755P, G776S/LC, G776VC/VC, A775_G776insYVMA, P780_Y781insGSP, S779_P780insVGS | ERBB2† | Small-molecule TKI, monoclonal antibody | Trastuzumab,* lapatinib* | Afatinib (BIBW2992), dacomitinib (PF-00299804) | ||
| FGFR-1 | 2 | S125L, P252T | Small-molecule TKI | BGJ398, FP1039 (HSG1036), ponatinib (AP24534) | ||||
| FGFR-3 | 5 | G370C, Y373C, A391E, K650Q/E, K650T/M | Small-molecule TKI | BGJ398, FP1039 (HSG1036), ponatinib (AP24534) | ||||
| FLT-3 | 2 | I836del, D835H/Y | ||||||
| JAK-2 | 1 | V617F | ||||||
| KIT | 27 | D52N, Y503_F504insAY, W557R/R/G, V559D/A/G, V559I, V560D/G, K550_K558del, K558_V560del, K558_E562del, V559del, V559_V560del, V560del, Y570_L576del, E561K, L576P, P585P, D579del, K642E, D816V, D816H/Y, V825A, E839K, M552L, Y568D, F584S, P551_V555del, Y553_Q556del | Small-molecule TKI | Imatinib, nilotinib, sunitinib, sorafenib, axitinib | ||||
| MET | 5 | R970C, T992I, Y1230C, Y1235D, M1250T | MET† | Small-molecule TKI, monoclonal antibody | MetMAb, crizotinib, ARQ197, XL184, XL 880, GSK1363089, SCH900105, JNJ38877605 | |||
| PDGFRa | 11 | V561D, T674I, F808L, D846Y, N870S, D1071N, D842_H845del, I843_D846del, S566_E571>K, I843_S847>T, D842V | Small-molecule TKI | Imatinib, nilotinib, sunitinib, sorafenib | ||||
| PIK3CA | 13 | R88Q, N345K, C420R, P539R, E542K, E545K, Q546K, H701P, H1047R/L, H1047Y, R38H, C901F, M1043I | PIK3CA | 4 | E542K, E545K, E545Q, H1047R | Small-molecule TKI | BKM120, BEZ235, PX-866, GDC-0941, SAR245408 | |
| KRAS | 12 | G12C, G12R, G12S, G12V, G12D, G12A, G12F, G13V/D, A59T, Q61E/K, Q61L/R/P, Q61H/H | KRAS (13 in common) | 16 | G12C, G12R, G12S, G12V, G12D, G12A, G13C, G13S, G13R, G13A, G13D, Q61K, Q61L/R, Q61H/H | Small-molecule TKIs: MEK inhibitor; MET inhibitor; AKT inhibitor | Multiple MEK inhibitors (eg, GSK1120212, selumetinib); MET inhibitors (eg, tivantinib); AKT inhibitors (eg, MK2206, GSK2141795) | |
| HRAS | 6 | G12V/D, G13C/R/S, Q61H/H, Q61L/R/P, Q61K | ||||||
| NRAS | 8 | G12V/A/D, G12C/R/S, G13V/A/D, G13C/R/S, A18T, Q61L/R/P, Q61H, Q61E/K | NRAS | 3 | Q61L, Q61K, Q61R | Small-molecule TKIs: MEK inhibitor; MET inhibitor; AKT inhibitor | Multiple MEK inhibitors (eg, GSK1120212, selumetinib); MET inhibitors (eg, tivantinib); AKT inhibitors (eg, MK2206, GSK2141795) | |
| RET | 6 | C634R, C634W, C634Y, E632_L633del, M918T, A664D | ||||||
| None | MEK1 (MAP2K1) | 3 | Q56P, K57N, D67N | Small-molecule TKI | MEK162, GDC-0973, GSK1120212, selumetinib (AZD6244) | |||
| None | PTEN | 1 | R233§ | |||||
| None | ROS1† | Small-molecule TKI | Crizotinib | |||||
Abbreviation: TKI, tyrosine kinase inhibitor.
Approved in other tumor types.
Detection by fluorescent in situ hybridization.
Detection by sizing assays.
This mutation results in a premature stop codon.
Sequenom Oncogenotype Mutational Analysis
The Sequenom platform is an array-based system that combines PCR with matrix-assisted laser desorption/ionization time of flight mass spectrometry for rapid multiplexed nucleic acid analysis.53 The Sequenom OncoCarta V1.0 kit uses multiplex PCR amplifications of a minimum of 500 ng tumor DNA per sample (ie, 20 ng DNA per multiplex reaction) for a total of 238 somatic mutations of oncogenes across 19 different genes commonly associated with cancer (Table 2).53 The PCR reactions are purified and subjected to matrix-assisted laser desorption/ionization time of flight mass spectrometry on the Sequenom MassArray. Specific amplicons (and the mutations) are assayed and quantitated. The assay can use tumor samples from fresh, frozen, or FFPE samples and/or cell lines. It can detect and quantify mutation frequencies from at least 10% of mutation-positive cells. The main advantages of the Sequenom oncogene mutation genotyping platform include the commercially available kit with technical support for optimizing each multiplex PCR reaction, easy operability, and the readily interpretable data report form. The turnaround time could be similar to that of single gene–based assay. Disadvantages include the requirement for the purchase of Sequenom equipment and the need for the vendor's involvement in modifying and updating the gene list of targeted mutations.
SNaPshot Oncogenotype Mutational Analysis
The SNaPshot platform from Applied Biosystems consists of multiplex PCR and single base extension reactions that generate fluorescently labeled probes designed to interrogate more than 50 hotspot mutation sites in eight to 14 key cancer genes. The gene list might vary slightly between different laboratories. Up to 10 single nucleotide polymorphisms from different amplicons can be interrogated in a single base extension reaction. It has increased sensitivity (approximately 10%) compared with standard sequencing, allowing detection of a single–base pair difference in each test tube. The SNaPshot products are then resolved and analyzed using capillary electrophoresis on several models of ABI Genetic Analyzers (Applied Biosystems) generally available at major academic institutions.54–56 Compared with the Sequenom platform, the list of hotspot mutations and oncogenes included in the SNaPShot platform is narrowed down to high-prevalence genetic abnormalities detected in NSCLC (Table 2). Although SNaPshot has improved molecular testing over conventional DNA-based tests (which have typically focused on EGFR and KRAS sequencing only), it is labor intensive and typically requires a 2- to 3-week turnaround time. It also requires more genomic DNA for testing compared with the Sequenom platform.
Clinical Applications of Multiplex Genotyping of Known Hotspot Oncogene Mutations
A number of individual institutions and collaborative groups have begun to apply genomic profiling to therapeutic decision making for patients with NSCLC.54–56 The Lung Cancer Mutation Consortium initiated a US collaborative genotyping effort among 14 academic centers (ClinicalTrials.gov identifier: NCT01014286), with the goal of genotyping 10 driver mutations in 1,000 patients with lung adenocarcinoma using the SNapShot platform as described earlier, together with the FDA-approved fluorescent in situ hybridization analysis for ALK gene rearrangement and MET amplification.57 In a preliminary report, at least one actionable driver mutation was present in 54% of the first 516 tumors tested, including KRAS mutation (22%), EGFR mutation (17%), and EML4-ALK rearrangement (7%).57 Almost all of these mutations (97%) were mutually exclusive for the tested genetic abnormalities. Consistent with previous single-institution experience,54,55 genotyping results changed the therapy in 20% to 40% of patients with NSCLC in the data set identified for one of several early-phase clinical trials evaluating the safety and efficacy of novel molecularly targeted agents against individual oncogene mutations.57,58 In the future, a comprehensive genetic annotation of NSCLC tumors may be appended to the gene and mutation list in Sequenom and SNaPshot panels.
HIGH-THROUGHPUT GENOME-WIDE UNBIASED NGS
NGS technologies offer novel and rapid ways for genome-wide characterization of DNA, mRNA, transcription factor regions, miRNA, chromatin structure, and DNA methylation patterns.20,22 They include several sequencing platforms for whole-genome, whole-exome, whole-transcriptome (RNA sequencing), and whole-epigenome analysis, using “sequencing-by-synthesis, addition and detection of the incorporated base by reversible terminator nucleotides” without the need for gels and prior knowledge of the genome sequence.20 Each sequencing platform has its unique features, and the platforms could be used to compliment each other for cancer genomic data of individual tumors, if affordable. There are several NGS platforms from Illumina (San Diego, CA), 454 Life Sciences (Branford, CT; part of Roche Applied Science), Helicos BioSciences (Cambridge, MA), and Applied Biosystems. They all generate an abundance of low-cost, high-volume sequencing data. The Illumina-Solexa NGS (RNA sequencing) technology (Illumina) was first commercialized in 2006, and the Illunima-Solexa genome analyzer is currently the most commonly used sequencer. Depending on the desired depth of sequencing resolution, the massively parallel sequencing requires 0.1 to 3 μg of nucleic acids to generate DNA, RNA, and microRNA sequences for point mutations, single nucleotide polymorphism, copy number variation, and importantly novel fusion genes that are unbiased (unprimed) and more fully representative of the entire transcriptome. One big advantage of NGS technology is that its coverage, which generally refers to the average number of sequencing reads that align to each base within the sample DNA, is highly adjustable. For instance, a whole genome sequenced at 20× coverage means that, on average, each base in the genome is covered by 20 sequencing reads, which can detect a base change at a frequency of at least 5%; and a whole genome sequenced at 100× coverage means that, on average, each base in the genome is covered by 100 sequencing reads, which can detect a base change at low frequency of at least 1%. NGS can also be multiplexed to sequence more than five human genomes in a single run. Several new NGS platforms, such as the Illumima HiSeq 2500 and Ion Torrent Proton (Ion Torrent Systems, Guilford, CT), are being developed to sequence the human genome in 1 day, which would further accelerate the clinical application of cancer genomics.
NGS technologies have been rapidly applied to clinical settings in almost all tumor types as reported at the recent 2012 annual meeting of the American Society of Clinical Oncology. They are being used as research tools for understanding of tumor molecular mechanisms, discovery of novel drug targets, and screening candidate patients for clinical trials. Currently, it takes approximately 1 week to generate sequencing data and at least 2 weeks for data analysis. It costs approximately $3,500 to $5,000 for the reagents needed for all three NGS tests,59 although the price could decrease further. One major challenge is the complexity of data generated and the need for robust bioinformatics tools to fully understand the functional impact of each of the many, simultaneously identified genomic abnormalities. The situation is best described as “$1000 genomic test and $100,000 genomic analysis.”60 Several targeted NGS approaches have been explored to simplify the data extraction by scaling down sequence coverage and multiplexing multisample analysis (eg, use of a targeted, massively parallel sequencing approach to detect tumor genomic changes in cancer-related genes only,61 or to focus on tyrosine kinase fusion genes only62,63). The reproducibility of these methods in large-scale studies and validation of their clinical utility remain to be evaluated.
Early experience of applying NGS technologies in NSCLC and other tumors suggests that, on average, more than 100 to 200 genomic abnormalities are identified for each tumor specimen,59,61 which is higher than 50 to 100 variants observed in inherited disorders.31,64 In addition to known hotspot oncogenic mutations or gene rearrangements in NSCLC, NGS has also identified genetic abnormalities that are previous known in other cancer types as well and uncovered many novel genetic abnormalities without knowledge of their biologic functions. A pathway-based, integrative systems biology approach has been used to interpret the data in the context of known and emerging hallmarks of cancer.17 Even for gene alterations known to have prognostic and/or predictive value in other cancer types, in many cases, their roles in NSCLC remain to be determined in rigorous clinical trial settings. The newly discovered genetic abnormalities could serve as potential drug targets and predictive biomarkers, as well as genetic variants that affect drug metabolism and cancer prognosis. Increasingly, these novel oncogenic molecular biomarkers have been discovered in rare subsets of patients. It is critical to sort out driver genomic abnormalities from passenger abnormalities for targeted treatment. Ideally, prospective, simultaneous multiple biomarker-driven therapeutics trials are needed to assess the clinical feasibility and efficacy of individualized cancer care in patients with advanced NSCLC. One reasonable interim approach for data collection and information exchange is to develop a publically accessible database for collecting clinical information on patients' NSCLC tumor responses harboring rare or new, single or multiple genetic and genomic abnormalities to different molecularly targeted agents, as suggested for rare EGFR mutations.65
CHALLENGES AND OPPORTUNITIES IN CLINICAL APPLICATION OF GENOTYPING AND GENOMIC TESTING
Translation of the state-of-the-art cancer genomics to routine clinical application demands new translational research platforms for selecting and validating clinically relevant drug target(s) and associated biomarker assay(s). Furthermore, when these assays are integrated into clinical practice, they must be broadly available to practicing clinicians, applicable to small tumor biopsies, and affordable to patients, and the turnaround time for test information to be returned to the treating physician must be short, usually defined as a maximum of 2 weeks. Currently, a high priority is to develop systematic testing algorithms to identify genomically defined subsets of patients with NSCLC for whom effective drug therapies are available either commercially or through clinical trials. Such a potential paradigm change in patient care has raised many new challenges.
First, NSCLC is well recognized as diverse based on interpatient tumor heterogeneity. More recently, an added layer of complexity related to intrapatient tumor heterogeneity has been observed, particularly relevant to the clonal evolution of somatic mutations from the primary tumor to metastatic lesions and the mixed tumor response to treatment with a molecularly targeted agent in different tumor sites.66 The role this phenomenon plays in the development of acquired drug resistance and biomarker testing is likely to be highly variable from one individual patient to the next or perhaps from one metastatic site to the next in the same patient.
Second, dynamic change within the cancer genome during the disease course is now being recognized as an additional challenge because the tumor genetic makeup may undergo substantial alteration during disease progression or in response to treatment. Current experience suggests that although most driver mutations are maintained in resistant tumors, additional actionable genetic/genomic abnormalities may emerge.67,68 Furthermore, EGFR mutations, ALK gene rearrangements, and KRAS mutations rarely coexist in treatment-naive NSCLC tumors, but they can coexist in rebiopsied tumor specimens from patients with refractory NSCLC.67,68 The clinical significance of this phenomenon remains to be defined. However, it does support obtaining serial biopsy specimens to assess real-time changes in histomorphology and cancer genomics during the disease course, which can be feasible and safe in patients with lung cancer.67,69
Third, both quantity and quality of tumor tissue are critical for all genetic and genomic testing. Several professional and regulatory agencies have established guidelines to address both the regulatory and quality control requirements to ensure that preanalytic variables for sample collection and processing could be tracked and controlled.49,70 The need to improve regulatory systems to ensure the quality of conducting genetic and genomic testing in humans has been recognized. Because cancer is a global health hazard, a key element to more effective oversight is to allow for more collaboration among regulatory agencies domestically and globally.
Fourth, although whole-genome sequencing holds unprecedented potential for personalized cancer therapy, a current challenge is how to analyze the huge amount of genomic data for clinical relevant drug targets and pharmacogenomic variants. As discussed in the previous section, scaling down sequencing coverage for specific cancer genes and multiplexing of several samples per NGS reaction are being actively explored as strategies to improve clinical applicability. In 2011 alone, several remarkable advances in genome technology have improved our ability to edit and analyze the genome using novel techniques, such as genome targeting editing,71 search-and-replace editing techniques,72,73 mapping structure variation using short reads,74 and multiplexed automated genome engineering.75 Notably, multiplexed automated genome engineering is an in vivo method using synthetic oligonucleotides to enable the rapid generation of mutants at high efficiency and specificity and can be implemented at the genome scale.
Last, but not least, clinical implementation of genotyping and genomic tests in NSCLC demands a close collaboration between multidisciplinary health care professionals, including, but not limited to, surgeons, pulmonologists, radiologists, pathologists, translational scientists, medical oncologists, insurers, and regulatory agencies. It is also vitally important to engage patients with NSCLC, the prime target of personalized cancer therapy, to help them understand the growing importance of molecular testing and to motivate them to participate in the process in appropriate ways.
SUMMARY AND PERSPECTIVES
In summary, detection of gain-of-function tyrosine kinase–activating EGFR mutations and ALK gene rearrangements in NSCLC tumors (predominantly lung adenocarcinomas) by modern molecular technologies has been used in routine clinical practice to select distinct subsets of patients with NSCLC for first-line therapy with EGFR TKIs since 2009 and for an ALK TKI since 2011, respectively. Many additional molecularly targeted therapies are being developed for small (< 5%) subsets of patients with NSCLC. In parallel, development and validation of predictive biomarkers are being incorporated into early phases of clinical trials for these drugs. This new paradigm change in both the drug development process and clinical care has created new hope, many opportunities, and many challenges at the same time, for all stakeholders in the fight against lung cancer. Currently, several multiplex genotyping platforms for actionable hotspot oncogene mutations or gene amplification/rearrangements are being evaluated in research settings with promising results and are progressing to widespread clinical use among oncology practices. However, whether broad-based genotyping approaches will improve clinical outcomes of patients with NSCLC has yet to be proven by rigorous, prospective clinical evaluation. Moving forward, an integrated, genome-wide, molecular annotation of individual NSCLC tumors using scalable and multiplex NGS technologies holds great promise for advancing personalized cancer treatment, with the goal of maximizing efficacy and minimizing toxicity. The biggest challenge in translating discoveries in cancer genomics to improvements in clinical care is to understand the biologic relevance of the genomic aberrations within the context of evolution of an individual patient's lung cancer over time. Although there are still many barriers to overcome, recent advances in genomic technologies and drug development and the resulting outpouring of genomic information and novel new drugs are making molecular-based and personalized lung cancer therapy no longer just a dream.
Acknowledgment
We are grateful to Joel Kugelmass for editorial assistance and Krysteena Tolentino for help with preparation of the figures.
Footnotes
T.L. is supported by Grant No. UL1 RR024146 from the National Center for Research Resources; University of California Davis Comprehensive Cancer Center Developmental Award (National Institutes of Health/National Cancer Institute Grant No. P30CA093373); a research fund from the Division of Hematology and Oncology, Department of Internal Medicine, School of Medicine, University of California Davis; and the Hope Foundation (Southwest Oncology Group Young Investigator Award).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Tianhong Li, Astellas (C), Genentech (C), Daiichi Sankyo (C); Philip C. Mack, Boehringer Ingelheim (C); David R. Gandara, Caris Life Sciences (C), Response Genetics (C) Stock Ownership: None Honoraria: Philip C. Mack, Boehringer Ingelheim Research Funding: Tianhong Li, Astellas, Eli Lilly Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Data analysis and interpretation: Tianhong Li, Philip C. Mack, David R. Gandara
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 2.Larsen JE, Minna JD. Molecular biology of lung cancer: Clinical implications. Clin Chest Med. 2011;32:703–740. doi: 10.1016/j.ccm.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun S, Schiller JH, Spinola M, et al. New molecularly targeted therapies for lung cancer. J Clin Invest. 2007;117:2740–2750. doi: 10.1172/JCI31809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 6.Gandara DR, Mack PC, Li T, et al. Evolving treatment algorithms for advanced non-small-cell lung cancer: 2009 looking toward 2012. Clin Lung Cancer. 2009;10:392–394. doi: 10.3816/CLC.2009.n.074. [DOI] [PubMed] [Google Scholar]
- 7.Gandara DR, Li T, Lara PN, Jr, et al. Algorithm for codevelopment of new drug-predictive biomarker combinations: Accounting for inter- and intrapatient tumor heterogeneity. Clin Lung Cancer. 2012;13:321–325. doi: 10.1016/j.cllc.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 9.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 10.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 11.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 12.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 13.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.US Food and Drug Administration: FDA approves Xalkori with companion diagnostic for a type of late-state lung cancer. http://www.accessdata.fda.gov/drugsatfda_docs/appletter/2011/202570s000ltr.pdf.
- 15.Pickl M, Ruge E, Venturi M. Predictive markers in early research and companion diagnostic developments in oncology. N Biotechnol. 2012;29:651–655. doi: 10.1016/j.nbt.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 16.National Cancer Institute: Dictionary of cancer terms. http://www.cancer.gov/dictionary?CdrID=561717.
- 17.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Chin L, Andersen JN, Futreal PA. Cancer genomics: From discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 19.Macconaill LE, Garraway LA. Clinical implications of the cancer genome. J Clin Oncol. 2010;28:5219–5228. doi: 10.1200/JCO.2009.27.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 23.International Human Genome Sequencing Consortium: Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 24.Levy S, Sutton G, Ng PC, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 26.Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee W, Jiang Z, Liu J, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 28.Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145:19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 29.Forbes S, Clements J, Dawson E, et al. COSMIC 2005. Br J Cancer. 2006;94:318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes SA, Bindal N, Bamford S, et al. COSMIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.1000 Genomes Project Consortium: A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research Network: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Field JK, Brambilla C, Caporaso N, et al. Consensus statements from the Second International Lung Cancer Molecular Biomarkers Workshop: A European strategy for developing lung cancer molecular diagnostics in high risk populations. Int J Oncol. 2002;21:369–373. doi: 10.3892/ijo.21.2.369. [DOI] [PubMed] [Google Scholar]
- 34.Von Hoff DD, Stephenson JJ, Jr, Rosen P, et al. Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010;28:4877–4883. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- 35.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 36.National Comprehensive Cancer Network: Non-Small Cell Lung Cancer. Version 3.2012. NCCN Clinical Practice Guidelines in Oncology. http://www.nccn.com. [DOI] [PubMed]
- 37.Keedy VL, Temin S, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: Epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol. 2011;29:2121–2127. doi: 10.1200/JCO.2010.31.8923. [DOI] [PubMed] [Google Scholar]
- 38.Paik PK, Hasanovic A, Wang L, et al. Multiplex testing for driver mutations in squamous cell carcinomas of the lung. J Clin Oncol. 2012;30(suppl):481s. abstr 7505. [Google Scholar]
- 39.Govindan R, Hammerman PS, Hayes DN, et al. Comprehensive genomic characterization of squamous cell carcinoma of the lung. J Clin Oncol. 2012;30(suppl):453s. abstr 7006. [Google Scholar]
- 40.Roberts PJ, Stinchcombe TE, Der CJ, et al. Personalized medicine in non-small-cell lung cancer: Is KRAS a useful marker in selecting patients for epidermal growth factor receptor-targeted therapy? J Clin Oncol. 2010;28:4769–4777. doi: 10.1200/JCO.2009.27.4365. [DOI] [PubMed] [Google Scholar]
- 41.Janne PA, Shaw AT, Pereira JR, et al. Phase II double-blind, randomized study of selumetinib (SEL) plus docetaxel (DOC) versus DOC plus placebo as second-line treatment for advanced KRAS mutant non-small cell lung cancer (NSCLC) J Clin Oncol. 2012;30(suppl):480s. abstr 7503. [Google Scholar]
- 42.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 45.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 46.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 47.Engstrom PF, Bloom MG, Demetri GD, et al. NCCN molecular testing white paper: Effectiveness, efficiency, and reimbursement. J Natl Compr Canc Netw. 2011;9(suppl 6):S1–S16. doi: 10.6004/jnccn.2011.0138. [DOI] [PubMed] [Google Scholar]
- 48.Jennings L, Van Deerlin VM, Gulley ML. Recommended principles and practices for validating clinical molecular pathology tests. Arch Pathol Lab Med. 2009;133:743–755. doi: 10.5858/133.5.743. [DOI] [PubMed] [Google Scholar]
- 49.College of American Pathologists: Lung cancer biomarkers guideline: Draft recommendations. http://www.cap.org/apps/docs/membership/transformation/new/lung_public_comment_supporting_materials.pdf.
- 50.Andre F, Nowak F, Arnedos M, et al. Biomarker discovery, development, and implementation in France: A report from the French National Cancer Institute and cooperative groups. Clin Cancer Res. 2012;18:1555–1560. doi: 10.1158/1078-0432.CCR-11-2201. [DOI] [PubMed] [Google Scholar]
- 51.Persson K, Hamby K, Ugozzoli LA. Four-color multiplex reverse transcription polymerase chain reaction: Overcoming its limitations. Anal Biochem. 2005;344:33–42. doi: 10.1016/j.ab.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 52.MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 54.Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol. 2011;22:2616–2624. doi: 10.1093/annonc/mdr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su Z, Dias-Santagata D, Duke M, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: A clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kris MG, Johnson BE, Kwiatkowski DJ, et al. Identification of driver mutations in tumor specimens from 1,000 patients with lung adenocarcinoma: The NCI's Lung Cancer Mutation Consortium (LCMC) J Clin Oncol. 2011;29(suppl):477s. abstr CRA7506. [Google Scholar]
- 58.Zander T, Heukamp LC, Bos MCA, et al. Regional screening network for characterization of the molecular epidemiology of non-small cell lung cancer (NSCLC) and implementation of personalized treatment. J Clin Oncol. 2012;30(suppl):663s. abstr CRA10529. [Google Scholar]
- 59.Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: A pilot study. Sci Transl Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mardis ER. The $1,000 genome, the $100,000 analysis? Genome Med. 2010;2:84. doi: 10.1186/gm205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chmielecki J, Peifer M, Jia P, et al. Targeted next-generation sequencing of DNA regions proximal to a conserved GXGXXG signaling motif enables systematic discovery of tyrosine kinase fusions in cancer. Nucleic Acids Res. 2010;38:6985–6996. doi: 10.1093/nar/gkq579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chmielecki J, Peifer M, Viale A, et al. Systematic screen for tyrosine kinase rearrangements identifies a novel C6orf204-PDGFRB fusion in a patient with recurrent T-ALL and an associated myeloproliferative neoplasm. Genes Chromosomes Cancer. 2012;51:54–65. doi: 10.1002/gcc.20930. [DOI] [PubMed] [Google Scholar]
- 64.1000 Genomes: Homepage. http://www.1000genomes.org.
- 65.Yatabe Y, Pao W, Jett JR. Encouragement to submit data of clinical response to EGFR-TKIs in patients with uncommon EGFR mutations. J Thorac Oncol. 2012;7:775–776. doi: 10.1097/JTO.0b013e318251980b. [DOI] [PubMed] [Google Scholar]
- 66.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18:1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim ES, Herbst RS, Wistuba II, et al. The BATTLE trial: Personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9(suppl 5):S1–S33. doi: 10.6004/jnccn.2011.0137. [DOI] [PubMed] [Google Scholar]
- 71.Wood AJ, Lo TW, Zeitler B, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dewey FE, Chen R, Cordero SP, et al. Phased whole-genome genetic risk in a family quartet using a major allele reference sequence. PLoS Genet. 2011;7:e1002280. doi: 10.1371/journal.pgen.1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Isaacs FJ, Carr PA, Wang HH, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y, Zheng H, Luo R, et al. Structural variation in two human genomes mapped at single-nucleotide resolution by whole genome de novo assembly. Nat Biotechnol. 2011;29:723–730. doi: 10.1038/nbt.1904. [DOI] [PubMed] [Google Scholar]
- 75.Wang HH, Church GM. Multiplexed genome engineering and genotyping methods applications for synthetic biology and metabolic engineering. Methods Enzymol. 2011;498:409–426. doi: 10.1016/B978-0-12-385120-8.00018-8. [DOI] [PubMed] [Google Scholar]