Abstract
Florigen, a protein encoded by the FLOWERING LOCUS T (FT) in Arabidopsis and Heading date 3a (Hd3a) in rice, is the universal flowering hormone in plants. Florigen is transported from leaves to the shoot apical meristem and initiates floral evocation. In shoot apical cells, conserved cytoplasmic 14-3-3 proteins act as florigen receptors. A hexameric florigen activation complex (FAC) composed of Hd3a, 14-3-3 proteins, and OsFD1, a transcription factor, activates OsMADS15, a rice homolog of Arabidopsis APETALA1, leading to flowering. Because FD is a key component of the FAC, we characterized the FD gene family and their functions. Phylogenetic analysis of FD genes indicated that this family is divided into two groups: (i) canonical FD genes that are conserved among eudicots and non-Poaceae monocots; and (ii) Poaceae-specific FD genes that are organized into three subgroups: Poaceae FD1, FD2 and FD3. The Poaceae FD1 group shares a small sequence motif, T(A/V)LSLNS, with FDs of eudicots and non-Poaceae monocots. Overexpression of OsFD2, a member of the Poaceae FD2 group, produced smaller leaves with shorter plastochrons, suggesting that OsFD2 controls leaf development. In vivo subcellular localization of Hd3a, 14-3-3 and OsFD2 suggested that in contrast to OsFD1, OsFD2 is restricted to the cytoplasm through its interaction with the cytoplasmic 14-3-3 proteins, and interaction of Hd3a with 14-3-3 facilitates nuclear translocation of the FAC containing OsFD2. These results suggest that FD function has diverged between OsFD1 and OsFD2, but formation of a FAC is essential for their function.
Keywords: FD, Florigen activation complex (FAC), Flowering, Hd3a, Plant transcription factor, Rice
Introduction
Florigen is a mobile flowering signal in plants, produced in leaves, and is transported through phloem tissue to the shoot apex where it initiates flowering (Zeevaart 2008, Matsoukas et al. 2012). The molecular nature of florigen has been revealed to be a protein encoded by Heading date 3a (Hd3a) in rice and its ortholog FLOWERING LOCUS T (FT) in Arabidopsis, both of which have a globular structure with a molecular mass of 22 kDa (Tsuji et al. 2011, Andres and Coupland 2012). Hd3a/FT protein moves through leaf phloem tissues, reaches the shoot apical meristem and triggers expression of floral meristem identity genes (Corbesier et al. 2007, Tamaki et al. 2007, Notaguchi et al. 2008, Yoo et al. 2012). Rice has two florigen genes, Hd3a and RFT1, and expression of both genes is controlled by the complex genetic network that integrates light signaling and circadian clock information (Itoh et al. 2010, Ishikawa et al. 2011, Matsubara et al. 2012, Saito et al. 2012) When expression of both genes is knocked down, the plant does not flower, suggesting that florigen is essential for flowering in rice (Komiya et al. 2008, Komiya et al. 2009). More recently, the molecular mechanism of florigen function in shoot apical cells was revealed in rice. Hd3a florigen interacts with 14-3-3 proteins in the cytoplasm and forms a ternary complex with OsFD1 in the nucleus. The ternary complex is known as the florigen activation complex (FAC), which activates OsMADS15, a MADS-domain transcription factor that regulates flowering (Taoka et al. 2011, Kobayashi et al. 2012).
FD is a basic leucine zipper (bZIP)-containing transcription factor, first identified in Arabidopsis, and its loss-of-function mutants are late flowering (Abe et al. 2005, Wigge et al. 2005). The C-terminus of FD contains a short motif targeted by calcium-dependent protein kinases (CDPKs), and an alanine substitution of a serine/threonine residue within this motif disrupts FD function (Abe et al. 2005). This phosphorylation motif is required for interaction of 14-3-3 proteins with FD in rice, supporting the importance of the phosphorylation-dependent 14-3-3 protein interaction for FD function. The crystal structure of the FAC suggests that FD acts to tether the protein complex on the target promoter DNA (Taoka et al. 2011). The FD function seems to be conserved among higher plants. Maize DELAYED FLOWERING1 (DLF1) and wheat FDL2/FDL6, which are homologs of Arabidopsis and rice FDs, can interact with maize ZCN8 and wheat TaFT florigen proteins, respectively (Muszynski et al. 2006, Li and Dubcovsky 2008, Meng et al. 2011). Interestingly, all these FD homologs share 14-3-3 protein interaction motifs at their C-terminus, suggesting that participation of FD in the FAC is conserved.
The FAC model provides insight into the conserved nature of florigen function because the three proteins comprising the FAC are conserved among seed plants (Ferl et al. 2002, Karlgren et al. 2011, Taoka et al. 2011). This model also raises the possibility that transcription factors such as FD are exchangeable in the FAC because the transcription factors containing the 14-3-3 protein interaction motif can potentially interact with 14-3-3 proteins through a canonical mode of 14-3-3–phosphoserine interaction. Thus, an interesting hypothesis is that FAC function can be modulated depending on the transcription factor bound to the 14-3-3 protein in the FAC. FD homologs are potential candidates for testing this hypothesis; however, detailed characterization of FD homologs is limited for all species (Hanano and Goto 2011). Here, we describe the molecular analysis of the rice FD genes to answer three questions arising from the FAC model. (i) Are FD genes conserved among plants? (ii) Do FD homologs form FACs? (iii) Does the function of the FAC change depending on the FDs incorporated in the FAC? Our results suggest that OsFD2, a rice FD homolog, potentially forms a FAC and regulates leaf development. These results suggest the functional diversification of OsFD2 compared with the role of OsFD1 in flowering, and supports the hypothesis that FAC activity can be modified by FDs.
Results
Phylogenetic analysis of the FD gene family
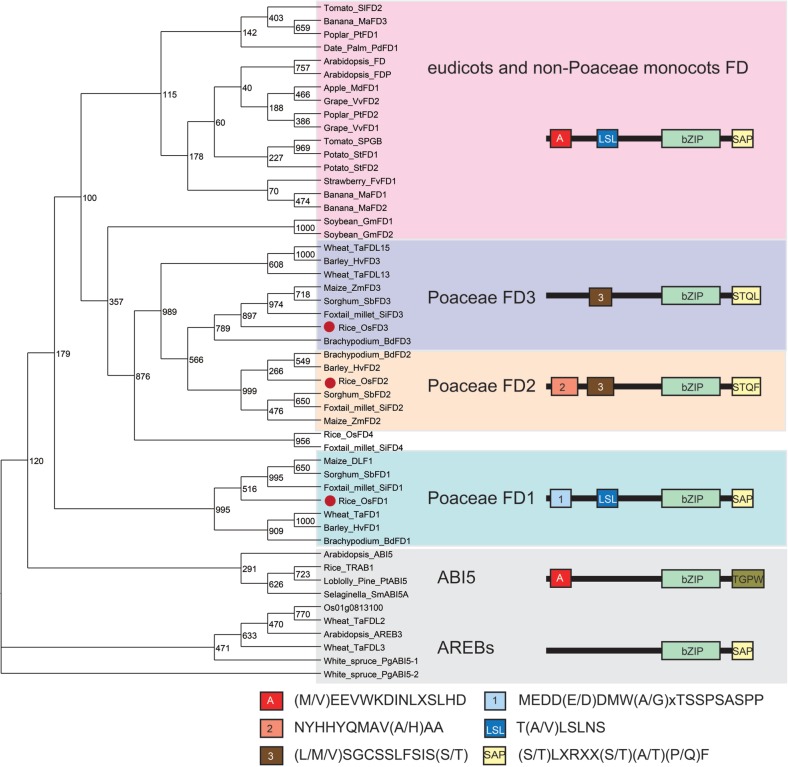
We identified five new members of the FD gene family in rice, designated OsFD2–OsFD6, whose protein products share homology with the bZIP motif and C-terminal phosphorylation motif (SAP motif) of OsFD1 and other known FD proteins (Supplementary Table S1). The OsFD2 gene (Os06g0720900) is triplicated in the genome, giving rise to OsFD5 (Os06g0724000) and OsFD6 (Os06g0195000), the latter encoding a mutant bZIP protein that has a truncated C-terminal region and is thus inferred to be a pseudogene.
To analyze the phylogenetic relationship of the FD gene family, we first identified 47 FD genes from diverse plant species by searching public databases of genomic sequences and expressed sequence tags (ESTs) (Supplementary Table S1, Supplementary text). Conserved amino acid motifs and their combinations in the predicted proteins were identified from these sequences using the SALAD database (Mihara et al. 2010) and by visual inspection. Finally we created a phylogenetic tree using the region spanning the bZIP motif to the C-terminal SAP motif. The structure of this phylogenetic tree matched the classification of proteins according to the combinations of amino acid motifs (Fig. 1). From this phylogenetic analysis, we found three interesting features about the FD gene family. First, eudicot and non-Poaceae monocots share FD genes that contain a conserved motif arrangement, whereas genes encoding this type of FD are absent from Poaceae genomes. Eudicot FDs share a conserved motif arrangement comprised of motif A [(M/V)EEVWKDINLSSLHD], LSL [T(A/V)LSLN], bZIP and SAP [(S/T)LXRX(S/T)(A/T)(P/Q)F] (Fig. 1; Supplementary Figs. S1–S3). The name of the SAP motif follows according to the definition from our previous characterization of OsFD1 (Taoka et al. 2011). This group of FD genes is found in two monocot species, banana (Musa acuminate) and date palm (Phoenix dactylifera), whose genome sequences were recently reported, suggesting conservation of FD (Paterson et al. 2009, Wei et al. 2009, Al-Dous et al. 2011, D’Hont et al. 2012). In contrast, this type of FD was not identified from Poaceae genomes. Genomic sequences of rice, maize, brachypodium and foxtail millet, and EST databases of barley and wheat do not contain this type of FD sequence (Tanaka et al. 2008, Paterson et al. 2009, Schnable et al. 2009, Wei et al. 2009, International Brachypodium Initiative 2010, Zhang et al. 2012). Secondly, three groups of Poaceae-specific FD genes were identified. The three groups were designated as Poaceae FD1, FD2 and FD3, respectively, according to their phylogenetic relationships and motif arrangements. The Poaceae FD1 group includes rice OsFD1 and maize DLF1, both of which were shown to participate in the activation of AP1/FUL homologs and promotion of flowering (Muszynski et al. 2006, Taoka et al. 2011). The characteristic feature of the motif combination in this group is the presence of motif 1 [MEDD(E/D)DMW(A/G)XTSSPSASPP], LSL, bZIP and SAP. The Poaceae FD2 group contains motif 2 [NYHHYQMAV(A/H)AA] and motif 3 [(L/M/V)SGCSSLFSIS(S/T)] with bZIP and a partially modified SAP motif. Motif 3 and SAP are partially shared with Poaceae FD3, suggesting that these two groups had the same evolutionary origin. Thirdly, the Poaceae FD1 group and eudicot/non-Poaceae monocot FDs share the LSL motif at the N-terminus of the sequences, although neither group shows strong similarity in the entire arrangement of motifs.
Fig. 1.
Phylogenetic tree of predicted FD proteins and arrangements of amino acid motifs in each FD group. The phylogenetic tree was constructed with Neighbor–Joining methods using regions from bZIP to the C-terminus of the deduced amino acid sequences of FDs. AREBs/ABI5s are included as an outgroup. Red dots beside the protein name denote rice FD1, FD2 or FD3. The motif arrangement of FD proteins is schematically presented, with boxes and lines representing the conserved motifs identified in this study and other protein regions, respectively. The consensus amino acid sequences are presented below the phylogenetic tree.
Organ-specific expression of rice FDs
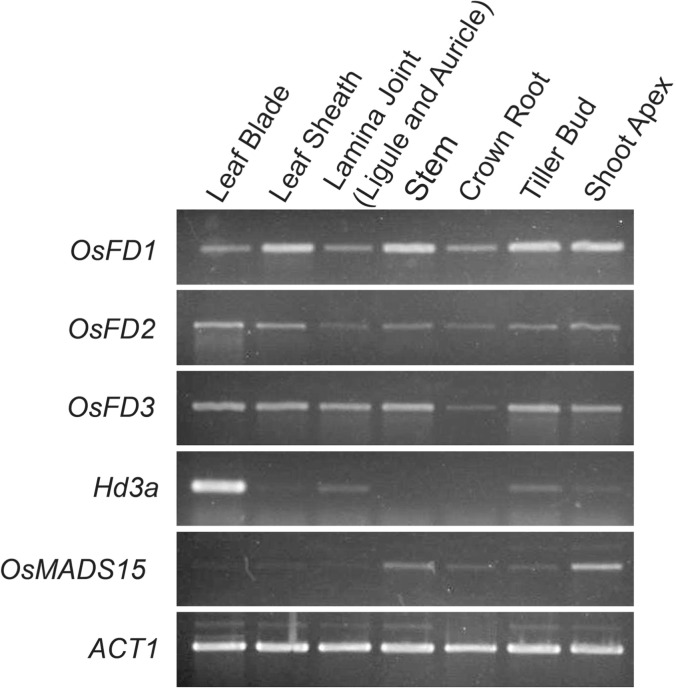
The accumulation of OsFD1, OsFD2 and OsFD3 mRNAs in various organs was examined by reverse transcription–PCR (RT–PCR) (Fig. 2). All three transcripts accumulated in all of the organs tested: leaf blades, leaf sheaths, lamina joint regions connecting the leaf blade and leaf sheath, stems of vegetative phase plants, crown roots, tiller buds and shoot apices of vegetative phase plants. Lamina joints included ligules and auricles. Hd3a was specifically expressed in the leaf blade, with faint expression in lamina joints, tiller buds and shoot apices. OsMADS15 was weakly expressed in shoot apices and stems.
Fig. 2.
Expression of rice FD genes in various organs. Rice FD gene family members are expressed in all organs tested, whereas Hd3a and OsMADS15 are expressed specifically in leaf blades and shoot apices, respectively. The ACT1 gene was used as the control for cDNA amplification.
Effect of OsFD2 overexpression on plant development
To study the function of OsFD2 in plants, we generated transgenic plants overexpressing OsFD2 or the OsFD2 S164A construct, in which an alanine residue was introduced into the putative phosphorylation site within the SAP motif to disrupt interaction with 14-3-3 proteins (Supplementary Fig. S4A; see Figs. 5 and 6) under the constitutive ubiquitin promoter (pUbq). In the vegetative stage, no obvious phenotypes were observed among three genotypes (Fig. 3A–C). Flowering time was not affected in these transgenic plants (Fig. 3D–F, J). In the reproductive stage, about 10% of the panicles that emerged from pUbq:OsFD2 plants showed a dense panicle phenotype, and S164A mutation suppressed this phenotype (Fig. 3G–I). These results suggest that OsFD2 can delay the transition from inflorescence branch meristem to floral (or spikelet) meristem in the panicle branch, because the number of lateral organs in the inflorescence branch is determined by the timing of the transition from the inflorescence branch meristem to the spikelet meristem. The inflorescence meristem develops lateral spikelet meristems or branch meristems until the inflorescence meristem itself turned into the spikelet meristem (Nakagawa et al. 2002). The delay in this transition allows the longer period of lateral meristem development, and in consequence the more plentiful spikelets or secondary branches, to produce the dense panicle phenotype (Nakagawa et al. 2002).
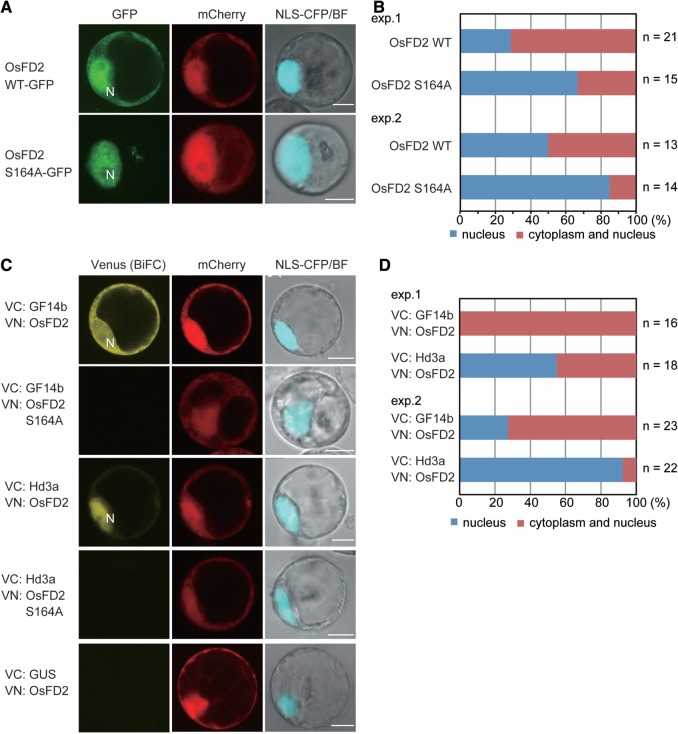
Fig. 5.
Subcellular localization of OsFD2 and interaction among OsFD2, GF14b and Hd3a in rice cells. (A) Confocal images of cells expressing GFP–OsFD2 and GFP–OsFD2 S164A. Nuclear marker proteins (NLS–CFP) and mCherry protein were co-expressed. (B) Quantification of the subcellular localization of GFP–OsFD2 and GFP–OsFD2 S164A. (C) BiFC assays showing interactions of GF14b–OsFD2, GF14b–OsFD2 S164A, Hd3a–OsFD2 and Hd3a–OsFD2 S164A. Venus fluorescence in cells expressing the indicated proteins tagged with the N- or C-terminal halves of Venus is shown. Nuclear marker proteins (NLS–CFP and/or mCherry protein) were co-expressed. (D) Quantification of subcellular localization of the BiFC signal arose from interactions of GF14b–OsFD2 and Hd3a–OsFD2.
Fig. 6.
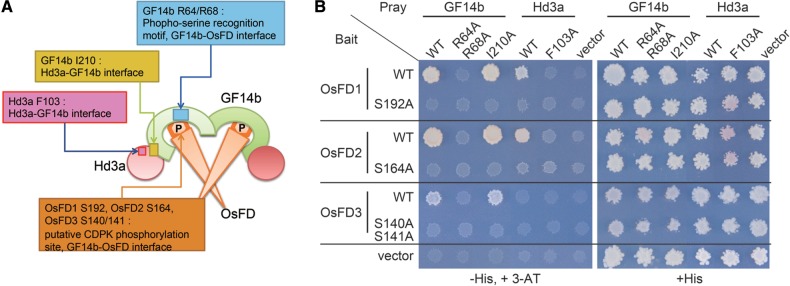
Yeast two-hybrid assays. (A) A model for the FAC composed of Hd3a–GF14–OsFD, highlighting the locations of residues critical for protein–protein interactions. P represents phosphorylation at the SAP motif of OsFDs. (B) Yeast two-hybrid assay of interactions between OsFD proteins and GF14b or Hd3a. The effects of alanine substitutions for the amino acids essential for the formation of FAC were examined.
Fig. 3.

The effect of overexpressing OsFD2 on flowering and inflorescence development. (A–C) Gross morphology of WT (A), pUbq:OsFD2 (B) and pUbq:OsFD2 S164A (C) plants in the vegetative stage, showing no apparent difference among the three genotypes. (D–F) Gross morphology of WT (D), pUbq:OsFD2 (E) and pUbq:OsFD2 S164A (F) plants in the reproductive stage. pUbq:OsFD2 develops abnormal branch shoots with small leaves (E), whereas pUbq:OsFD2 S164A shows normal development (F). (G–I) Panicles of WT (G) and pUbq:OsFD2 (H). pUbq:OsFD2 produces more spikelets to form a dense panicle architecture (H). Scale bars are 5 cm in (A, B, C, G and H) and 10 cm in (D, E and F). (J) Flowering times of transgenic plants of the T0 generation under SD conditions. WT indicates plants regenerated from non-transformed calli. Statistical significance compared with the WT was calculated using Student’s t-test.
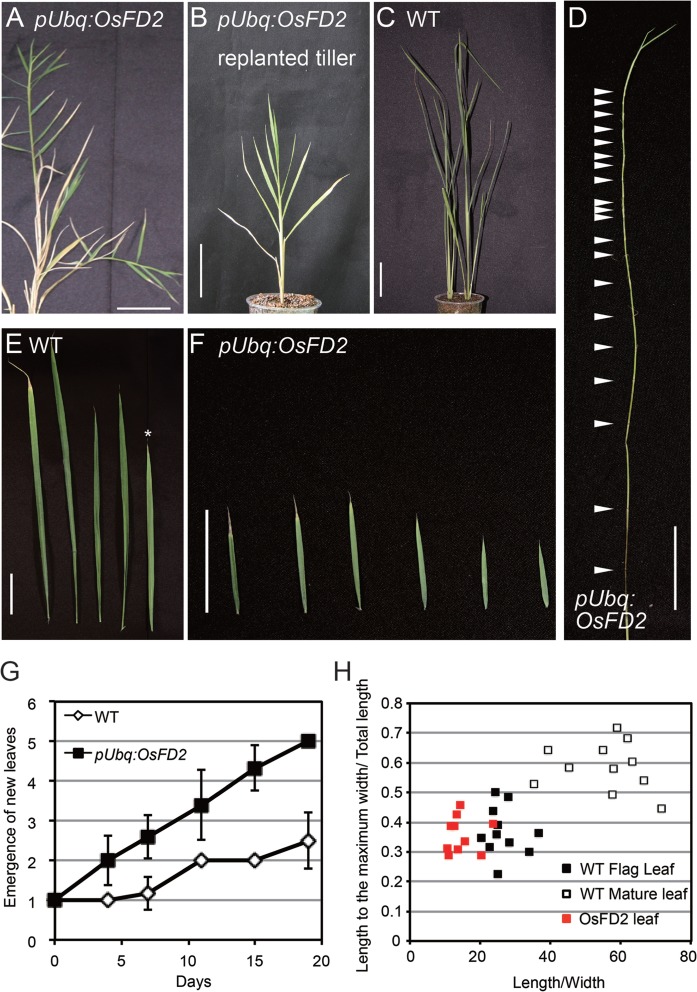
At the reproductive stage, pUbq:OsFD2 plants showed a striking phenotype in the leaves of branch shoots (Fig. 4A–D; Supplementary Fig. S4B, C). pUbq:OsFD2 plants elongated abnormal branch shoots from several branch buds that are dormant in the wild-type (WT) plants (Fig. 4A; Supplementary Fig. S4B, C). The majority of these abnormal shoots never developed panicles and iterated leaf development, occasionally producing panicles similar to the main culm (Fig. 3H). These shoots can be detached, replanted in soil and grown for several weeks to develop leaves or, occasionally, panicles (Fig. 4B; Supplementary Fig. S4B, C). The abnormal shoots iterated the development of small leaves and short internodes (Fig. 4D–F). The rate of leaf initiation (plastochron) is shortened in pUbq:OsFD2 plants compared with the WT shoots (Fig. 4G), contributing to the generation of many phytomers that can elongate at the internodes.
Fig. 4.
The effect of overexpressing OsFD2 on leaf development. (A) Branch shoots developing small leaves grew out from nodes on the elongating stem internodes in pUbq:OsFD2 plants at the late reproductive stage. (B) A branch shoot detached from a pUbq:OsFD2 plant replanted in soil. (C) WT plants transplanted at the same timing of replantation of the pUbq:OsFD2 branch shoot in (B). (D) Stem of a growing branch shoot from pUbq:OsFD2. Reiteration of leaf development and internode elongation produced numerous nodes (arrowheads). (E and F) Leaves of a WT plant (E) and pUbq:OsFD2 branch shoot (F). The asterisk indicates a flag leaf, the last leaf that develops before flowering. Scale bars in (A–F) = 5 cm. (G) Emergence of new leaves from tillers of WT (open diamonds) and branch shoots of pUbq:OsFD2 plants (filled squares). Leaf number was counted after 90 d since transplantation, when the pUbq:OsFD2 plants produced abnormal shoots. (H) A comparison of leaf morphology among flag leaves (filled squares) and mature leaves (open squares) of WT plants and the small leaves that developed in the branch shoots of pUbq:OsFD2 plants (red squares). The x-axis indicates the length/width ratio and the y-axis indicates the ratio of the positions at which the leaves reached the maximum width, to the total length of the leaves.
The SAP-like motif of OsFD2 at its C-terminus is similar to the canonical mode-I type binding motif of 14-3-3 proteins (RXXSTQF in OsFD2, compared with the RXXSAPF in OsFD1), previously shown to be required for OsFD1 to interact with 14-3-3 proteins in rice cells. The serine residue within the SAP motif is probably phosphorylated by unknown CDPK(s) (Abe et al. 2005), and this phosphorylation is essential for the recognition of OsFD1 by 14-3-3 proteins (Taoka et al. 2011). Consistent with the role of the SAP motif in OsFD1 function, an alanine substitution in S164, the putative phosphorylation site within the SAP motif of OsFD2, attenuated the interaction of OsFD2 with a 14-3-3 isoform GF14b in rice cells and yeast (see Figs. 5, 6; Supplementary Fig. S5). None of the typical leaf phenotypes observed in pUbq:OsFD2 WT plants was observed in pUbq:OsFD2 S164A plants, indicating that the OsFD2 C-terminal serine residue in the SAP motif was required for OsFD2 function. These data suggest that the interaction of OsFD2 with 14-3-3 proteins plays a role in OsFD2 function in leaf development (Supplementary Fig. S4D, E).
We next characterized the small leaves produced by pUbq:OsFD2 plants. In rice, the last leaf produced before panicle formation is called the flag leaf, and its shape is different from that of the other leaves when evaluated by the length/width ratio and the position where the leaf reaches its maximum width (Fig. 4H). Because the abnormal leaves of pUbq:OsFD2 plants were produced at and after flag leaf development, we measured leaf morphology parameters for the abnormal leaves and compared them with those for the WT flag leaves and mature leaves. We found that the morphology of pUbq:OsFD2 leaves was more similar to that of flag leaves than to that of mature leaves (Fig. 4H), suggesting that the abnormal leaves share characteristics with flag leaves. These results suggest that OsFD2 controls leaf development. Although we tried to produce OsFD2 suppression lines by RNA interference (RNAi) and artificial microRNA methods, we were not able to obtain transgenic plants with significant reductions in OsFD2 expression among >50 independent transgenic plants (data not shown).
Subcellular localization and in vivo interaction of Hd3a, 14-3-3 and OsFD2
Previous work indicated that OsFD1 accumulated predominantly in the nuclei of rice cells (Taoka et al. 2011). In contrast, when the green fluorescent protein (GFP)–OsFD2 fusion protein was expressed in rice cells, clear GFP fluorescence was observed in nuclei and the cytoplasm (Fig. 5A, B). Because some transcription factors are anchored in the cytoplasm through 14-3-3 protein binding (Igarashi et al. 2001, Ishida et al. 2004, Bai et al. 2007, Gampala et al. 2007, Wang et al. 2011), and a 14-3-3 protein recognition motif within the SAP motif of OsFD2 was present (Fig. 1), we hypothesized that accumulation of OsFD2 in the cytoplasm was changed by its interaction with 14-3-3 proteins. To test this hypothesis, we introduced a construct containing the S164A mutation to disrupt interaction between OsFD2 and its corresponding 14-3-3 protein (see Figs. 5C, 6; Supplementary Fig. S5B). GFP–OsFD2 S164A was localized exclusively in nuclei (Fig. 5A), and the ratio of nuclear localization was much higher with OsFD2 S164A than in OsFD2 WT (Fig. 5B), suggesting that OsFD2 was excluded from nuclei through 14-3-3 protein binding. This result was in contrast to OsFD1 that normally accumulates in nuclei. The alanine substitution for the serine residue within the SAP motif of OsFD1 had no effect on its nuclear accumulation (see Discussion) (Taoka et al. 2011).
Next, the interaction between OsFD2 and a 14-3-3 protein (GF14b) was monitored by bimolecular fluorescent complementation (BiFC) assays (Fig. 5C). When constructs encoding N-terminal or C-terminal halves of Venus (VN or VC, respectively) were used to tag GF14b and OsFD2 and were expressed in rice cells, a Venus signal was detected mainly in the cytoplasm and very weakly in nuclei (Fig. 5C, D; Supplementary Fig. S5). This result is consistent with the hypothesis that 14-3-3 proteins bind OsFD2 and anchor it in the cytoplasm. The OsFD2 S164A mutant could not interact with a 14-3-3 protein (GF14b) in vivo, indicating the importance of the SAP motif for 14-3-3–FD interaction (Fig. 5C; Supplementary Fig. S5B). We then tested the interaction of Hd3a and OsFD2, which is possibly mediated by an endogenous 14-3-3 protein to form the FAC (FAC-OsFD2). Interestingly, the Hd3a–OsFD2 BiFC signal was mainly detected in nuclei and very weakly in the cytoplasm, whereas the GF14b–OsFD2 interaction was predominantly detected in the cytoplasm (Fig. 5C, D). OsFD2 S164A could not interact with Hd3a (Fig. 5C; Supplementary Fig. S5C), indicating the 14-3-3 binding to OsFD2 is essential for the interaction of OsFD2 with Hd3a. Collectively, these results suggest that OsFD2 can potentially form a FAC in rice cells. Furthermore, FAC-OsFD2 may be a nuclear–cytoplasmic shuttling complex, and its localization may be controlled by 14-3-3 protein and Hd3a.
Protein interactions among Hd3a, 14-3-3 protein and OsFDs
To test for the formation of a FAC containing OsFD2, we analyzed interactions among Hd3a, 14-3-3 protein and OsFD2 by yeast two-hybrid analysis. The outline of protein–protein interactions among the three proteins of the FAC is shown in Fig. 6A, taking Hd3a, GF14b (a 14-3-3 protein) and OsFD1 as an example (Taoka et al. 2011). The FAC is a hetero-hexamer composed of two molecules each of Hd3a, 14-3-3 protein and OsFD1. Two Hd3a proteins cover both sides of a 14-3-3 protein dimer, and the OsFD1 dimer is located at the center of the complex through the interaction with the 14-3-3 protein’s phosphoserine-binding pocket.
The 14-3-3 protein has a phosphoserine-binding pocket (Fig. 6A, blue box in the center of GF14b) that recognizes the phosphoserine in R/K-X-X-pS/pT-X-P, and the phosphorylated form of the SAP motif of OsFD1 is inserted into this pocket (Fig. 6A, orange hexagon labeled as P at the center of the complex). GF14b R64 and R68 contribute to the structure of the phosphoserine-binding pocket, and R64 forms a hydrogen bond with the phosphorylated S192 in the OsFD1 SAP motif. Thus, GF14b R64/R68 and OsFD1 S192 are essential for GF14b–OsFD1 interaction. Alanine substitutions of GF14b R64/R68 or OsFD1 S192 abolished GF14b–OsFD1 interaction (Fig. 6B). Hd3a does not contact OsFD1 directly, and these two proteins interact with each other through interaction with the 14-3-3 protein.
The 14-3-3 protein interacts with Hd3a through the wide surface of the C-terminal region of the 14-3-3 protein (Taoka et al. 2011) (Fig. 6A, magenta and yellow boxes inside Hd3a and GF14b, respectively), which is composed of a hydrophobic cavity and an acidic loop on the surface of GF14b. GF14b I210 is located within this hydrophobic cavity and interacts with the hydrophobic side chain of Hd3a F103, indicating that both GF14b I210 and Hd3a F103 are essential for Hd3a–GF14b interaction (Taoka et al. 2011) (Fig. 6A, magenta and yellow boxes inside Hd3a and GF14b, respectively). On the other hand, GF14b I210 is dispensable for interaction of GF14b with OsFD1 because GF14b I210 is distantly located from the site of interaction with OsFD1 (Fig. 6A, compare yellow and blue boxes in GF14b). Thus, the GF14b I210A mutation specifically disrupts the interaction with Hd3a, but does not affect the interaction with OsFD1 (Fig. 6B, upper right panel). The 14-3-3 protein bridges the interaction between Hd3a and OsFD1; thus Hd3a F103A lost its ability to interact with the 14-3-3 protein and, in consequence, with OsFD1 (Fig. 6B) (Taoka et al. 2011). OsFD1 S192A also disrupts the interaction with the 14-3-3 protein, and, consequently, with Hd3a (Fig. 6B) (Taoka et al. 2011). In our yeast two-hybrid assays, Hd3a and OsFD1 interaction can be bridged by endogenous 14-3-3 proteins in yeast cells since yeast 14-3-3 proteins have conserved the structural requirements for Hd3a–14-3-3 and 14-3-3–OsFD1 interactions (Taoka et al. 2011).
The above information indicates that the yeast two-hybrid assay using mutations at essential amino acids can be used to examine whether OsFD2 and OsFD3 can form a FAC (Fig. 6B). OsFD2 interacted with GF14b, and alanine substitutions of GF14b R64/R68 abolished GF14b–OsFD2 interaction, suggesting that this interaction is phosphoserine dependent (Fig. 6B). Consistent with this result, an alanine substitution in OsFD2 S164, located at the putative phosphorylation site in the OsFD2 C-terminal SAP-like motif, abolished interaction between OsFD2 and GF14b. On the other hand, the GF14b I210A mutation, known to disrupt specifically the interaction of GF14b with Hd3a, did not affect the interaction of GF14b with OsFD2. This finding suggests that GF14b interacts with Hd3a and OsFD2 via distinct regions, similar to the model for OsFD1 interaction. Next, we performed yeast two-hybrid assays using Hd3a and OsFD2. OsFD2 interacted with Hd3a and, when the Hd3a–14-3-3 interaction was disrupted by the Hd3a F103A mutation (Taoka et al. 2011), Hd3a lost its ability to interact with OsFD2. The OsFD2 S164A mutation disrupted the interaction of OsFD2 with GF14b and, in consequence, the interaction with Hd3a (Fig. 6B). These data suggest that OsFD2 can form a FAC with Hd3a and GF14b in a similar manner to OsFD1 (Taoka et al. 2011).
In contrast to OsFD2, we could not detect any interaction between Hd3a and OsFD3, whereas we observed an interaction between GF14b and OsFD3 by the canonical 14-3-3 protein and phosphoserine binding system. These results suggest that there may be technical difficulties in detecting interactions in yeast, or the presence of an unknown mechanism inhibiting FAC formation by OsFD3 in yeast cells. To examine the function of OsFD3, we generated OsFD3 RNAi plants; however, these plants showed no changes in morphology or flowering time (Supplementary Fig. S6).
Discussion
Evolution of FD in plants
Phylogenetic analysis of FD in plants suggested unique evolutionary aspects of FD genes in the Poaceae family (Fig. 1). Three groups of Poaceae-specific FD genes were identified, but canonical FD genes are absent from the Poaceae genome. Although the entire sequence context is not strongly conserved, at least two deduced proteins, OsFD1 and OsFD2, can form FACs (Fig. 6), suggesting conservation of FAC formation in different groups of FD proteins. We found that the small LSL motif was well conserved between the Poaceae-specific OsFD1 group and the eudicot/non-Poaceae monocot FD group (Fig. 1). Interestingly, both groups contribute to the promotion of flowering; thus, the presence of the LSL motif may define the FD proteins capable of activating the AP1/FUL clade of MADS box genes for flowering (Abe et al. 2005, Wigge et al. 2005, Li and Dubcovsky 2008, Taoka et al. 2011).
We found FD genes from diverse species of angiosperm plants, but not from the moss (bryophyte) Physcomitrella patens (Rensing et al. 2008l Hauser et al. 2011) or the spike moss (lycophyte, basal vascular plant) Selaginella moellendorffii (Banks et al. 2011), suggesting that FACs containing FD proteins may not have occurred before the emergence of seed plants and evolved after the emergence of angiosperms. Although searches of the available databases did not detect FD genes in gymnosperms, further genome sequence analysis may help our understanding of the evolution of FACs in land plants.
Diversification of FD functions in rice
Phylogenetic analysis of FDs in plants suggested that the FD gene family is divided into two groups, the eudicot/non-Poaceae FDs and the Poaceae-specific FDs (Fig. 1). We studied the functions of OsFD1 and OsFD2, two members of the Poaceae-specific FDs in rice, and found that the function of FD has diverged between these homologs. OsFD1 functions in the activation of AP1/FUL genes and promotion of flowering (Taoka et al. 2011), and OsFD2 functions in leaf development (Figs. 3, 4). Although the precise mechanism for the functional difference between OsFD1 and OsFD2 is unclear, modifications of the motif arrangement and DNA-binding domains could contribute to these differences (Supplementary Fig. S7). Two amino acids in the basic regions of OsFD1 and OsFD2 are different, and this slight difference may change the target genes that are regulated by these transcription factors. OsMADS15 is not the target of OsFD2, because OsFD2 and Hd3a co-expression could not induce OsMADS15 expression in our transient assay using protoplasts (data not shown).
The recent discovery of FAC explains the molecular mechanism by which florigen Hd3a, 14-3-3 protein and OsFD1 promote flowering through activation of downstream target genes (Taoka et al. 2011). Hd3a interacts with 14-3-3 proteins and OsFD1 to form the ternary transcriptional complex, FAC-OsFD1, to activate OsMADS15 expression. Here, we show that OsFD2 can form a FAC with Hd3a and the 14-3-3 isoform GF14b (Figs. 5, 6). The OsFD2 S164A mutation disrupted the interaction of OsFD2 with GF14b and Hd3a, and abolished the function of OsFD2 in leaf development. These data suggest that OsFD2 is a potential component of the FAC, and FAC formation is essential for OsFD2 function to control leaf development in rice.
Our model of FAC that includes OsFD1 and OsFD2 suggests that FAC function is modified depending on the transcription factors recruited through the 14-3-3 protein (Fig. 7A). In this model, the Hd3a–14-3-3 subcomplex constitutes a common backbone of the FACs, and transcription factors interacting with 14-3-3 proteins determine the function of the FAC. Our results suggest that if OsFD1 is recruited, FAC-OsFD1 acts in the promotion of flowering, and if OsFD2 is recruited, FAC-OsFD2 functions in rice leaf development (Figs. 3, 4, 7A). Our model offers a molecular basis for the participation of florigen in multiple developmental processes other than flowering, including stomatal opening in Arabidopsis (Kinoshita et al. 2011), panicle morphology in rice (Endo-Higashi and Izawa 2011), leaf morphology and inflorescence architecture in Arabidopsis and tomato (Teper-Bamnolker and Samach 2005, Lifschitz et al. 2006, Krieger et al. 2010, Hiraoka et al. 2013), tuber formation in potato (Navarro et al. 2011) and growth cessation in tree species (Bohlenius et al. 2006, Hsu et al. 2011). In potato tuberization, for example, SP6A, a Hd3a homolog, moves from the leaves to the stolon to initiate tuber formation. SP6A may form a FAC with tuberization-specific transcription factors to activate downstream gene expression in potato (Navarro et al. 2011). Exploring the participation of FACs in developmental processes other than flowering is a promising direction for further study of the multifunctionality of florigen.
Fig. 7.
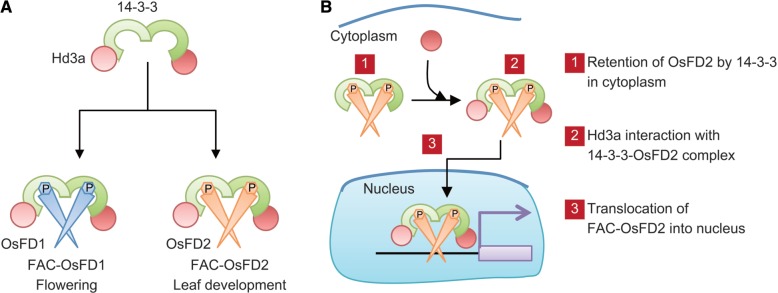
Model of FAC function converted by OsFD1 or OsFD2 (A) and the mechanism of FAC formation including OsFD2 (B). (A) The Hd3a–14-3-3 protein subcomplex serves as the basic component of the FAC. When OsFD1 enters the complex, the resultant FAC-OsFD1 promotes flowering. OsFD2 is the proposed component of FAC to form FAC-OsFD2 that putatively controls leaf development. In this model, the function of FAC can be converted, depending on the function of the OsFDs recruited into the FAC. (B) A part of the OsFD2 protein is localized in the cytoplasm by the interaction with 14-3-3 proteins. When Hd3a interacts with the 14-3-3–OsFD2 complex in the cytoplasm to form a FAC, the complex enters the nucleus to regulate gene expression.
FAC formation and nuclear translocation
Subcellular localization analysis and in vivo interaction studies suggested a novel mechanism for FAC-OsFD2 formation in cell nuclei (Fig. 7B). OsFD2 can shuttle between the cytoplasm and the nucleus, and 14-3-3 proteins are involved in this shuttling by facilitating the cytoplasmic translocation of OsFD2 (Fig. 5A, B). A mutant version of OsFD2 that had lost its ability to interact with a 14-3-3 protein, as a result of alanine substitution of OsFD2 S164 in the conserved 14-3-3 binding motif, localized exclusively to nuclei. However, the nuclear translocation of OsFD2 is not sufficient for OsFD2 function because overexpression of this mutant version in plants did not show any phenotype (Figs. 3, 4; Supplementary Fig. S4), suggesting that FAC formation is necessary for OsFD2 function. The BiFC experiments indicated interaction between 14-3-3 protein and OsFD2 in the cytoplasm, whereas interaction between Hd3a and OsFD2, which is mediated by 14-3-3 protein, occurred in the nuclei (Fig. 5C, D). This finding suggested that the presence of Hd3a in the FAC changed its subcellular localization. In our model, OsFD2 is localized in the cytoplasm through its binding with 14-3-3 protein, and Hd3a interacts with the C-terminal region of a 14-3-3 protein to form a FAC. This interaction initiates the nuclear translocation of the FAC into the nucleus (Fig. 7B).
The molecular mechanism for this nuclear translocation is an open question, but inhibition of the nuclear exclusion signal (NES) within the 14-3-3 protein by Hd3a could contribute to this mechanism. The crystal structure of FACs indicated that the interface for Hd3a and 14-3-3 interaction is located in the region that overlaps with the NES region within the 14-3-3 protein, and Hd3a binding on the 14-3-3 protein covers the entire NES (Taoka et al. 2011). This binding may inhibit the interaction of the evolutionarily conserved nuclear exclusion machinery including exportin proteins onto the 14-3-3 NES (Rittinger et al. 1999). Subsequently, the activity of the OsFD2 nuclear localization signal (NLS) can cause the translocation of the entire FAC complex into the nucleus. Detailed observations about the cellular processes during FAC formation will be valuable for understanding this mechanism. In contrast, GFP–OsFD1 localized to nuclei exclusively in the presence of 14-3-3 protein interaction (Taoka et al. 2011), suggesting that different FACs may behave differently in their formation and nuclear translocation. The precise mechanisms generating these differences remain unknown, but differences in important amino acids in the NLS in OsFD1 and OsFD2 may contribute to these differences (Supplementary Fig. S7). OsFD1 and OsFD2 contain a bipartite NLS in the basic region of their bZIP motifs, but the more N-terminal region of the bipartite NLS contains different amino acids: RRKR in OsFD1 and RTIR in OsFD2. This slight difference in OsFD2 may affect nuclear accumulation because the corresponding region of the NLS in opaque2 (O2), the bZIP transcription factor of maize, is essential for its NLS activity (Varagona and Raikhel 1994). RKRK in WT O2 accumulates in nuclei, whereas a mutated sequence RTNR, which is similar to the OsFD2 NLS, accumulates both in the cytoplasm and in the nucleus (Varagona and Raikhel 1994).
Materials and Methods
Plant materials and growth conditions
Rice (Oryza sativa L. subspecies japonica) variety Norin 8 was used as the WT. pUbq:OsFD1 transgenic rice plants were described previously (Taoka et al. 2011). pUbq:OsFD1 ΔLSL, pUbq:OsFD2 and pUbq:OsFD2 S164A rice plants were generated using Agrobacterium-mediated transformation of rice calli, as previously described (Hiei et al. 1994). Hygromycin-resistant plants were regenerated from the transformed callus. Transgene integration was further confirmed by PCR amplification of the hygromycin phosphotransferase gene in genomic DNA extracted from regenerated plants. Plants were grown in climate chambers at 70% humidity, under short-day (SD) conditions with daily cycles of 10 h of light at 30°C and 14 h of dark at 25°C. Light was provided by fluorescent white lights. Flowering time was measured as the number of days to the heading stage after T0 transgenic plants were transferred to SD conditions. For flowering time measurement, the tillers were removed to save space (Ohnishi et al. 2011). Rice suspension-cultured cells were maintained as described previously (Taoka et al. 2011). The leaf morphology and plastochron were measured at 90 d after transplantation when the majority of the pUbq:OsFD2 plants showed characteristic leaf phenotypes on their abnormally outgrowing branch shoots.
Phylogenetic analysis
Databases listed in Supplementary Table S1 were searched for DNA sequences encoding FD proteins using the Arabidopsis and rice FDs as the queries.
Predicted FD amino acid sequences were used for phylogenetic and motif analyses. Conserved motifs and their arrangements were extracted from FDs with interactive SALAD analysis from the SALAD database (Mihara et al. 2010), with the parameters of 10 for ‘maximum number of motifs to find’ and 1e-2 for ‘expect threshold’. The extracted motifs were then manually curated and aligned with the T-Coffee program and displayed with Boxshade software (Di Tommaso et al. 2011). Phylogenetic trees using the regions spanning bZIP to the C-terminal SAP motif were constructed based on the alignment from CLUSTALW using the Neighbor–Joining method. We obtained a phylogenetic tree with a similar shape from the T-Coffee program using the same bZIP-SAP region and interactive SALAD analysis using the entire motif architecture of FDs.
Protoplast transformation
Transformation of rice Oc protoplasts was performed as described previously (Taoka et al. 2011, Kim et al. 2012). For transient expression analysis, 8 µg of Hd3a expression vectors and 16 µg of OsFD1 expression vectors were introduced into 500 µl of a protoplast suspension at a concentration of 2×107 protoplasts ml−1 by the polyethylene glycol (PEG)-mediated transformation method. After 16 or 48 h incubations at 30°C, the protoplast suspension was centrifuged and the cell pellet was frozen at −80°C for RNA extraction.
RNA extraction and real-time RT–PCR analysis
Total RNA from protoplasts was extracted using TRizol reagent (Invitrogen) according to the manufacturer’s protocol. cDNA was synthesized from 0.1–1.0 µg of total RNA, using a 21 nucleotide oligo(dT) primer and Superscript II reverse transcriptase (Invitrogen). cDNA (1 µl) was used for quantitative analysis of gene expression using SYBR Green PCR master mix (Life Technologies). Data were collected using the StepOnePlus sequence detection system in accordance with the manufacturer’s instruction manual. The sequences of primers used in this study are listed in Supplementary Table S2.
Subcellular localization and bimolecular fluorescent complementation
The OsFD2 and OsFD2 S164A coding regions were cloned into fluorescent protein expression vectors or BiFC vectors and purified using the Purelink Plasmid Midiprep kit (Invitrogen). We co-transformed 5 µg of GFP–OsFD2 or GFP–OsFD2 S164A expression plasmids with both 5 µg of mCherry and 10 µg of NLS–cyan fluorescent protein (CFP) expression plasmids. For BiFC experiments, 5 µg of VN- or VC-tagged protein expression vector was co-transformed with both 5 µg mCherry and 10 µg of NLS–CFP expression plasmids as markers. Protein–protein interactions from BiFC experiments were quantified as described previously, with some modifications (Taoka et al. 2011). Briefly, we calculated the ratio of Venus/mCherry from each of cells in the BiFC experiment and could recognize reliable BiFC signals in cells showing Venus/mCherry ratios >0.83 (experiment 1), 0.53 (experiment 2) for the OsFD2–GF14b and OsFD2–Hd3a interaction, 0.37 (experiment 3) and 0.38 (experiment 4) for the OsFD2–GF14b and OsFD2 S164A–GF14b interaction, and 0.29 (experiment 5) and 0.34 (experiment 6) for the OsFD2–Hd3a and OsFD2 S164A–Hd3a interaction. The number of cells showing ratios exceeding these values was recorded.
We examined the degree of nuclear accumulation of GFP fusion proteins and the BiFC signal arising from GFP–OsFD2, OsFD2–GF14b and OsFD2–Hd3a. First, we measured the fluorescence intensities of Venus and mCherry in the nuclei and cytoplasm of transformed cells. Next, we calculated values for (Venus in nucleus/mCherry in nucleus)/(Venus in cytoplasm/mCherry in cytoplasm) that indicates a measure of nuclear accumulation of Venus. Finally, we compared these values and the corresponding confocal images from each cell to determine the nuclear accumulation of the fluorescent proteins.
Yeast two-hybrid assay
Gateway destination vectors pBTM116-GW and pVP16-GW were used to construct the bait and pray vectors by LR recombination reactions. Yeast cells were grown at 30°C for 5 d on SC medium without uracil, tryptophan, leucine and histidine, or containing added histidine or 1–10 mM 3-amino-1,2,4-triazole (3-AT). The concentration of 3-AT was determined by the bait–prey combination (Purwestri et al. 2009).
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by Grants-in-Aid for Scientific Research [to H.T and K.S.]; Grants-in-Aid for Scientific Research on Priority Areas [to K.S.]; the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry from Bio-oriented Research Advancement Institution (BRAIN) [to H.T.].
Supplementary Material
Acknowledgments
We thank S. Takayama for the BiFC vectors. We also thank E. Kawano, M. Kanda, S. Toyoda and Y. Mitsubayashi for technical assistance; Y. Tamaki, Y. Konomi and J. Naritomi for rice transformation; and members of the Laboratory of Plant Molecular Genetics at Nara Institute of Science and Technology (NAIST) for discussions.
Glossary
Abbreviations
- BiFC
bimolecular fluorescence complementation
- bZIP
basic leucine zipper
- CDPK
calcium-dependent protein kinases
- CFP
cyan fluorescent protein
- EST
expressed sequence tag
- FAC
florigen activation complex
- FT
FLOWERING LOCUS T
- GFP
green fluorescent protein
- Hd3a
Heading date 3a
- NES
nuclear exclusion signal
- NLS
nuclear localization signal
- RNAi
RNA interference
- RT–PCR
reverse transcription–PCR
- SD
short day
- WT
wild type.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Al-Dous EK, George B, Al-Mahmoud ME, Al-Jaber MY, Wang H, Salameh YM, et al. De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera) Nat. Biotechnol. 2011;29:521–527. doi: 10.1038/nbt.1860. [DOI] [PubMed] [Google Scholar]
- Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, et al. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl Acad. Sci. USA. 2007;104:13839–13844. doi: 10.1073/pnas.0706386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, dePamphilis C, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- D’Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488:213–217. doi: 10.1038/nature11241. [DOI] [PubMed] [Google Scholar]
- Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–W17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo-Higashi N, Izawa T. Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice. Plant Cell Physiol. 2011;52:1083–1094. doi: 10.1093/pcp/pcr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl RJ, Manak MS, Reyes MF. The 14-3-3s. Genome Biol. 2002;3:REVIEWS3010. doi: 10.1186/gb-2002-3-7-reviews3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S, Goto K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell. 2011;23:3172–3184. doi: 10.1105/tpc.111.088641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F, Waadt R, Schroeder JI. Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 2011;21:R346–R355. doi: 10.1016/j.cub.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Hiraoka K, Yamaguchi A, Abe M, Araki T. The florigen genes FT and TSF modulate lateral shoot outgrowth in Arabidopsis thaliana. Plant Cell Physiol. 2013;54:352–368. doi: 10.1093/pcp/pcs168. [DOI] [PubMed] [Google Scholar]
- Hsu CY, Adams JP, Kim H, No K, Ma C, Strauss SH, et al. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl Acad. Sci. USA. 2011;108:10756–10761. doi: 10.1073/pnas.1104713108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y. 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell. 2001;13:2483–2497. doi: 10.1105/tpc.010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Ishida S, Fukazawa J, Yuasa T, Takahashi Y. Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. Plant Cell. 2004;16:2641–2651. doi: 10.1105/tpc.104.024604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Aoki M, Kurotani K-i, Yokoi S, Shinomura T, Takano M, et al. Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol. Genet. Genomics. 2011;285:461–470. doi: 10.1007/s00438-011-0621-4. [DOI] [PubMed] [Google Scholar]
- Itoh H, Nonoue Y, Yano M, Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 2010;42:635–638. doi: 10.1038/ng.606. [DOI] [PubMed] [Google Scholar]
- Karlgren A, Gyllenstrand N, Kallman T, Sundstrom JF, Moore D, Lascoux M, et al. Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiol. 2011;156:1967–1977. doi: 10.1104/pp.111.176206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Oikawa T, Kyozuka J, Wong HL, Umemura K, Kishi-Kaboshi M, et al. The bHLH Rac immunity1 (RAI1) is activated by OsRac1 via OsMAPK3 and OsMAPK6 in rice immunity. Plant Cell Physiol. 2012;53:740–754. doi: 10.1093/pcp/pcs033. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Ono N, Hayashi Y, Morimoto S, Nakamura S, Soda M, et al. Flowering locus T regulates stomatal opening. Curr. Biol. 2011;21:1232–1238. doi: 10.1016/j.cub.2011.06.025. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, et al. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell. 2012;24:1848–1859. doi: 10.1105/tpc.112.097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;136:3443–3450. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 2010;42:459–463. doi: 10.1038/ng.550. [DOI] [PubMed] [Google Scholar]
- Li CX, Dubcovsky J. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 2008;55:543–554. doi: 10.1111/j.1365-313X.2008.03526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, et al. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl Acad. Sci. USA. 2006;103:6398–6403. doi: 10.1073/pnas.0601620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsoukas IG, Massiah AJ, Thomas B. Florigenic and antiflorigenic signaling in plants. Plant Cell Physiol. 2012;53:1827–1842. doi: 10.1093/pcp/pcs130. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Ogiso-Tanaka E, Hori K, Ebana K, Ando T, Yano M. Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 2012;53:709–716. doi: 10.1093/pcp/pcs028. [DOI] [PubMed] [Google Scholar]
- Meng X, Muszynski MG, Danilevskaya ON. The FT-Like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell. 2011;23:942–960. doi: 10.1105/tpc.110.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Itoh T, Izawa T. SALAD database: a motif-based database of protein annotations for plant comparative genomics. Nucleic Acids Res. 2010;38:D835–D842. doi: 10.1093/nar/gkp831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou Z, Bruggemann E, et al. delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 2006;142:1523–1536. doi: 10.1104/pp.106.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Shimamoto K, Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002;29:743–750. doi: 10.1046/j.1365-313x.2002.01255.x. [DOI] [PubMed] [Google Scholar]
- Navarro C, Abelenda JA, Cruz-Oro E, Cuellar CA, Tamaki S, Silva J, et al. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature. 2011;478:119–122. doi: 10.1038/nature10431. [DOI] [PubMed] [Google Scholar]
- Notaguchi M, Abe M, Kimura T, Daimon Y, Kobayashi T, Yamaguchi A, et al. Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 2008;49:1645–1658. doi: 10.1093/pcp/pcn154. [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Yoshino M, Yamakawa H, Kinoshita T. The biotron breeding system: a rapid and reliable procedure for genetic studies and breeding in rice. Plant Cell Physiol. 2011;52:1249–1257. doi: 10.1093/pcp/pcr066. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Purwestri YA, Ogaki Y, Tamaki S, Tsuji H, Shimamoto K. The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant Cell Physiol. 2009;50:429–438. doi: 10.1093/pcp/pcp012. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Rittinger K, Budman J, Xu J, Volinia S, Cantley LC, Smerdon SJ, et al. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- Saito H, Ogiso-Tanaka E, Okumoto Y, Yoshitake Y, Izumi H, Yokoo T, et al. Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol. 2012;53:717–728. doi: 10.1093/pcp/pcs029. [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuom S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y, Numa H, et al. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 2008;36:D1028–D1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K-i, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Teper-Bamnolker P, Samach A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell. 2005;17:2661–2675. doi: 10.1105/tpc.105.035766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Taoka K-i, Shimamoto K. Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 2011;14:45–52. doi: 10.1016/j.pbi.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Varagona MJ, Raikhel NV. The basic domain in the bZIP regulatory protein Opaque2 serves two independent functions: DNA binding and nuclear localization. Plant J. 1994;5:207–214. doi: 10.1046/j.1365-313x.1994.05020207.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang C, Zhang C, Wang N, Lu D, Wang J, et al. Dual role of BKI1 and 14-3-3s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell. 2011;21:825–834. doi: 10.1016/j.devcel.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Wei F, Zhang J, Zhou S, He R, Schaeffer M, Collura K, et al. The physical and genetic framework of the maize B73 genome. PLoS Genet. 2009;5:e1000715. doi: 10.1371/journal.pgen.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Hong SM, Jung HS, Ahn JH. The cotyledons produce sufficient FT protein to induce flowering: evidence from cotyledon micrografting in Arabidopsis. Plant Cell Physiol. 2012;54:119–128. doi: 10.1093/pcp/pcs158. [DOI] [PubMed] [Google Scholar]
- Zeevaart JA. Leaf-produced floral signals. Curr. Opin. Plant Biol. 2008;11:541–547. doi: 10.1016/j.pbi.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 2012;30:549–554. doi: 10.1038/nbt.2195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.