Abstract
Purpose
MicroRNAs (miRNAs), a class of noncoding small RNAs that regulate gene expression, are involved in numerous physiologic processes in normal and malignant cells. Our in vivo study measured miRNA and gene expression changes in human blood cells in response to ionizing radiation, to develop miRNA signatures that can be used as biomarkers for radiation exposure.
Methods and Materials
Blood from 8 radiotherapy patients in complete remission 1 or 2 was collected immediately before and 4 hours after total body irradiation with 1.25 Gy x-rays. Both miRNA and gene expression changes were measured by means of quantitative polymerase chain reaction and microarray hybridization, respectively. Hierarchic clustering, multidimensional scaling, class prediction, and gene ontology analysis were performed to investigate the potential of miRNAs to serve as radiation biomarkers and to elucidate their likely physiologic roles in the radiation response.
Results
The expression levels of 45 miRNAs were statistically significantly upregulated 4 hours after irradiation with 1.25 Gy x-rays, 27 of them in every patient. Nonirradiated and irradiated samples form separate clusters in hierarchic clustering and multidimensional scaling. Out of 223 differentially expressed genes, 37 were both down-regulated and predicted targets of the upregulated miRNAs. Paired and unpaired miRNA-based classifiers that we developed can predict the class membership of a sample with unknown irradiation status, with accuracies of 100% when all 45 upregulated miRNAs are included. Both miRNA control of and gene involvement in biologic processes such as hemopoiesis and the immune response are increased after irradiation, whereas metabolic processes are underrepresented among all differentially expressed genes and the genes controlled by miRNAs.
Conclusions
Exposure to ionizing radiation leads to the upregulation of the expression of a considerable proportion of the human miRNAome of peripheral blood cells. These miRNA expression signatures can be used as biomarkers of radiation exposure.
Keywords: miRNA expression, Gene expression, Peripheral blood cells, Radiotherapy patients, Biodosimetry
INTRODUCTION
MicroRNAs (miRNAs) are a class of small noncoding RNAs that have been identified as potent regulators of gene expression. Sequencing and functional analysis show that miRNAs control the expression of more than 50% of human genes by mRNA destabilization and translational repression (1). At the same time, in vivo and in vitro studies have found that miRNA control is essential for the proper execution of many processes active in normal cells, including cell metabolism, cell differentiation, and cell signaling (2). Moreover, numerous analyses of miRNA expression signatures in tumors have found that they are highly specific and can be correlated with tumor state and tumor prognosis (3, 4). miRNA dysregulation has also been identified in many other diseases (5–9).
Recently, it has been shown that ionizing radiation can induce changes in miRNA expression profiles in normal human fibroblasts (10) and immortalized cell lines (11, 12). Inasmuch as exposure to medical sources of radiation (13, 14) or to radiation emitted by an improvised radiologic or nuclear device (15) poses a health risk to the exposed population, we investigated to what degree human blood miRNA signatures can be used as biomarkers for radiation exposure.
In the present work we studied the effects of ionizing radiation on the expression levels of miRNAs and their predicted target genes in blood cells of patients exposed to ionizing radiation. We found that radiation induces significant changes in both miRNA and gene expression signatures. Importantly, despite the individual differences between the patients, we found 45 miRNAs that were statistically significantly upregulated 4 hours after treatment with 1.25 Gy x-rays, compared with pretreatment control samples. The expression of 27 of these miRNAs was induced by radiation treatment in all patients.
We used these miRNA signatures to develop class prediction classifiers for samples with unknown radiation status. We also used the miRNA target genes with downregulated expression levels after irradiation to acquire more knowledge of the potential functional involvement of miRNAs in the radiation response.
METHODS AND MATERIALS
Radiotherapy patients
Eight patients undergoing total body irradiation (TBI) at the Memorial Sloan-Kettering Cancer Center in preparation for stem cell transplantation were recruited for this study. All experiments had been approved by the institutional review boards of Memorial Sloan-Kettering Cancer Center and Columbia University Medical Center and conformed to the principles of the Declaration of Helsinki; all patients had declared their informed consent to participate in the study. The patients received 1.25 Gy x-ray TBI, delivered as a single fraction. Peripheral blood (2.5 mL) was collected before and 4 hours after irradiation.
RNA isolation
Blood from radiotherapy patients was drawn directly into PAX-gene blood RNA tubes (PreAnalytiX GmbH, Hombrechtikon, Switzerland) for RNA extraction. The PAXgene Blood RNA Kit (Qiagen Inc., Valencia, CA) was used for RNA extraction according to the manufacturer’s instructions with the exception of the dilution of buffers BR3 and BR4. These buffers were mixed with ethanol at a proportion of 1:1 before use because this allows a more efficient isolation of small RNA species. RNA concentration was measured on a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and RNA integrity was determined using the Agilent 2100 Bioanalyzer microelectrophoretic system (Agilent Technologies, Santa Clara, CA).
Quantitative reverse-transcription polymerase chain reaction for miRNA expression
To determine miRNA expression in the patients’ blood samples, 50 ng total RNA was reverse-transcribed with miRNA specific primers, and the resultant cDNA was loaded on a 384-well low-density TaqMan human miRNA expression array (array A) for quantitative real-time polymerase chain reaction (PCR) according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA).
Analysis of miRNA expression data
The real-time PCR data were imported into RQ Manager v. 1.2 (Applied Biosystems) to determine CT values. The data were then exported to Excel (Microsoft Corporation, Redmond, WA) for preprocessing, which included the removal of the miRNAs that did not have detectable expression levels (CT = 40) in ≥50% of both the treatment samples (samples obtained after radiation therapy) and the control samples (samples obtained before radiation therapy). This way of preprocessing ensures that miRNAs that are not expressed in at least half of both the control and the treatment samples are excluded from the analysis, whereas miRNAs that are consistently expressed in only one of the two conditions (either before or after radiation therapy) are retained. The preprocessed data were then imported into RealTime StatMiner v. 4.1 software (Integromics, Madrid, Spain) for normalization and statistical testing. The average CT values of two endogenous control small nucleolar RNAs included on the arrays, RNU6B and RNU48, were used for normalization of the expression data. A t test for paired samples with false discovery rate (FDR) adjustment according to the method of Benjamini and Hochberg (16) was used to determine miRNAs that had statistically significantly changed expression levels after radiation therapy. miRNAs with an FDR <0.08 were considered to be differentially expressed.
Microarray hybridization for gene expression
Whole genome microarrays (G4112A; Agilent Technologies) containing 44,794 probes were used for microarray hybridization according to the manufacturer’s instructions. Before labeling for microarray analysis, α- and β-globin mRNA was removed from the RNA samples using the GLOBIN clear kit (Ambion Inc., Austin, TX). The RNA was labeled with the One-Color Quick Amp labeling kit (Agilent Technologies). The microarray slides were scanned on an Agilent G2404B Scanner. The scanned images were extracted with Feature Extraction v. 9.1 software (Agilent Technologies) using default parameters for background correction and flagging of nonuniform outliers.
Analysis of gene expression data
Background-corrected fluorescence intensity values were imported into BRB-ArrayTools v. 3.8.0 (16), log2-transformed, and normalized to the global array median. Nonuniform outliers, features not significantly above background intensity in ≥50% of the samples, and features changing ≤1.5-fold in ≥20% of the samples were filtered out. A random-variance t test was used to determine genes differentially expressed between treatment and control samples (17). Genes with p values of <0.001 (which corresponded to an FDR of <0.035) were considered statistically significant. The microarray data were deposited in the gene expression omnibus database and can be found under accession number GSE23393 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23393).
Hierarchic clustering and multidimensional scaling
BRB-ArrayTools v. 4.1.0 software (18) was used for hierarchic clustering and multidimensional scaling (MDS) of the miRNA expression data. Both clustering methods are based on −ΔCT values. Specifically for this analysis, we standardized the average of the array means of any two pairs of arrays corresponding to blood samples from the same patient before and after irradiation to the global array mean across all samples. A Euclidean distance metric and average-linkage clustering were used for hierarchic clustering. For MDS, a Euclidean distance metric was used to compute a distance matrix, and the first three principal components of miRNA expression were used as the axes for the MDS plot.
Class prediction
BRB-ArrayTools v. 4.1.0 (18) was also used to perform miRNA-based class prediction using the ΔCT values of the differentially expressed miRNAs. The prediction methods used were compound covariate predictor, diagonal linear discriminant analysis, K-nearest neighbors (for K = 1 and 3), nearest centroid, and support vector machines. The ΔCT values were paired among samples collected from the same patient (i.e., the ability of the classifier to correctly assign each member of a pair to the correct irradiation condition, before or after radiation therapy, was tested). In addition, we constructed unpaired classifiers; these classifiers were normalized to the global array means within groups (before and after radiation therapy) and used the same prediction methods as the paired classifiers. The robustness of each predictor was assessed by the leave-one-out cross-validated misclassification error rate with 10,000 random permutations, which produced error p values of less than 0.02 for all predictors.
Gene ontology analysis
Predicted target genes of the differentially expressed miRNAs were determined in the downregulated gene pool, obtained by the gene expression analysis, using TargetScanHuman v. 5.1 (19). Targets included both phylogenetically conserved and poorly conserved targets with context scores of ≤−0.3, corresponding to log2 gene expression ratios of ≤−0.3, as determined by multivariate linear regression fitting of gene expression microarray data (19). The differentially expressed downregulated genes that were predicted targets of the differentially expressed miRNAs, and all differentially expressed genes, were uploaded to the PANTHER GO database v. 7.0 (20), which classifies genes according to GO terms, using published scientific experimental evidence. Gene set enrichment analysis was performed for the biologic process category, with all genes expressed above fluorescence background level serving as the reference set.
RESULTS
Patient characteristics, irradiation conditions, and blood collection
All patients were scheduled to receive myeloablative fractionated radiation before autologous or allogenic stem cell transplants. They were in complete remission 1 or 2 at the time of irradiation and had blood cell counts within the normal limits. No patient had been treated with radiation before. All patients had a history of leukemia or lymphoma, and chemotherapy treatment regimens had been completed more than a month before TBI (Table 1). Blood was collected directly in fixing solution before and 4 hours after radiation treatment with the first fraction of 1.25 Gy x-rays. This method of blood collection preserves the in vivo miRNA expression signatures in the collected samples.
Table 1.
Patient information
| Diagnosis | Age (y) | Sex | WBC differential (%) before beginning of radiation therapy
|
||||
|---|---|---|---|---|---|---|---|
| Neut | Lymph | Mono | Eos | Baso | |||
| MCL | 53 | F | 70.1 | 17.5 | 5.4 | 4.4 | 0.1 |
| AML | 57 | M | 56.6 | 18.6 | 7.5 | 12.9 | 0.8 |
| AML | 19 | M | 61.4 | 12.3 | 14.0 | 6.8 | 1.3 |
| ALL | 33 | M | 71.8 | 12.1 | 5.6 | 6.6 | 0.7 |
| AML | 28 | F | 71.9 | 16.8 | 5.6 | 4.2 | 0.5 |
| AML | 31 | M | 76.1 | 9.7 | 6.4 | 5.5 | 0.6 |
| AML | 54 | M | 71.2 | 12.8 | 6.1 | 7.2 | 0.5 |
| AML | 53 | M | 63.4 | 20.6 | 9.5 | 3.6 | 0.2 |
Abbreviations: WBC = white blood cell; Neut = neutrophils; Lymph = lymphocytes; Mono = monocytes; Eos = eosinophils; Baso = basophils; MCL = mantle cell lymphoma; AML = acute myelogenous leukemia; ALL = acute lymphoblastic leukemia.
miRNA expression analysis and clustering
A total of 195 miRNAs (out of 377 on the array) were expressed in the patient blood cells, using our filtering criteria. The PCR data were subjected to thorough statistical analysis, which included four steps: (1) normalization of the expression data for all arrays, (2) calculation of the differences in ΔCT values between samples collected before and after radiation treatment for all patients, (3) comparison of the ΔCT difference scores and calculation of p values by means of a paired t test, and (4) calculation of the FDR for the results from Step 3.
Normalization is a crucial issue in gene (including miRNA) expression studies, and different normalization methods can lead to different results and conclusions (21, 22). For this reason, we evaluated multiple normalization methods. These include normalization with any of the endogenous control genes supplied with the array (RNU6B, RNU44, and RNU48) and combinations thereof, normalization with any of the five most stably expressed miRNAs (those that had the lowest standard deviations between CT values across arrays: hsa-miR-331-3p, miR-494, miR-320, miR-532-3p, and miR-191) and combinations thereof, and normalization to the RNA loading amount. All tested normalization methods produced similar results. Finally, we chose the two most stably expressed endogenous controls for normalization because they are better applicable to cross-comparison analyses between different experiments. A total of 94 miRNAs with p values <0.05 were identified. The expressed miRNAs were subjected to an additional FDR analysis to account for multiple comparisons. Only miRNAs with an FDR <0.08 were considered to be differentially expressed, which means that only 8% of the miRNAs declared as differentially expressed were expected to be false positives. Moreover, we selected only miRNAs with an absolute fold change of more than two. Applying these criteria, we identified 45 differentially expressed miRNAs. These miRNAs and their predicted target genes among the statistically significantly downregulated genes (see below) are shown in Table 2. Notably, all detected differentially expressed miRNAs had increased expression levels after irradiation. We did not detect any statistically significantly downregulated miRNAs, although we found downregulated miRNAs in every patient after irradiation. Very importantly, 27 of these miRNAs were upregulated in all patients. This number increased to 39 when 1 patient with a distinctively different miRNA expression profile was removed from the analysis. Normalized expression levels of all expressed miRNAs are shown in Table E1.
Table 2.
miRNAs upregulated after radiation therapy and their predicted target genes
| miRNA name | FC | −ΔΔCT* | SEM† | Predicted miRNA target genes‡ |
|---|---|---|---|---|
| hsa-miR-143§ | 16.442 | 4.039 | 1.3488 | HLA-DOA, SPIB |
| hsa-miR-570 | 11.281 | 3.496 | 1.1493 | BACH2, CCNB1, CENPF, KIAA1407, PASK, TPD52 |
| hsa-miR-548d-3p§ | 9.842 | 3.299 | 0.9426 | BACH2, CCR6, CDKN1C, BIRC5, HIP1R, MYC, NCAPG, RRAS2 |
| hsa-miR-376a§ | 9.584 | 3.261 | 0.9302 | KIAA1407, TPD52 |
| hsa-miR-590-5p | 5.840 | 2.546 | 0.7749 | AGPAT5, BCL7A, NCAPG |
| hsa-miR-190§ | 5.166 | 2.369 | 0.5568 | AK5, BACH2 |
| hsa-miR-101§ | 4.743 | 2.246 | 0.5648 | AP3M2, CEP55, STMN1 |
| hsa-miR-598 | 3.866 | 1.951 | 0.4307 | N/A¶ |
| hsa-miR-140-5p§ | 3.606 | 1.851 | 0.5054 | AGPAT5 |
| hsa-miR-199a-3p§ | 3.492 | 1.804 | 0.4475 | N/A¶ |
| hsa-miR-29c§ | 3.476 | 1.798 | 0.5258 | BACH2, FCRLA |
| hsa-miR-21§ | 3.439 | 1.782 | 0.4347 | AGPAT5, BCL7A, NCAPG |
| hsa-miR-142-5p§ | 3.384 | 1.759 | 0.4119 | CCNB1, CCR6, NR1D2 |
| hsa-miR-301a§ | 3.232 | 1.692 | 0.4824 | APCDD1, FAM129C, JAKMIP1 |
| hsa-let-7f | 3.212 | 1.683 | 0.5007 | E2F5 |
| hsa-miR-454 | 3.202 | 1.679 | 0.5101 | APCDD1, FAM129C, JAKMIP1 |
| hsa-miR-126 | 3.142 | 1.652 | 0.5111 | N/A¶ |
| hsa-miR-106b§ | 3.135 | 1.648 | 0.5209 | APCDD1, E2F5, PDCD1 |
| hsa-miR-19a | 3.132 | 1.647 | 0.5129 | AGPAT5, CD96, FAM129C, JAKMIP1, SEMA4C |
| hsa-miR-148a | 3.092 | 1.628 | 0.4486 | HLA-DOA, STAP1 |
| hsa-miR-660§ | 3.022 | 1.595 | 0.4928 | AGPAT5 |
| hsa-miR-221 | 3.002 | 1.586 | 0.5171 | AQP3, MS4A1, MYC, PDCD1, STMN1 |
| hsa-miR-374a§ | 2.962 | 1.567 | 0.4561 | AGPAT5, AK5, APCDD1, BACH2, KIAA1407, PDCD1, TPD52 |
| hsa-miR-17§ | 2.930 | 1.551 | 0.4232 | APCDD1, E2F5, PDCD1 |
| hsa-miR-20a | 2.919 | 1.545 | 0.4861 | APCDD1, E2F5, PDCD1 |
| hsa-miR-27a§ | 2.889 | 1.531 | 0.3449 | NR1D2 |
| hsa-miR-340§ | 2.828 | 1.500 | 0.4540 | AGPAT5, NR1D2 |
| hsa-miR-106a§ | 2.794 | 1.482 | 0.4696 | APCDD1, E2F5, PDCD1 |
| hsa-miR-20b | 2.637 | 1.399 | 0.4225 | APCDD1, E2F5, PDCD1 |
| hsa-miR-502-5p§ | 2.627 | 1.394 | 0.3727 | BACH2, CCR6 |
| hsa-miR-142-3p | 2.604 | 1.381 | 0.4016 | E2F5, NR1D2 |
| hsa-miR-16 | 2.547 | 1.349 | 0.4487 | CEP55, PDCD1 |
| hsa-miR-26b | 2.522 | 1.334 | 0.3743 | AGPAT5 |
| hsa-let-7g§ | 2.518 | 1.333 | 0.3483 | E2F5 |
| hsa-miR-130a | 2.486 | 1.314 | 0.4343 | APCDD1, FAM129C, JAKMIP1 |
| hsa-miR-29a§ | 2.479 | 1.310 | 0.4175 | BACH2, FCRLA |
| hsa-miR-195 | 2.412 | 1.270 | 0.4078 | CEP55, PDCD1 |
| hsa-miR-93§ | 2.409 | 1.268 | 0.4132 | APCDD1, E2F5, PDCD1 |
| hsa-miR-185§ | 2.303 | 1.204 | 0.3238 | EHMT2, ITM2C, MS4A1, NCAPG, TPD52 |
| hsa-miR-24 | 2.228 | 1.155 | 0.3269 | HIP1R, PRSS33, SCAMP5 |
| hsa-miR-145§ | 2.211 | 1.145 | 0.2897 | BACH2 |
| hsa-miR-148b | 2.161 | 1.112 | 0.3640 | HLA-DOA, STAP1 |
| hsa-miR-103§ | 2.116 | 1.081 | 0.3657 | CDKN1C, PDCD1, TPD52 |
| hsa-miR-362-5p§ | 2.080 | 1.057 | 0.2430 | N/A¶ |
| hsa-miR-222§ | 2.050 | 1.036 | 0.3153 | AQP3, MS4A1, MYC, PDCD1, STMN1 |
Abbreviations: FC = fold change; SEM = standard error of the mean; N/A = not applicable.
−ΔΔCT equals log2(FC).
SEM of individual −ΔΔCT values = − [(ΔCT after irradiation) − (ΔCT before irradiation)].
Predicted miRNA target genes among the genes statistically significantly downregulated after radiotherapy; miRNAs belonging to the same family have the same targets.
These miRNAs were upregulated in all 8 patients.
These miRNAs do not have any predicted targets among the genes statistically significantly downregulated after radiotherapy. These miRNAs might control protein synthesis by means of translational repression, rather than mRNA destabilization and downregulation.
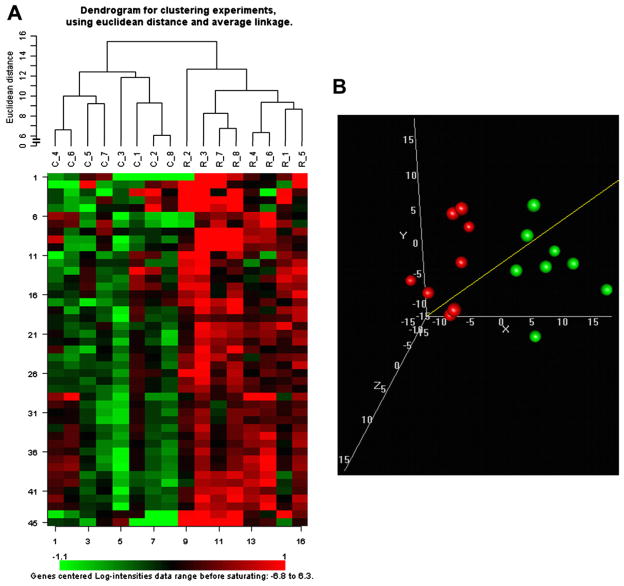
The different samples were subjected to hierarchic clustering (Fig. 1A). This clustering method grouped the patient samples according to irradiation condition, which confirms that despite individual differences between patients, radiation induces similar changes in the expression levels of specific miRNAs. We also performed an MDS analysis, which is a classification method that visualizes global differences between samples (Fig. 1B). The results show that irradiated and nonirradiated samples occupy separate subspaces in the three-dimensional graphic representation. This demonstrates again the significance of the differences of the miRNA signatures between the two conditions. Clustering results, including all expressed miRNAs, regardless of statistical significance, are shown in Fig. E1.
Fig. 1.
Significant changes in miRNA signatures after irradiation. (A) Hierarchic clustering. A heat map shows miRNA expression levels for all samples (columns) and miRNAs (rows). All patient control samples (labels starting with C) form a separate cluster from the samples obtained after radiation therapy (labels starting with R). (B) Multidimensional scaling. Control samples (shown in green) and samples that were collected after total body irradiation (displayed in red) form two separate clusters.
Gene expression analysis
Out of the 10,009 features that were expressed above background intensity and that fulfilled the filtering criteria, as described in Methods and Materials, 8,763 were unique. Of these, 275 had a p value <0.001 comparing preirradiation expression levels with expression 4 hours after TBI; this corresponds to an FDR of <0.035, calculated according to the method of Benjamini and Hochberg (16) (Table E2). These genes were considered to be differentially expressed. Out of the differentially expressed genes, 223 could be assigned a HUGO gene symbol (http://www.genenames.org) (23).
Class prediction
The miRNA ΔCT values were used to build class prediction classifiers to test to what degree class prediction is able to indicate the irradiation status of patients’ blood samples based on their miRNA expression profiles. The leave-one-out cross-validation method was used to test the robustness of the classifiers.
Two types of classifiers were constructed. One contained all 45 differentially expressed miRNAs. For the other classifier, only the six differentially expressed miRNAs that showed an expression fold change of >5 after irradiation (hsa-miR-143, 570, 548d-3p, 376a, 590-5p, and 190) were used.
Both paired and unpaired classifiers were constructed. A pair consisted of the pre- and postexposure samples from the same patient. Both types of paired classifiers performed with an accuracy of 100% in all classification methods used (i.e., they correctly determined the irradiation status before or after radiation therapy of all paired samples).
The unpaired classifiers were normalized to the global array means within groups (before and after radiation therapy) to improve the performance of the classifiers and minimize the effect of outlier samples. The classifiers containing all 45 upregulated miRNAs performed with an accuracy of 100% in all classification methods used. However, the classifiers built from the six miRNAs with the highest fold changes had accuracies between 81% and 88%, depending on the classification method used.
We did not build gene expression–based classifiers in this study because a larger study submitted for publication describes the development of such classifiers (24).
Prediction of miRNA target genes
A two-step procedure was used for miRNA target determination: (1) identification of the potential miRNA target genes by gene expression analysis and (2) software-assisted identification of the miRNA targets in the pool of downregulated genes. The 98 genes found to be statistically significantly downregulated in our gene expression analysis were selected as potential miRNA targets. To determine the targets of the 45 upregulated miRNAs in this gene pool, we used TargetScanHuman v. 5.1 software (2, 19), which uses sequence- and motif-based algorithms for miRNA target prediction. Only high-probability target genes with context scores of ≤−0.3 and at least one conserved or poorly conserved miRNA target site, as defined by phylogenetic tree analysis, were selected. According to this analysis, 37 downregulated genes were predicted targets of the differentially expressed miRNAs (Tables 2 and 3). These genes were used for the subsequent gene ontology (GO) analysis.
Table 3.
Genes that are both downregulated and predicted targets of upregulated miRNAs
| Gene symbol | Expression ratio | SEM |
|---|---|---|
| FCRLA | 0.1118 | 0.03063 |
| MS4A1 | 0.1759 | 0.04833 |
| E2F5 | 0.2886 | 0.01194 |
| BCL7A | 0.3363 | 0.05564 |
| FAM129C | 0.3515 | 0.03859 |
| SPIB | 0.3988 | 0.09082 |
| STAP1 | 0.4022 | 0.05387 |
| TPD52 | 0.4524 | 0.03860 |
| KIAA1407 | 0.4601 | 0.04552 |
| PRSS33 | 0.4638 | 0.07454 |
| HIP1R | 0.4793 | 0.08530 |
| SCAMP5 | 0.4805 | 0.02610 |
| BIRC5 | 0.4821 | 0.06725 |
| APCDD1 | 0.4958 | 0.06256 |
| CDKN1C | 0.5030 | 0.05420 |
| JAKMIP1 | 0.5052 | 0.07040 |
| CCR6 | 0.5126 | 0.04589 |
| ITM2C | 0.5150 | 0.05083 |
| BACH2 | 0.5172 | 0.04652 |
| CEP55 | 0.5241 | 0.05009 |
| AK5 | 0.5487 | 0.06124 |
| HLA-DOA | 0.5557 | 0.04204 |
| CCNB1 | 0.5592 | 0.06380 |
| AQP3 | 0.5673 | 0.05703 |
| NR1D2 | 0.5699 | 0.03268 |
| STMN1 | 0.5718 | 0.05456 |
| AGPAT5 | 0.5719 | 0.06280 |
| MYC | 0.5739 | 0.05366 |
| AP3M2 | 0.5757 | 0.05129 |
| SEMA4C | 0.5840 | 0.05398 |
| CENPF | 0.5893 | 0.04902 |
| EHMT2 | 0.5959 | 0.05342 |
| NCAPG | 0.5977 | 0.05996 |
| PASK | 0.6040 | 0.04210 |
| CD96 | 0.6052 | 0.06322 |
| RRAS2 | 0.6109 | 0.06773 |
| PDCD1 | 0.6586 | 0.04782 |
Abbreviation: SEM = standard error of the mean.
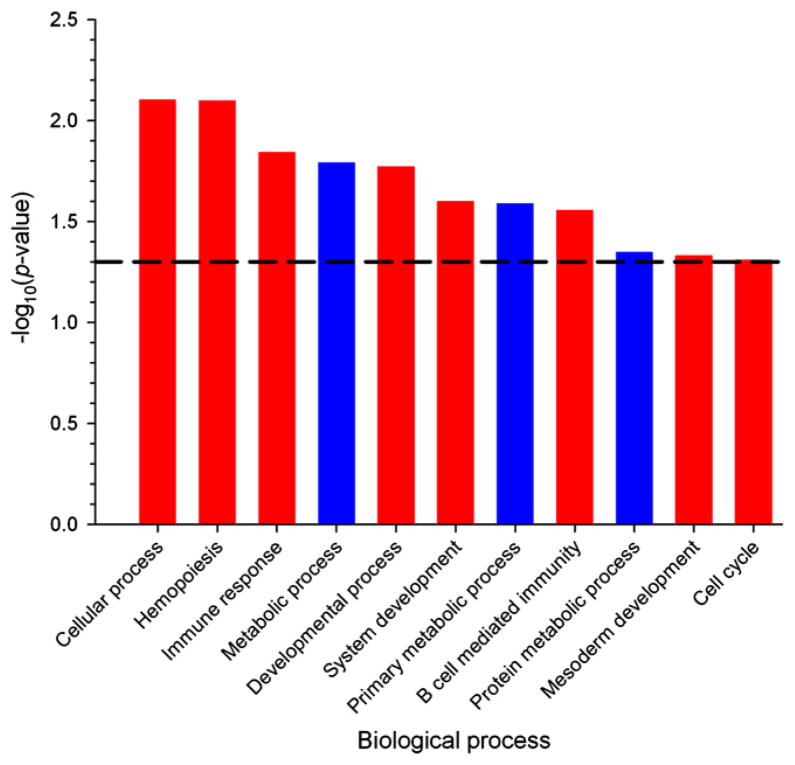
Gene ontology analysis of miRNA and gene expression
The goal of the GO analysis is to predict the effect of the upregulated miRNAs on cell functions. The downregulated genes predicted to be targeted by the radiation-induced miRNAs, and all differentially expressed genes, were uploaded to the PANTHER v. 7.0 GO database (20), and gene-set enrichment analysis was performed in the biologic process category. An overview of the biologic processes overrepresented or underrepresented in the pools of the differentially expressed genes and the genes targeted by miRNAs, compared with all genes expressed above fluorescence background level, is given in Table 4. Our results predict that miRNAs could control about 30% of the biologic processes that are regulated by the products of the radiation-induced genes. Specifically, the analysis suggests that TBI results in an increased miRNA control of genes involved in hemopoiesis and the immune response, whereas metabolic processes are underrepresented among the genes controlled by miRNAs (Fig. 2). A GO analysis of the genes differentially expressed upon radiation exposure showed that processes such as the immune response, signal transduction, the response to stress, the cell cycle, apoptosis, and hemopoiesis are enriched in these genes, but, as for the miRNA target analysis, the differentially expressed genes are underrepresented in metabolic processes, compared with the reference gene set (Fig. E2).
Table 4.
Number of biologic processes over- or underrepresented in relevant gene sets*
| Gene set | Overrepresented processes† | Underrepresented processes† |
|---|---|---|
| Differentially expressed genes‡ | 33 | 3 |
| Putatively targeted genes§‡ | 8 | 3 |
Gene set enrichment analysis was performed using the biologic process category of the PANTHER database (23).
Out of a total of 131 processes in the biologic process category.
All genes expressed above fluorescence background level were used as the reference set.
Genes that were both downregulated and predicted by Target-Scan (22) to be miRNA targets.
Fig. 2.
Regulation of several biologic processes by miRNAs after total body irradiation. Gene ontology analysis of the predicted miRNA target genes revealed the biologic processes in which the predicted target genes significantly responding to radiation are overrepresented (red columns) or underrepresented (blue columns). Processes above the horizontal dashed line show statistically significant over- or underrepresentation at a p value of <0.05.
DISCUSSION
Recently, high-throughput assays have been successfully used for radiation biodosimetry. The power of gene expression profiles to discriminate irradiated from nonirradiated samples and low-dose exposure from high-dose exposure has been demonstrated in ex vivo– and in vivo–irradiated blood samples (25–27). In parallel, miRNA-related studies have shown that miRNA expression signatures define cell functions and cell types with high precision and in some cases better than gene expression. miRNA expression profiles correctly classified poorly differentiated tumors (28), different types of breast cancers (29), lung cancers (30), and many other diseases, including those not related to cancer (5). Moreover, ionizing radiation changes the miRNA expression profiles of normal human fibroblasts (10) and immortalized cell lines (11, 12). However, to date no study has characterized miRNA expression signatures in humans irradiated in vivo as far as we are aware. Given that blood cells are among the most radiation-sensitive cells in the human body, we investigated the potential of radiation-induced miRNA expression profiles in peripheral blood cells to provide high-resolution biomarkers for radiation exposure. Such biomarkers could be used to monitor the presence and duration of radiation effects in individuals exposed to ionizing radiation in therapeutic or diagnostic settings.
We measured miRNA expression profiles in blood cells from 8 patients in complete remission 1 or 2. We found that 45 miRNAs (23% of all miRNAs detected in blood cells) showed statistically significant changes in expression levels 4 hours after irradiation with a dose of 1.25 Gy x-rays when compared with preirradiation control samples. This result indicates an extensive shift in miRNA expression, with important consequences for cell functions. Notably, all differentially expressed miRNAs showed increased expression levels after irradiation. We did detect underexpressed miRNAs in every patient, but they were not statistically significantly downregulated across all patients. The predominance of upregulated miRNAs could be a characteristic of the early radiation response in blood cells. Importantly, 27 of the 45 radiation-induced miRNAs were upregulated in every patient. When 1 patient who had a markedly different miRNA expression profile from those of the other patients was excluded from the analysis, the number of miRNAs upregulated across all patients increased to 39. The high consistency of miRNA expression changes in human blood cells across individuals emphasizes the great value of miRNA signatures as radiation biomarkers. Although we identified a large number of radiation-induced miRNAs, a limitation of our study is that we do not have results for different doses and time points after irradiation because of the difficulties associated with obtaining human samples. Another limitation is the relatively small number of patients we studied. This limits the statistical power of our analysis and might explain why we did not detect any statistically significantly downregulated miRNAs. It also limits the data available for developing class prediction classifiers. Moreover, the microarray gene expression data were not replicated; however, a larger radiotherapy-induced gene expression study, based on a substantially larger patient population, has been submitted for publication by our group (24).
A major concern for any comparative analysis of data derived from total blood cell populations before and after irradiation is that blood cell subsets may change after irradiation because of the radiation sensitivity of white blood cells. According to estimates of the effect of irradiation on blood cell counts, the lymphocyte depletion rate constant is very low for doses of up to 1 Gy, and the decrease in the number of white blood cells has been estimated to be 6% 12 hours after irradiation (31, 32). On the basis of these data, we assumed that a dose of 1.25 Gy would not induce a significant change in the numbers of blood cells for the 4-hour time point that we used. This implies that the miRNA and gene expression changes we observed were due to functional changes in the irradiated cells and were not the consequence of cell subset depletion. Although workable biomarkers can be developed for class prediction even when blood cell subsets change in response to irradiation, the postulation of subset constancy substantially facilitates the interpretation of the results of the GO analysis.
We attempted to cross-validate our results with other studies. We did not find any other published work describing radiation-induced miRNA expression changes in blood cells for comparison. A comparison of our findings with radiation-induced miRNA expression profiles in other cell types shows very little overlap, probably because of cell type differences (10–12). Additionally, far fewer miRNAs were induced by ionizing radiation in non–blood cells, which could be explained by differences in radiation sensitivity or the molecular milieu of transformed cells, which generally express lower numbers of miRNAs (28).
Because of the high number of differentially expressed miRNAs identified in our study, we developed radiation classifiers that can predict the irradiation status of blood samples. Both paired and unpaired classifiers constructed from all differentially expressed miRNAs performed with an accuracy of 100%. The robust performance of the unpaired classifiers is especially important because their functioning does not require pre-exposure control samples. These results encourage the pursuit of additional studies that further investigate the potential of miRNAs to serve as reliable biomarkers of radiation exposure.
We estimated the effects of the radiation-induced miR-NAs on cell processes by combining gene expression, software-assisted miRNA target analysis, and GO data analysis. A strength of our study is that we determined radiation-induced gene expression changes in addition to miRNA profiles. Consequently, we possess information on physiologically downregulated genes, and we do not solely rely on bioinformatics-based predictions of miRNA targets. Overall, 38% of the downregulated genes were predicted targets of the upregulated miRNAs, using our context-score cutoff criterion. It should be noted that only downregulated, not translationally repressed, mRNAs are included in this number because the detection of translationally repressed mRNAs requires different methods. Our GO analysis showed that miRNA target genes are involved in hemopoiesis, the immune response, B cell–mediated immunity, and other processes. Interestingly, miRNA control of genes involved in metabolic processes is decreased, most likely because of underrepresentation of genes involved in metabolic processes among the differentially expressed genes.
In summary, our study shows that radiation induces a robust miRNA expression response in peripheral blood cells. The results of our study expand our knowledge of the processes activated in blood cells after radiation exposure and can be used as the foundation for the further development of reliable radiation biomarkers.
Supplementary Material
Acknowledgments
Supported in part by NASA, Grant # NNX07AT41G, and by the Center for High-Throughput Minimally-Invasive Radiation Biodosimetry, National Institute of Allergy and Infectious Diseases, Grant # U19 AI067773.
Footnotes
Supplementary material for this article can be found at www.red-journal.org.
Conflict of interest: none.
References
- 1.Shkumatava A, Stark A, Sive H, et al. Coherent but overlapping expression of microRNAs and their targets during vertebrate development. Genes Dev. 2009;23:466–481. doi: 10.1101/gad.1745709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 5.Taft RJ, Pang KC, Mercer TR, et al. Non-coding RNAs: Regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 6.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 7.Nana-Sinkam SP, Hunter MG, Nuovo GJ, et al. Integrating the MicroRNome into the study of lung disease. Am J Respir Crit Care Med. 2009;179:4–10. doi: 10.1164/rccm.200807-1042PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerr TA, Davidson NO. Therapeutic RNA manipulation in liver disease. Hepatology. 2010;51:1055–1061. doi: 10.1002/hep.23344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Simone NL, Soule BP, Ly D, et al. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One. 2009;4:e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin S, Cha HJ, Lee EM, et al. Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. Int J Oncol. 2009;35:81–86. [PubMed] [Google Scholar]
- 12.Chaudhry MA. Real-time PCR analysis of micro-RNA expression in ionizing radiation-treated cells. Cancer Biother Radiopharm. 2009;24:49–56. doi: 10.1089/cbr.2008.0513. [DOI] [PubMed] [Google Scholar]
- 13.Becker GJ, Bosma J, Hendee W. Radiation exposure from medical imaging procedures. N Engl J Med. 2009;361:2289–2290. author reply 2291–2282. [PubMed] [Google Scholar]
- 14.Hall EJ, Brenner DJ. Cancer risks from diagnostic radiology. Br J Radiol. 2008;81:362–378. doi: 10.1259/bjr/01948454. [DOI] [PubMed] [Google Scholar]
- 15.Weinstock DM, Case C, Jr, Bader JL, et al. Radiologic and nuclear events: Contingency planning for hematologists/oncologists. Blood. 2008;111:5440–5445. doi: 10.1182/blood-2008-01-134817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 17.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 18.Simon R, Lam A, Li MC, et al. Analysis of gene expression data using BRB-array tools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 19.Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 22.Pradervand S, Weber J, Thomas J, et al. Impact of normalization on miRNA microarray expression profiling. RNA. 2009;15:493–501. doi: 10.1261/rna.1295509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruford EA, Lush MJ, Wright MW, et al. The HGNC database in 2008: A resource for the human genome. Nucleic Acids Res. 2008;36:D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul S, Barker CA, Turner HC, et al. Prediction of In Vivo Radiation Dose Status in Radiotherapy Patients using Ex Vivo and In Vivo Gene Expression Signatures. Radiat Res. 2011 doi: 10.1667/RR2420.1. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meadows SK, Dressman HK, Daher P, et al. Diagnosis of partial body radiation exposure in mice using peripheral blood gene expression profiles. PLoS One. 2010;5:e11535. doi: 10.1371/journal.pone.0011535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys. 2008;71:1236–1244. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dressman HK, Muramoto GG, Chao NJ, et al. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med. 2007;4:e106. doi: 10.1371/journal.pmed.0040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 29.Khoshnaw SM, Green AR, Powe DG, et al. MicroRNA involvement in the pathogenesis and management of breast cancer. J Clin Pathol. 2009;62:422–428. doi: 10.1136/jcp.2008.060681. [DOI] [PubMed] [Google Scholar]
- 30.Wu X, Piper-Hunter MG, Crawford M, et al. MicroRNAs in the pathogenesis of lung cancer. J Thorac Oncol. 2009;4:1028–1034. doi: 10.1097/JTO.0b013e3181a99c77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waselenko JK, MacVittie TJ, Blakely WF, et al. Medical management of the acute radiation syndrome: Recommendations of the Strategic National Stockpile Radiation Working Group. Ann Intern Med. 2004;140:1037–1051. doi: 10.7326/0003-4819-140-12-200406150-00015. [DOI] [PubMed] [Google Scholar]
- 32.Dainiak N. Hematologic consequences of exposure to ionizing radiation. Exp Hematol. 2002;30:513–528. doi: 10.1016/s0301-472x(02)00802-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.