SUMMARY
Chemical inhibitors can help analyze dynamic cellular processes, particularly when probes are active in genetically tractable model systems. Although fission yeast has served as an important model system, which shares more cellular processes (e.g., RNAi) with humans than budding yeast, its use for chemical biology has been limited by its multidrug resistance (MDR) response. Using genomics and genetics approaches, we identified the key transcription factors and drug-efflux transporters responsible for fission yeast MDR and designed strains sensitive to a wide-range of chemical inhibitors, including commonly used probes. We used this strain, along with acute chemical inhibition and high-resolution imaging, to examine metaphase spindle organization in a “closed” mitosis. Together, our findings suggest that our fission yeast strains will allow the use of several inhibitors as probes, discovery of new inhibitors, and analysis of drug action.
INTRODUCTION
Cell-permeable chemical inhibitors can be powerful tools to examine dynamic cellular processes, such as cell division (Lampson and Kapoor, 2006; Peterson and Mitchison, 2002;Weiss et al., 2007). In many cases, these inhibitors can block target function within minutes (or seconds), allowing the time-scales of the perturbation to match that of the underlying cellular mechanisms. When the inhibitors are reversible, relief from inhibition can also be used to activate target function. In addition to serving as useful research tools, chemical inhibitors can also provide good starting points for developing new chemotherapeutic agents (Bergnes et al., 2005). In the last two decades, chemical probe discovery has become more efficient, in large part due to the numerous advances in chemical library design and high-throughput screening technology (Mayr and Bojanic, 2009). However, identifying the physiological targets and confirming specificity of chemical inhibitors remains very difficult, and therefore the use and further development of many chemical probes and candidate drugs has been restricted (Burdine and Kodadek, 2004).
We envisioned that a model system, which is compatible with a wide array of genetic manipulations, could be developed to address some of the challenges in chemical biology. In such a system, a range of strategies, such as analysis of drug resistance mechanisms, can be used to reveal a chemical inhibitor’s physiological target and address its specificity. In addition, if basic cellular processes, for example, cell division, DNA replication, RNA interference, and heterochromatin assembly, are conserved between the model system and human cells, chemical tools to analyze these processes could be developed. Furthermore, if detailed phenotypic analysis was also readily accessible, the inhibitor could be used to analyze complex and dynamic cellular processes. These criteria are met by Schizosaccharomyces pombe (fission yeast), in which several basic cellular mechanisms are more closely related to human cells than Saccharomyces cerevisiae (budding yeast) (Roguev et al., 2008; Wood et al., 2002), a more widely used model system for chemical biology. For example, fission yeast, like human cells, has the RNA interference pathway and epigenetically determines its centromere position (White and Allshire, 2008). In contrast, S. cerevisiae lacks RNA interference and defines centromere position based on DNA sequence (Cheeseman et al., 2002). However, the use of fission yeast for chemical probe discovery has been very limited, in large part due to fission yeast’s robust multidrug resistance (MDR) mechanisms (Arita et al., 2011; Wolfger et al., 2001).
Our understanding of the MDR mechanisms in fungi are mainly based on studies in budding yeast (Moye-Rowley, 2003). In current models, the MDR response involves overexpression of two types of drug efflux pumps, the ATP-binding cassette (ABC) family (Higgins, 1992) and the major facilitator superfamily (MFS) (Sá-Correia et al., 2009). The expression of these pumps is believed to be regulated by zinc-finger and AP-1 transcription factors (Moye-Rowley, 2003). In fission yeast, Bfr1 and Pmd1 have been shown to be the key ABC family transporters (Arita et al., 2011; Iwaki et al., 2006), but the MFS transporters involved remain unclear. Pap1, an AP-1 like transcription factor, has been shown to have important roles in MDR (Toda et al., 1991; Toone et al., 1998), but the zinc-finger transcription factors remain uncharacterized. Therefore, to develop fission yeast as a model system for chemical probe discovery and chemical biology, it is important to analyze these mechanisms and suppress the MDR response.
Here, we report a systematic analysis of MDR in fission yeast using microarray, gene deletion, and gene overexpression approaches. We identified key transcription factors and drug-efflux transporters, and functionally characterized Mfs1, an MFS transporter, and Prt1, a fission yeast zinc-finger transcription factor that is a homolog of budding yeast Pdr1/3. Guided by these data, we engineered a fission yeast strain that is sensitive to a wide-range of chemical inhibitors, including several commonly used chemical probes. Finally, we use chemical probes and high-resolution microscopy-based phenotypic analyses to examine mechanisms underlying metaphase spindle assembly.
RESULTS
Analysis of Fission Yeast’s Basal and Drug-Induced MDR Response
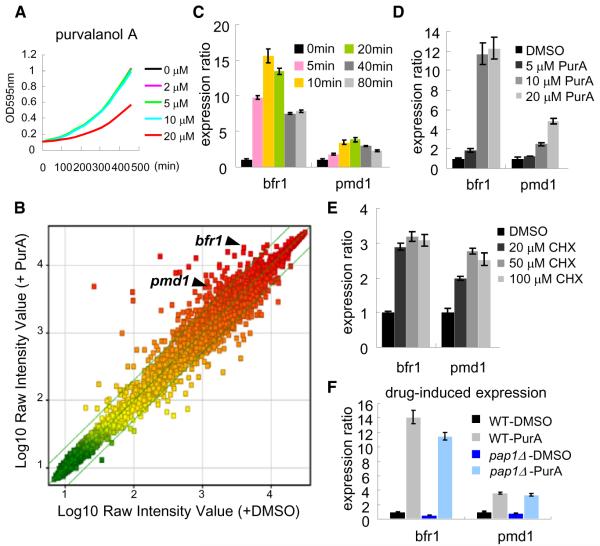
To examine fission yeast’s transcriptional response to drug treatment, we used microarray-based analysis. Purvalanol A, which inhibits the well-conserved cyclin-dependent kinases (Gray et al., 1998), was selected for these studies as we had observed that cell growth was only partly inhibited, even at relatively high doses (20 μM) (Figure 1A), possibly due to MDR mechanisms. We found that purvalanol A (20 μM) treatment induced, within minutes, the expression of ~100 genes (Figure 1B; Table S3 available online), including bfr1+ and pmd1+, which represent only two of the possible 11 ABC transporters in fission yeast. Six, of the potential 49 MFS transporters, were also upregulated (Table S3).
Figure 1. Analysis of Fission Yeast’s Transcriptional Response to Drug Treatment.
(A) Growth of wild-type cells (in YE4S medium at 32°C) in the presence or absence of purvalanol A (PurA).
(B) Microarray analysis of mRNA levels in exponentially growing wild-type cells that were treated for 20 min with 20 μM PurA (or DMSO). Scatter plot is color-coded for expression levels (green, low; red, high). The lines show ± 2-fold change in response to drug (n = 2 independent experiments, average is shown). The list of genes that were upregulated (>2-fold) by PurA treatment are provided in Table S3.
(C–E) The expression levels of bfr1+ and pmd1+ genes in chemical inhibitor-treated exponentially growing wild-type cells analyzed by RT-qPCR (n = 3). (C) Cells were treated with PurA (20 μM, 0 min), and total RNA was purified at the indicated time points. (D and E) Cells were treated with the indicated concentration of PurA (D) or cycloheximide (CHX) (E) for 20 min, after which the total RNA was purified.
(F) Exponentially growing wild-type (WT) or pap1Δ cells were treated with 20 μM PurA (or DMSO) for 20 min, after which the total RNA was purified (n = 3). In all RT-qPCR experiments, the histograms show the ratio of the genes (value of WT was defined as one) with respect to the signals obtained for ACT1, used as a normalization control. Error bars indicate SD.
See also Table S3.
We next focused on transcription factors involved in regulating the expression of these different pumps that may mediate fission yeast’s MDR response. As a readout of the drug-induced MDR response, we focused on the transcription levels of two ABC transporters, Bfr1 and Pmd1, for which the transcriptional response was rapid and dose-dependent (Figures 1C and 1D) and was also induced by cycloheximide, a protein synthesis inhibitor (Figure 1E). Pap1, an AP-1 like transcription factor, is needed for the oxidative stress response in fission yeast and has been shown to have important roles in MDR (Toda et al., 1991; Toone et al., 1998). We found that Pap1 controls the basal, but not the drug-induced expression, of the bfr1+ and pmd1+ genes (Figure 1F), suggesting that another transcription factor is likely to be required for the drug-induced MDR response.
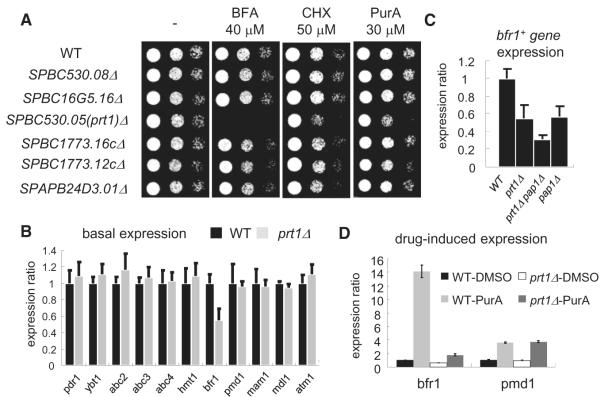
To identify the fission yeast transcription factor responsible for the drug-induced expression of the ABC transporters, we focused on other transcription factors implicated in the fungal MDR response. Studies in S. cerevisiae and Candida albicans have shown that the zinc-finger transcription factor ScPdr1/3 (or CaTac1) mediates MDR through transcriptional activation of drug efflux pumps (Coste et al., 2004; Kolaczkowska et al., 2008; Thakur et al., 2008). To characterize the fission yeast homologs of ScPdr1, we combined bioinformatics and phenotypic analyses of strains from a genome-wide gene-deletion library (Kim et al., 2010). Of the six proteins that were most similar to ScPdr1, our data showed that only the deletion of a previously uncharacterized gene, SPBC530.05 enhanced sensitivity to cytotoxic drugs (Figure 2A). We named this transcription factor Prt1 (Pdr1-related transcription factor 1).
Figure 2. Analysis of Transcription Factors Regulating Fission Yeast MDR Response.
(A) Serial dilutions of the indicated strains were spotted onto YE4S plates, or YE4S plates containing indicated drugs, and incubated at 29°C.
(B and C) The expression levels of ABC transporters were measured by RT-qPCR in the indicated strains (n = 5). Total RNA was purified from asynchronous cultures.
(D) Exponentially growing wild-type or prt1Δ cells were treated with 20 μM PurA (or DMSO) for 20 min, after which total RNA was purified (n = 3). Expression ratios are calculated as in Figure 1. Error bars indicate SD.
See also Figure S2.
We next examined if Prt1 controls the drug-induced expression of drug efflux pumps in fission yeast. Of the 11 ABC transporters, only the basal expression of the bfr1+ gene was reduced in prt1Δ cells (Figure 2B). The levels of the bfr1+ gene transcripts were further reduced in prt1Δ pap1Δ cells (Figure 2C). Interestingly, drug-induced expression of the bfr1+ gene, but not the pmd1+ gene, was largely suppressed in prt1Δ cells (Figure 2D). These data indicate that the Pap1 and Prt1 transcription factors together control the basal expression levels of this important drug efflux pump, whereas Prt1 is responsible for the induced expression of the pump in response to drug treatment.
Analysis of Prt1-Dependent Regulation of Fission Yeast MDR
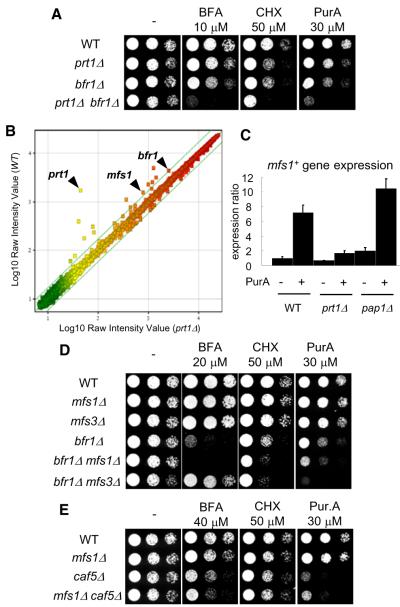
Cells lacking the ABC transporter Bfr1 were less drug sensitive than cells lacking this transporter and the transcription factor Prt1 (prt1Δ bfr1Δ) (Figure 3A), suggesting that Prt1 likely regulates the expression of other genes required for the MDR response. To identify these genes, we used microarray analysis to compare gene expression profiles between wild-type and prt1Δ cells. In addition to Bfr1, two previously uncharacterized genes, which based on bioinformatics analyses were expected to be MFS transporters, SPAC17C9.16c (hereafter, mfs1+) and SPBC36.03c (hereafter, mfs3+), were found to be regulated by Prt1. Interestingly, although the drug-induced expression of these MFS transporters was largely reduced in prt1Δ, their expression was not dependent on Pap1 (Figures 3B, 3C, and S1).
Figure 3. Analysis of the MFS Transporters Contributing to the Fission Yeast MDR Response.
(A, D, and E) Serial dilutions of the indicated strains were spotted onto YE4S plates, or YE4S plates containing indicated drugs, and incubated at 29°C.
(B) Microarray analysis of mRNA levels in exponentially growing wild-type and prt1Δ cells. Scatter plot is color-coded for expression levels (green, low; red, high). The lines show ± 1.7-fold change in response to drug (n = 2 independent experiments, average is shown).
(C) The expression level of mfs1+ gene was measured by RT-qPCR in the indicated strains (n = 3). Total RNA was purified after treatment with 20 μM PurA (or DMSO) for 20 min. Expression ratios are calculated as in Figure 1. Error bars indicate SD.
We next examined if these two MFS transporters contribute to the fission yeast MDR response. Although single gene deletions of mfs1+ or mfs3+ did not increase drug sensitivity, deletions of bfr1+ and mfs1+ together did (Figure 3D). Interestingly, bfr1Δ mfs3Δ increased sensitivity toward cycloheximide and purvalanol A, but not brefeldin A, when compared to the bfr1Δ strain (Figure 3D), suggesting that Mfs3 may contribute to drug influx as well as drug efflux. Taken together, our findings indicate that Prt1 regulates MDR through activating the expression of at least two drug efflux pumps, an ABC transporter (Bfr1) and an MFS transporter (Mfs1).
Using Gene Overexpression to Identify Other Components of the Fission Yeast MDR Response
To complement the transcriptional profiling and gene-deletion-based analyses of MDR mechanisms, we analyzed genes whose overexpression could confer drug resistance in cells lacking both the transcription factors we characterized and also the ABC transporter Bfr1 (prt1Δ pap1Δ bfr1Δ). As bfr1 mutants confer sensitivity to brefeldin A, we used this compound for the screen. Approximately 107 transformants derived from a S. pombe cDNA library were screened and 44 brefeldin A-resistant clones were isolated (Table S4). Consistent with our transcpritional profiling and gene deletion data, Pap1 and Mfs1 overexpressing clones were isolated. In addition, we isolated clones in which another MFS transporter, Caf5, was overexpressed. Caf5 overexpression was reported to confer caffeine resistance (Benko et al., 2004). To examine the Caf5’s contribution to fission yeast MDR, we tested sensitivity to brefeldin A, cycloheximide, and purvalanol A. Analysis of caf5Δ cells revealed multidrug sensitivity that was greater than that of wild-type cells (Figure 3E), indicating that Caf5 is also involved in the fission yeast MDR response. In addition, dual deletion of Mfs1 and Caf5 further increase sensitivity to brefeldin A but not significantly to the other two drugs tested, consistent with drug composition being a key determinant of MDR efficiency.
Engineering a Fission Yeast Strain with Increased Drug-Sensitivity
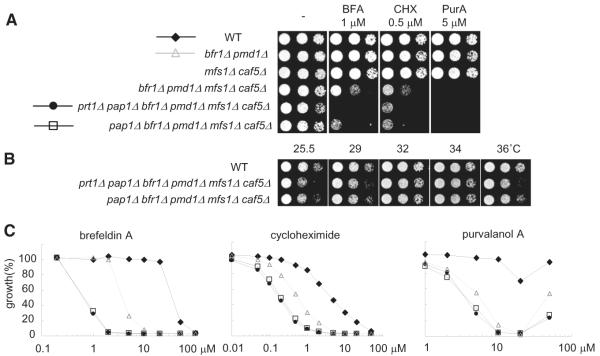
The fission yeast strain with maximum drug sensitivity, reported thus far, has two ABC pumps deleted bfr1Δ pmd1D (Arita et al., 2011). We used our findings to design fission yeast strains that we anticipated could be much more sensitive to a wide range of drugs than the bfr1Δ pmd1Δ strain. We first generated a fission yeast strain lacking the two ABC transporters and the two MFS transporters we characterized (bfr1Δ pmd1Δ mfs1D caf5Δ) and tested sensitivity to brefeldin A, cycloheximide, and purvalanol A. Importantly, the deletion of the MFS pumps significantly increased drug sensitivity of fission yeast relative to the bfr1Δ pmd1Δ strain (Figure 4A). Combined deletions of the four drug-pumps and Pap1 (pap1Δ bfr1Δ pmd1Δ mfs1D caf5Δ) further enhanced drug sensitivity (Figures 4A and 4C). In addition, sensitivity to these drugs is similar in both YE-based complete medium and EMM-based minimal medium (Figures S3F–S3J). To further characterize this strain, we measured growth at different temperatures (25°C, 32°C, and 36°C), cell-cycle progression, and spore formation in meiosis (Figures S3A–S3E). We found that these parameters were similar to that measured for wild-type cells. A strain (prt1Δ pap1Δ bfr1Δ pmd1Δ mfs1Δ caf5Δ), in which prt1+ gene was also deleted, did not significantly further enhance drug sensitivity (Figures 4A and 4C), suggesting that Bfr1 and Mfs1 are the major targets of Prt1 for the MDR response. However, the deletion of these six genes resulted in weak temperature sensitivity in the absence of drug (Figure 4B). Therefore, we reasoned that the 5-gene-deleted strain is optimal for chemical biology studies and named it the “MDR-sup” (for MDR-suppressed) strain.
Figure 4. Construction of the “MDR-Sup” Fission Yeast Strain.
(A) Serial dilutions of the indicated strains were spotted onto YE4S plates, or YE4S plates containing indicated drugs, and incubated at 29°C.
(B) Serial dilutions of the indicated strains were spotted onto YE4S plate and incubated at the indicated temperature.
(C) Exponentially growing culture (OD = 0.5) of WT (diamond), bfr1Δ pmd1Δ (triangle), pap1Δ bfr1Δ pmd1Δ mfs1Δ caf5Δ (square), or prt1Δ pap1Δ bfr1Δ pmd1Δ mfs1Δ caf5Δ (circle) cells were diluted 50 times in YE4S medium, treated with indicated compounds at the indicated concentrations (μM), and incubated for 14 hours at 32°C. Growth (%) is presented relative to DMSO-treated cells.
See also Figure S3.
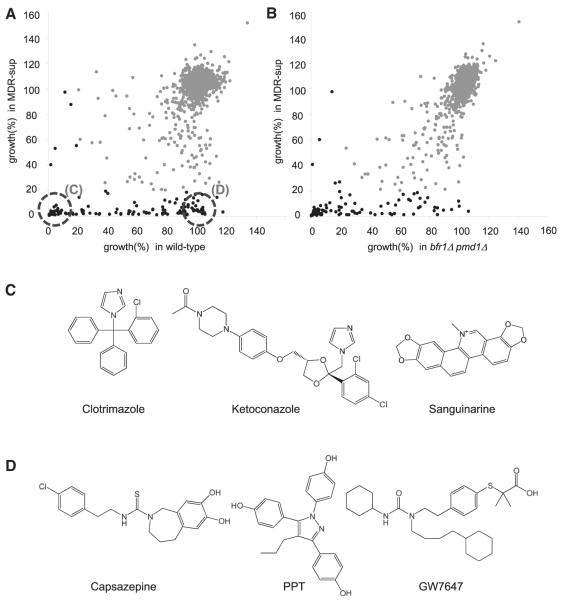
To better evaluate the MDR-sup strain we analyzed the toxicity of chemical inhibitors that are commonly used in yeast research, such as actin assembly inhibitor (latrunculin A) and benomyl-related tubulin poisons (MBC and benomyl), or other cell-cycle kinase inhibitors used to examine mammalian cell division, such as Aurora kinase (hesperadin) and Mps1 kinase (reversine) inhibitors. We found that sensitivity to latrunculin A, hesperadin, and reversine were significantly enhanced in the MDR-sup strain (Figure S4). The activity of benomyl-based compounds was similar in the MDR-sup strain and the wild-type strain, suggesting that these compounds are not likely to be pumped out by ABC and MFS transporters (Figure S4). We also examined the activity of compounds from a 1,280-member library of diverse bioactive small molecules (LOPAC1280 [Sigma-Aldrich, St. Louis, MO, USA], 20 μM). The number of compounds that inhibit growth by >80% in the wild-type, the strain lacking two ABC pumps (bfr1Δ pmd1Δ), and our MDR-sup strain was found to be 51, 92, and 132, respectively (Figures 5A and 5B; Table S5). As would be expected, known antifungal agents (e.g., azoles) were equally toxic to wild-type fission yeast and the MDR-sup strain (Figure 5C; Table S5). Enhanced toxicity, compared to the wild-type or the bfr1Δ pmd1Δ strain was observed for inhibitors of proteases, kinases, and topoisomerase (Table S5), suggesting that MDR mechanisms reduce the efficacy of these compounds in wild-type cells. Interestingly, several compounds were found whose toxicity to fission yeast would not be predicted based on their anticipated targets. These include capsazepine, a vanilloid receptor antagonist, PPT, an estrogen receptor-α agonist, and GW7647, a PPARα agonist (Figure 5D; Table S5). It will be important to determine the targets of these compounds as it is possible they may be unanticipated off-targets. It is likely that the MDR-sup strain will be useful for this analysis. Together, our data indicate that our engineered MDR-sup strain has enhanced drug sensitivity to a wide-range of chemical inhibitors.
Figure 5. The “MDR-Sup” Fission Yeast Strain Is Sensitive to a Wide Range of Chemical Inhibitors.
(A and B) Scatter plot shows growth of WT (A) or bfr1Δ pmd1Δ (B) strain (x axis) and MDR-sup strain (pap1Δ bfr1Δ pmd1Δ mfs1Δ caf5Δ) (y axis) treated with compounds in the LOPAC 1280 library (20 μM, Sigma-Aldrich), normalized to the growth measured in DMSO alone. Black circles indicate compounds that inhibit growth by >80%.
(C) Representative chemical structures of compounds that inhibit growth by >90% in both WT and MDR-sup strain.
(D) Representative chemical structures of compounds that inhibit growth by >90% in MDR-sup but by <10% in WT strain. The full list of compounds that inhibit growth >80% in either WT, bfr1Δ pmd1Δ, or MDR-sup strain is shown in Table S5.
See also Figure S4.
Analysis of the Mechanism-of-Action of Commonly Used Chemical Probes
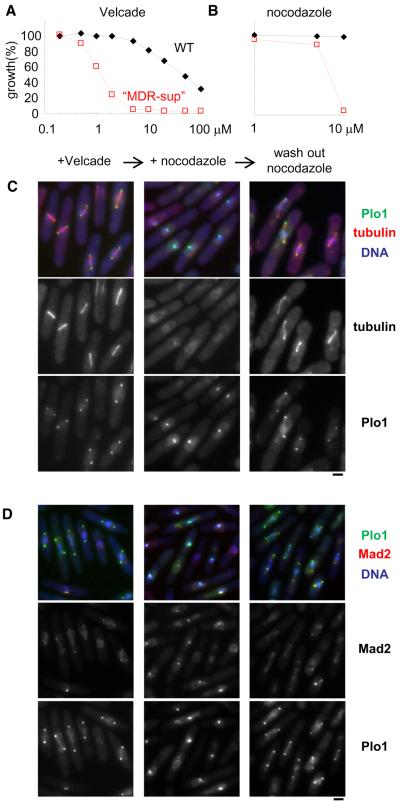
Many chemical inhibitors that are powerful probes of cell division dynamics in human cells are not very effective in wild-type fission yeast cells (Figures 6A and 6B). For example, nocodazole, an inhibitor of microtubule polymerization, has been found to be active in fission yeast only when a mutation in α- or β-tubulin is present (Umesono et al., 1983). There are also studies showing the activity of these compounds at extremely high concentrations. For example, Velcade has been shown to be active at a millimolar concentrations (Takeda et al., 2011), approximately 100,000-fold higher than its effective dose in human cells, raising concerns about off-target activity at these doses. Therefore, the use of the compounds has been limited in fission yeast and combining acute chemical perturbations with genetic manipulations and detailed phenotypic analysis has been greatly restricted. Importantly, we find that both nocodazole and Velcade are active in the MDR-sup strain at ~10 μM (Figures 6A and 6B), suggesting that they could be useful tools to dissect cell division mechanisms.
Figure 6. Examining Mitotic Mechanisms Using Nocodazole and Velcade in the “MDR-Sup” Strain.
(A and B) Exponentially growing culture (OD = 0.5) of WT (black diamond) and pap1Δ bfr1Δ pmd1Δ mfs1Δ caf5Δ (red square) cells were diluted 50 times in YE4S medium, treated with indicated compounds at the indicated concentrations (μM), and incubated for 14 hours at 32°C. Growth (%) is presented relative to DMSO-treated cells.
(C and D) Cells were blocked at S-phase using hydroxyurea, incubated for 30 min, then treated with Velcade (40 μM), and incubated for 60 min at 32°C (+ Velcade, left lanes). Then nocodazole (15 μM) was added, and incubated for 30 min at 32°C (+ nocodazole, middle lanes). After that, nocodazole was washed out, and incubated for 30 min at 32°C (wash out nocodazole, right lanes). Representative images of Mcherry-tubulin (C), Mad2-mcherry (D), and Plo1-mYFP (C and D) signals are shown. Scale bars, 2 μm.
See also Figure S5.
We next examined the cellular phenotypes associated with nocodazole and Velcade treatments. In the presence of Velcade MDR-sup fission yeast cells accumulated with separated spindle pole bodies (SPBs), short spindles, and condensed chromosome, as revealed by examining Plo1 (the fission yeast homolog of Polo-like kinase known to concentrate at SPBs in mitosis; Mulvihill et al., 1999) signals, tubulin distribution, and DAPI staining, respectively. This phenotype is consistent with the cells being arrested at metaphase with bipolar spindles (Figures 6C and 6D, left lanes), as would be expected upon proteasome inhibition. Subsequent addition of nocodazole resulted in the disruption of the microtubule cytoskeleton (Figure 6C, middle lanes, Figure S5A). The loss of spindle microtubules was also indicated by the presence of Mad2 signals at kinetochores (Figure 6D, middle lanes), as disruption of chromosome-spindle attachments results in recruitment of spindle assembly checkpoint components to kinetochores. Interestingly, the SPBs are frequently clustered upon treatment with nocodazole (Figures 6C and 6D, middle lanes, Figure S5). This is surprising as fission yeast undergoes a “closed” mitosis, such that the nuclear envelop does not breakdown at the G2-M transition. The SPBs are anchored in the nuclear envelope (Ding et al., 1997), and in principle their separation could be maintained as the nuclear envelope is not disrupted by nocodazole treatment. To exclude the possibility that these results are due to nocodazole having a target other than tubulin in the MDR-sup strain, we used another antimicrotubule agent MBC or cold-treatment to disrupt spindle apparatus (Gachet et al., 2008). In both conditions, reclustering of SPBs was observed (Figure S5B). Together, these data indicate that microtubules are needed not only for establishing SPB separation but also for maintaining SPB separation in fission yeast.
Finally, relief from nocodazole treatment restored metaphase-arrested cells with bipolar spindles (Figures 6C and 6D, right lanes) within 30 min, consistent with nocodazole being a reversible inhibitor. Together, these data indicate that the mechanism-of-action of nocodazole and Velcade are conserved in the MDR-sup fission yeast, and these chemical probes will be useful for acutely inhibiting, and even activating, key processes required for the stable propagation of genomes in eukaryotes.
DISCUSSION
Our analysis of MDR mechanisms in fission yeast have revealed key factors needed for this response. Our studies have led to the functional characterization of Mfs1, an MFS transporter, and Prt1, a zinc-finger transcription factor. We show that Prt1, which is likely to be a fission yeast homolog of S. cerevisiae PDR1 and C. albicans Tac1, regulates the MDR response mainly through drug-induced expression of an ABC transporter (Bfr1) and an MFS transporter (Mfs1). Sequence comparison of S. pombe, S. cerevisiae, and C. albicans proteins indicates that Prt1 has two highly conserved domains (Figure S2). One of these is a cysteine-rich motif at the N terminus and is likely to be involved in zinc-dependent binding to DNA. The second conserved domain is in the middle of the protein, is a part of the xenobiotic binding domain (XBD), and likely binds to drugs and xenobiotics (Thakur et al., 2008). Therefore, Prt1 may activate expression of drug efflux pumps and induction of MDR via direct binding to drugs.
Our analysis of the MDR mechanisms in fission yeast has led to the construction of a MDR-sup strain that is sensitive to a range of diverse chemical inhibitors. The MDR response in fungi is highly complex, and it is likely that it is not completely eliminated in the MDR-sup strains we have developed for at least two reasons. First, sequence analysis predicts 11 ABC and 49 MFS transporters in fission yeast. It is likely that some of these contribute, possibly redundantly, to the influx or efflux of chemical inhibitors. Second, several studies indicate that genes involved in ergosterol biosynthesis, vacuolar protein sorting, and vacuolar H+-ATPase function are involved in MDR mechanisms (Parsons et al., 2004, 2006) (Dawson et al., 2008). We note that although deletion of Erg6, a gene in ergosterol biosynthetic pathway, can increase drug sensitivity in budding yeast, fission yeast strains with erg6 deleted are sick, even in the absence of drugs (Iwaki et al., 2008). Therefore, we did not examine deletions of genes in ergosterol biosynthetic pathway to further enhance drug-sensitivity of the MDR-sup strain. Although further analysis is needed to more completely characterize the fission yeast MDR response, our findings do suggest that this response to a wide-range of chemical inhibitors can be suppressed by the deletion of two ABC transporters, two MFS pumps, and a transcription factor, indicating that these are likely to be the key mechanisms.
Our MDR-sup strain should be particularly useful for analyzing mechanisms of drug action. In addition to detailed phenotypic analyses, for which a wide-range of strains and reagents are available, classical yeast forward genetics can also be used to analyze drug resistance mechanisms and thereby identify the physiologically relevant drug targets. The MDR-sup strain provides an important advantage for a random mutagenesis-mediated selection of drug resistance. It has been show that a very most common mechanism of drug resistance involves the MDR response. For example, “activating” mutations in transcription factors can lead to the overexpression of drug efflux pumps (Moye-Rowley, 2003). As our MDR-sup (or combined with deletion of prt1 gene) strain lacks many of these genes, it is more likely that the drug-resistant clones isolated will have mutations in the direct drug target of cellular pathways. The MDR-sup strain is also likely to be useful for identifying new chemical probes as high-throughput screens can be carried out at lower compound concentrations than would be needed when using wild-type strains. In addition, powerful screens, such as “chemical synthetic lethality” screens, can be designed in fission yeast to select compounds that elicit genotype specific-effects (Nehil et al., 2007; Torrance et al., 2001). It is possible that such screens will lead to new probes for RNA interference and heterochromatin formation, processes that are conserved between fission yeast and humans.
The MDR-sup strain allows the use of chemical inhibitors to be combined with high-resolution imaging and other genetic manipulations to dissect dynamic cellular mechanisms. Our studies with Velcade and nocodazole suggest that maintaining the separation of spindle pole bodies (SPBs), key organizers of spindle microtubules in dividing fission yeast cells, depends on microtubules (Figure 6). This observation is surprising, as in current models the major pulling and pushing forces acting on the SPBs depend on microtubules and motor proteins (e.g., kinesin-5) (Dumont and Mitchison, 2009). Once separated, the SPBs could be kept apart by the nuclear envelop, in which the SPBs are embedded (Ding et al., 1997). We favor the model in which another, microtubule-independent, force brings the two SPBs together. However, at this stage, we cannot exclude other possibilities, such as the presence of microtubules that cannot be readily detected or if SPBs are clustered during microtubule disassembly, that is, being “reeled-in” by shrinking microtubules that somehow maintain attachments to chromosomes and SPBs. Characterizing the molecular basis of this SPB clustering mechanism is an important step for future studies, as it could shed new light on how metaphase spindles assemble and how the size of these structures is determined in different contexts.
Our MDR-sup strain should help analyze cellular processes that have been difficult to study using available approaches. For example, it is likely that our system will be useful to examine the first and second meiotic cell divisions, which occur sequentially after one round of DNA replication. The successful completion of these basic cellular processes is required to prevent pregnancy loss and developmental defects in humans (Hassold and Hunt, 2001). As high or low temperature severely affects meiotic progression in fission yeast, canonical temperature-sensitive genetic mutants have not been very useful. Moreover, as mechanisms of cell-cycle progression are likely to be conserved between meiosis I and II, genetic mutations of cell-cycle or chromosome segregation genes can affect meiosis I, making proper interpretations of any observed perturbations on meiosis II very difficult. We believe that the MDR-sup strain and a validated set of chemical inhibitors, which will allow acute inhibition, should help examine molecular mechanisms required for the first or the second meiotic divisions.
SIGNIFICANCE
Fission yeast is a genetically tractable model system that has provided valuable insights into cellular mechanism. Importantly, fission yeast shares more processes (e.g., RNAi and centromere specification) with human cells than budding yeast, another widely used model system. However, fission yeast has not been very useful for chemical biology, as many commonly used chemical probes are not active in these cells, in large part due to an effective multidrug resistance (MDR) response. With the goal to develop fission yeast for chemical biology, we systematically analyzed drug-pumps and transcription factors using microarray-based, gene deletion, and gene overexpression approaches. These studies led to two ABC transporters (Bfr1 and Pmd1), two MFS pumps (Mfs1 and Caf5), and two transcription factors (Prt1 and Pap1) as the major contributors to fission yeast’s MDR response. These findings represent the functional characterization of Mfs1, an MFS transporter, and Prt1, a zinc-finger transcription factor that is a homolog of budding yeast Pdr1/3. Guided by these data, we engineered the MDR-sup fission yeast strain, which has five of these MDR genes deleted. We show that this strain is sensitive to a wide-range of bioactive small molecules, including nocodazole and Velcade. We combined the use of chemical inhibitors, high-resolution imaging, genetic manipulations, and the MDR-sup strain to examine metaphase spindle assembly during mitosis. Our analysis suggests that microtubules are needed to maintain the normal separation of the two microtubule-organizing spindle pole bodies in a “closed mitosis,” when the nuclear membrane persists through M-phase and encapsulates the division apparatus. Together, our findings indicate that our MDR-sup strain will be useful for analyzing complex and dynamic cellular processes. In addition, our studies suggest that fission yeast should be a valuable genetically tractable model system for chemical inhibitor discovery and analysis of drug mechanism of action.
EXPERIMENTAL PROCEDURES
Schizosaccharomyces pombe Strains
All strains used are listed in Table S1. Standard growth conditions and methods were used (Moreno et al., 1991). Deletions of each gene in S. pombe were performed using the PCR-based gene-targeting method for S. pombe (Bähler et al., 1998). For the Figure 2A experiment, strains from the deletion library were used (Kim et al., 2010).
Chemical Compounds
Cycloheximide, nocodazole, MBC, and benomyl were purchased from Sigma-Aldrich. Brefeldin A and Velcade (bortezomib) were purchased from LC Laboratories (Woburn, MA, USA). Purvaranol A and latrunculin A was purchased from Tocris Bioscience (Ellisville, MO, USA). Reversine was purchased from Cayman Chemical (Ann Arbor, MI, USA). Hesperadin was synthesized in our laboratory. All chemicals were dissolved in DMSO, kept in −20°C, and used as 0.25%–1% DMSO solution.
Purification of Total RNA from Fission Yeast
Total RNA was isolated from S. pombe cells using a hot phenol method followed by phenol-chloroform extraction, precipitation, and purification using Qiagen RNeasy columns (Venlo, the Netherlands) (Lyne et al., 2003).
RT-qPCR Analysis
Total RNA (1 μg ) was used for RT reactions. RT reactions were carried out using the manufacturer’s protocol in the presence or absence of enzyme (SuperScript III First-Strand Synthesis System, Invitrogen, Carlsbad, CA, USA). For quantitative PCR (qPCR), SYBR Green (Applied Biosystems, Foster City, CA, USA) and primers were mixed, and the starting quantity of DNA was estimated from the number of cycles (Ct value) required to reach the threshold using Roche LightCycler 480 System (Indianapolis, IN, USA). Primers used in this study are listed in Table S2.
Microarray Analysis
RNA quality was assessed using the Agilent 2100 Bioanalyzer and the RNA 6000 Nano kit (Agilent Technologies, Santa Clara, CA, USA). Total RNA (200 ng) was used to prepare biotin-labeled RNA using Ambion MessageAmp Premier RNA Amplification Kit (Applied Biosystems). Briefly, 200 ng of total RNA was used to synthesize the first strand of cDNA using ArrayScript reverse-transcriptase and an oligo (dT) primer bearing a T7 promoter. The single-stranded cDNA was then converted into a double-stranded DNA (dsDNA) by DNA polymerase I in the presence of Escherichia coli RNase H and DNA ligase. The dsDNA was used as a template for in vitro transcription in a reaction containing biotin-labeled UTP, unlabeled NTPs, and T7 RNA polymerase. The amplified, biotin-labeled antisense RNA (aRNA) was purified, and its quality was assessed using the Agilent 2100 Bioanalyzer and the RNA 6000 Nano kit. The fragmented aRNA (4 μg) was fragmented and hybridized to Affymetrix Yeast Genome 2.0 arrays for 16 hr at 45°C as described in the manufacturer’s protocol (Affymetrix, Santa Clara, CA, USA). After hybridization, arrays were stained with streptavidin-phycoerythrin, followed by an antibody solution (antistreptavidin) and a second streptavidin-phycoerythrin solution, with all liquid handling performed by a GeneChip Fluidics Station 450. Gene Chips were then scanned with the Affymetrix GeneChip Scanner 3000 7G. The raw intensity data of Gene Chips was normalized and further analyzed in GeneSpring 11.0 (Agilent Technologies).
Overexpression Screen
A Gateway-compatible Lifetech library was constructed from total S. pombe RNA derived from mitotic, meiotic, and shmooing cells in a 2:1:1 ratio, within a Gateway-modified version of the ura4-based pRep4X vector (Fersht et al., 2007). The library was transformed into SAK31 cells. The transformed cells were plated out and left to grow at 32°C on EMM-Uri plates. The colonies were replica-plated onto EMM-Uri plates containing 10 μM brefeldin A and incubated at 32°C. We screened 1.5 × 105 colonies and identified 44 colonies as brefeldin A-resistant. The plasmids conferring brefeldin A resistance were sequenced.
Chemical Screen
SAK1, SAK27, and SAK84 strains were used for chemical screen. Logarithmically growing cells (OD = 0.2) were diluted 16 times, mixed with compounds (LOPAC1280, 20 μM), and incubated for 18 hr at 29°C (total volume: 50 ml per well). Multidrop Combi (Thermo Scientific, Waltham, MA, USA) was used to dispense the cells into wells of the 384-plate (Greiner clear, flat-bottom PS plate). The growth was measured by microtiter plate reader (Perkin-Elmer EnVision, 590 nm filter). For calculation of growth ratio, OD values of each well were divided by that of control well incubated with DMSO.
Microscopy
For methanol fixation, cell pellet from 1 ml culture is mixed with 800 μl chilled methanol and incubated for >12 hr at −20°C. The fixed cells are mixed with DAPI (4,6-diamidino-2-phenylindole), and cell pellet are dissolved in PEMS (100 mM PIPES [pH 6.9], 1 mM EGTA, 1 mM MgSO4, and 1 M Sorbitol). Images were acquired at room temperature on a microscope (Axioplan 2; Carl Zeiss, Inc., Maple Grove, MN), equipped with a CoolsnapHQ camera (Roper Scientific, Trenton, NJ, USA), and were processed with MetaMorph software (Molecular Devices, Sunnyvale, CA, USA). A Z-stack of about 3 μm thickness, with single planes spaced by 0.3 μm, was acquired and subsequently projected to a single image.
Supplementary Material
ACKNOWLEDGMENTS
We thank Fraser Glickman for the use of the Rockefeller University High Throughput Screening Resource Center; Connie Zhao for the use of the Rockefeller University Genomics Resource Center; Yoshinori Watanabe, Masamitsu Sato, and Silke Hauf for providing strains and plasmids. This work was supported by JSPS Postdoctoral Fellowships for Research Abroad (to S.A.K. and A.T.); by the Beast Cancer Research Foundation, the Wellcome Trust, and the Rockefeller University (to P.N.); and the National Institutes of Health/National Institute of General Medical Sciences (GM098579) (to T.M.K.).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes five figures and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.chembiol.2012. 06.008.
REFERENCES
- Arita Y, Nishimura S, Matsuyama A, Yashiroda Y, Usui T, Boone C, Yoshida M. Microarray-based target identification using drug hypersensitive fission yeast expressing ORFeome. Mol. Biosyst. 2011;7:1463–1472. doi: 10.1039/c0mb00326c. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Benko Z, Fenyvesvolgyi C, Pesti M, Sipiczki M. The transcription factor Pap1/Caf3 plays a central role in the determination of caffeine resistance in Schizosaccharomyces pombe. Mol. Genet. Genomics. 2004;271:161–170. doi: 10.1007/s00438-003-0967-3. [DOI] [PubMed] [Google Scholar]
- Bergnes G, Brejc K, Belmont L. Mitotic kinesins: prospects for antimitotic drug discovery. Curr. Top. Med. Chem. 2005;5:127–145. doi: 10.2174/1568026053507697. [DOI] [PubMed] [Google Scholar]
- Burdine L, Kodadek T. Target identification in chemical genetics: the (often) missing link. Chem. Biol. 2004;11:593–597. doi: 10.1016/j.chembiol.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Drubin DG, Barnes G. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 2002;157:199–203. doi: 10.1083/jcb.200201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell. 2004;3:1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson K, Toone WM, Jones N, Wilkinson CR. Loss of regulators of vacuolar ATPase function and ceramide synthesis results in multidrug sensitivity in Schizosaccharomyces pombe. Eukaryot. Cell. 2008;7:926–937. doi: 10.1128/EC.00037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr. Biol. 2009;19:R749–R761. doi: 10.1016/j.cub.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht N, Hermand D, Hayles J, Nurse P. Cdc18/CDC6 activates the Rad3-dependent checkpoint in the fission yeast. Nucleic Acids Res. 2007;35:5323–5337. doi: 10.1093/nar/gkm527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y, Reyes C, Courthéoux T, Goldstone S, Gay G, Serrurier C, Tournier S. Sister kinetochore recapture in fission yeast occurs by two distinct mechanisms, both requiring Dam1 and Klp2. Mol. Biol. Cell. 2008;19:1646–1662. doi: 10.1091/mbc.E07-09-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NS, Wodicka L, Thunnissen AM, Norman TC, Kwon S, Espinoza FH, Morgan DO, Barnes G, LeClerc S, Meijer L, et al. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Higgins CF. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Iwaki T, Giga-Hama Y, Takegawa K. A survey of all 11 ABC transporters in fission yeast: two novel ABC transporters are required for red pigment accumulation in a Schizosaccharomyces pombe adenine biosynthetic mutant. Microbiology. 2006;152:2309–2321. doi: 10.1099/mic.0.28952-0. [DOI] [PubMed] [Google Scholar]
- Iwaki T, Iefuji H, Hiraga Y, Hosomi A, Morita T, Giga-Hama Y, Takegawa K. Multiple functions of ergosterol in the fission yeast Schizosaccharomyces pombe. Microbiology. 2008;154:830–841. doi: 10.1099/mic.0.2007/011155-0. [DOI] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowska A, Kolaczkowski M, Goffeau A, Moye-Rowley WS. Compensatory activation of the multidrug transporters Pdr5p, Snq2p, and Yor1p by Pdr1p in Saccharomyces cerevisiae. FEBS Lett. 2008;582:977–983. doi: 10.1016/j.febslet.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Kapoor TM. Unraveling cell division mechanisms with small-molecule inhibitors. Nat. Chem. Biol. 2006;2:19–27. doi: 10.1038/nchembio757. [DOI] [PubMed] [Google Scholar]
- Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bähler J. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics. 2003;4:27. doi: 10.1186/1471-2164-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr LM, Bojanic D. Novel trends in high-throughput screening. Curr. Opin. Pharmacol. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Moye-Rowley WS. Transcriptional control of multidrug resistance in the yeast Saccharomyces. Prog. Nucleic Acid Res. Mol. Biol. 2003;73:251–279. doi: 10.1016/s0079-6603(03)01008-0. [DOI] [PubMed] [Google Scholar]
- Mulvihill DP, Petersen J, Ohkura H, Glover DM, Hagan IM. Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol. Biol. Cell. 1999;10:2771–2785. doi: 10.1091/mbc.10.8.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehil MT, Tamble CM, Combs DJ, Kellogg DR, Lokey RS. Uncovering genetic relationships using small molecules that selectively target yeast cell cycle mutants. Chem. Biol. Drug Des. 2007;69:258–264. doi: 10.1111/j.1747-0285.2007.00496.x. [DOI] [PubMed] [Google Scholar]
- Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Peterson JR, Mitchison TJ. Small molecules, big impact: a history of chemical inhibitors and the cytoskeleton. Chem. Biol. 2002;9:1275–1285. doi: 10.1016/s1074-5521(02)00284-3. [DOI] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá-Correia I, dos Santos SC, Teixeira MC, Cabrito TR, Mira NP. Drug:H+ antiporters in chemical stress response in yeast. Trends Microbiol. 2009;17:22–31. doi: 10.1016/j.tim.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Takeda K, Mori A, Yanagida M. Identification of genes affecting the toxicity of anti-cancer drug bortezomib by genome-wide screening in S. pombe. PLoS ONE. 2011;6:e22021. doi: 10.1371/journal.pone.0022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur JK, Arthanari H, Yang F, Pan SJ, Fan X, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E, et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature. 2008;452:604–609. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- Toda T, Shimanuki M, Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, Jones N. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 1998;12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW. Use of isogenic human cancer cells for high-throughput screening and drug discovery. Nat. Biotechnol. 2001;19:940–945. doi: 10.1038/nbt1001-940. [DOI] [PubMed] [Google Scholar]
- Umesono K, Toda T, Hayashi S, Yanagida M. Cell division cycle genes nda2 and nda3 of the fission yeast Schizosaccharomyces pombe control microtubular organization and sensitivity to anti-mitotic benzimidazole compounds. J. Mol. Biol. 1983;168:271–284. doi: 10.1016/s0022-2836(83)80018-7. [DOI] [PubMed] [Google Scholar]
- Weiss WA, Taylor SS, Shokat KM. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat. Chem. Biol. 2007;3:739–744. doi: 10.1038/nchembio1207-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SA, Allshire RC. RNAi-mediated chromatin silencing in fission yeast. Curr. Top. Microbiol. Immunol. 2008;320:157–183. doi: 10.1007/978-3-540-75157-1_8. [DOI] [PubMed] [Google Scholar]
- Wolfger H, Mamnun YM, Kuchler K. Fungal ABC proteins: pleiotropic drug resistance, stress response and cellular detoxification. Res. Microbiol. 2001;152:375–389. doi: 10.1016/s0923-2508(01)01209-8. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.